Perturbing Endothelial Biomechanics via Connexin 43 Structural Disruption
Summary
Here, we present a mechanics-based protocol to disrupt the gap junction connexin 43 and measure the subsequent impact this has on endothelial biomechanics via observation of tractions and intercellular stresses.
Abstract
Endothelial cells have been established to generate intercellular stresses and tractions, but the role gap junctions play in endothelial intercellular stress and traction generation is currently unknown. Therefore, we present here a mechanics-based protocol to probe the influence of gap junction connexin 43 (Cx43) has on endothelial biomechanics by exposing confluent endothelial monolayers to a known Cx43 inhibitor 2,5-dihydroxychalcone (chalcone) and measuring the impact this inhibitor has on tractions and intercellular stresses. We present representative results, which show a decrease in both tractions and intercellular stresses under a high chalcone dosage (2 µg/mL) when compared to control. This protocol can be applied to not just Cx43, but also other gap junctions as well, assuming the appropriate inhibitor is used. We believe this protocol will be useful in the fields of cardiovascular and mechanobiology research.
Introduction
The field that refers to the study of the effects of physical forces and of mechanical properties on cellular and tissue physiology and pathology is known as mechanobiology1. A few useful techniques that have been utilized in mechanobiology are monolayer stress microscopy and traction force microscopy. Traction force microscopy allows for the computation of tractions generated at the cell-substrate interface, while monolayer stress microscopy allows for the computation of intercellular stresses generated between adjacent cells within a monolayer2,3,4,5,6. Results yielded from previous methods have suggested that cell-derived mechanical stresses play a crucial role in determining the fate of a host of cellular processes3,4,5. For example, upon exposure to an external mechanical force, a group of cells migrating as a collective can alter their morphology and polarize their shape to align and migrate along the direction of applied force by, in part, generating tractions7,8. Tractions provide a metric that can be used to evaluate cell contractility and are calculated using traction force microscopy (TFM). Traction force microscopy (TFM) begins with the determination of cell-induced substrate deformations followed by the calculation of the traction field using a mathematically rigorous, mechanics-based computational approach. Since the ability to calculate tractions has been around for quite some time, researchers have utilized TFM to reveal the impact tractions have on a host of processes, including cancer9, wound healing10 and assessment of engineered cardiac tissue11.
Implementation of TFM and MSM together can be divided into three essential steps that must be executed in the following order: first, the hydrogel deformations produced by the cells are determined; second, tractions are recovered from hydrogel deformations; and third, a finite element approach is used to compute normal and shear intercellular stresses within the entire monolayer. To compute gel displacements, fluorescence bead images with cells were compared with the reference bead image (without cells) by using a custom-written particle image velocimetry (PIV) routine. The cross-correlation window size and overlap for PIV analysis were chosen to be 32 x 32 pixels and 0.5, respectively. At this time, pixel shifts were converted into microns by multiplying with a pixel-to-micron conversion factor (for our microscope, this conversion factor is 0.65) to obtain in-plane displacements. Errors associated with ignoring out-of-plane displacements are negligible12,13. After computation of gel displacements, there are two types of traction measurements that can be utilized, constrained tractions and unconstrained tractions8,14. Unconstrained tractions provide the traction field for the entire field of view (including regions with and without cells), while constrained tractions provide the traction field only for regions that include cells14. Then, intercellular stresses are calculated using monolayer stress microscopy (MSM), which is an extension of traction force microscopy. Implementation of MSM is based off the assumption that local tractions exerted by a monolayer of cells at the cell-substrate interface must be balanced by mechanical forces transmitted between cells at the cell-cell interface as demanded by Newton's laws7,12,13. A key assumption here is that the cell monolayer can be treated as a thin elastic sheet because the traction distribution in the monolayer is known and the force balance does not depend on cell material properties. Another key assumption is that the traction forces are balanced by local intercellular stresses within the optical field of view (within the monolayer) and the influence of this force balance is minimal in the distal region (outside of the monolayer)13. Therefore, the boundary conditions defined by intercellular stresses, displacements, or a combination of both at the monolayer boundary are crucial to perform MSM13.Taking into account the above information, we utilize MSM to perform a finite element analysis (FEM) to recover the maximum principal stress (σmax) and minimum principal stress (σmin) by rotating the stress plane at every point within the monolayer. These principal stresses are subsequently used to compute the 2D average normal intercellular stress [(σmax + σmin) /2] and 2D maximum shear intercellular stress [(σmax – σmin) /2] within the entire monolayer12,13. This procedure is described in more detail by Tambe et al.12,13
Monolayer stress microscopy (MSM) allows for the calculation of cell-cell intercellular stresses generated within a monolayer6,7,8,12,13. These intercellular stresses have been suggested to be important for tissue growth and repair, wound healing, and cancer metastasis12,15,16,17. In addition, intercellular stresses have been suggested to also be important in endothelial cell migration and endothelial barrier function17,18. While cell-cell junctions such as tight junctions and adherens junctions have both been suggested to play a critical role in endothelial intercellular stress generation and transmission, the role of gap junctions remains elusive. Gap junctions physically connect adjacent cells and provide a pathway for electrical current and molecules (<1 KDa) to pass between neighboring cells19,20,21. Although endothelial cells express Cx37, Cx40, and Cx43 gap junctions19,22, Cx43 is arguably the most important in terms of disease progression23. Evidence of Cx43's importance may be found in the fact that genetic deletion of Cx43 in mice results in hypotension24 and has adverse effects on angiogenesis25. In addition, Cx43 has been documented to be important for cell migration and proliferation and in the progression of atherosclerosis18,22,23,24,25.
In this protocol, we used TFM and MSM to investigate whether traction and intercellular stress generation within the confluent, endothelial monolayer would be impacted by the disruption of the endothelial gap junction Cx43. We disrupted Cx43 with 2,5-dihydroxychalcone (chalcone), a molecule documented to inhibit Cx43 expression26. Chalcone was used to disrupt Cx43 instead of siRNA as chalcone has been reported previously by Lee et al. to disrupt Cx43 expression26. In addition, we were particularly interested in chalcone's influence on the endothelium as it has also been reported to be an anti-inflammatory and anti-platelet compound that can potentially be used for the prevention and treatment of various vascular pathologies26. Chalcone treatments were performed an hour after the experiment onset, chalcone-treated monolayers were imaged for a total of six hours, and image processing was performed with a custom-written MATLAB code to determine tractions and subsequently intercellular stresses. Our results showed an overall decrease in tractions and intercellular stresses, suggesting Cx43 plays a key role in endothelial biomechanics.
Protocol
1. Making polyacrylamide (PA) gels
- Preparation of Petri dish
- Prepare bind silane solution by mixing 200 mL ultrapure water with 80 µL acetic acid and 50 µL of 3-(trimethoxysilyl)propyl methacrylate. Bind silane is a solution used to functionalize the glass bottom Petri dish surface for hydrogel attachment.
- Stir the bind silane solution on a stir plate for at least 1 h.
- Treat the center well of the Petri dish with bind silane solution for 45 min.
- Remove bind silane solution and rinse the Petri dish with ultra-pure water 2x-3x.
- Dry the Petri dish surface and store at room temperature for future use.
- Preparation of hydrogel solution
- Mix ultra-pure water, 40% acrylamide, and 2% bis-acrylamide in a 15 mL centrifuge tube according to Table 1.
- Add 80 µL of fluorescent beads to the hydrogel solution.
- Gently shake the tube to mix beads with the gel solution.
- Lightly tighten the tube cap on the centrifuge tube and place in a vacuum chamber.
- Degas the gel solution for at least 45 min in vacuum chamber.
- Hydrogel polymerization
- First, add 75 µL of 10% ammonium persulfate (dissolved in ultra-pure water) and then add 8 µL of TEMED (N,N,N',N'-tetramethylethane-1,2-diamine) to the hydrogel solution.
- Place 24 µL of hydrogel solution at the center of the Petri dish (see Table 2).
- Use an 18 mm coverslip to flatten the hydrogel. This will give a height of ~100 µm.
- Wait at least 30-40 min for gel polymerization.
- Submerge the polymerized hydrogel in ultra-pure water to prevent gel dehydration.
- Cover the Petri dish with aluminum foil to prevent photobleaching of fluorescent beads and store at 4 °C.
NOTE: Hydrogels can be stored a period of up to 3 months, but it is suggested that these gels be used no longer than 1 week after fabrication to obtain the best results.
2. Cell culture
- Culture human umbilical vein endothelial cells (HUVECs) in cell culture medium 200 (see Table of Materials) supplemented with 1% penicillin-streptomycin on 0.1% gelatin-coated flasks at 37 °C and 5% CO2.
3. Micropattern stencil preparation
- Cure a thin layer of polydimethylsiloxane (PDMS) in a 100 mm Petri dish by mixing the silicone base with the silicone curing agent at a ratio of 20:1 (base: curing agent).
NOTE: It is possible to use other base to curing agent ratios (ex. 10:1 or 30:1). However, a lower base to curing agent ratio will yield a stiffer pattern, while a higher base to curing agent ratio will produce a softer pattern. - Prepare a 20:1 (base:curing agent) PDMS solution and mix carefully in a 50 mL centrifuge tube. Invert the tube upside down and shake vigorously multiple times to ensure proper mixing of the PDMS solution, as nonuniform mixing will result in incomplete polymerization.
- Remove bubbles that have been introduced from the above step by centrifuging the PDMS solution for 1 min at 190 x g.
- Pour 5-6 mL of PDMS solution in the center of a 100 mm Petri dish and agitate the dish until the PDMS solution covers the entire Petri dish surface.
- Cure the PDMS solution overnight at 50-60 °C. PDMS can also be cured at room temperature.
- Remove a circular, 16 mm diameter PDMS stencil with a hole puncher.
- Use a biopsy punch to create small holes in the PDMS stencil. This protocol uses a 1.25 mm diameter biopsy punch.
- Sterilize PDMS stencils by first submerging them in 70% ethanol for 2-3 min, aspirating off excess ethanol and then placing under a UV light for 5 min.
4. Collagen-I hydrogel coating
- Remove the coverslip from the hydrogel and aspirate any excess liquid.
- Place the PDMS stencil on the hydrogel surface.
NOTE: Tweezers can be used to apply light pressure to PDMS stencil to ensure a water-tight seal between the PDMS stencil and hydrogel surface. Our PDMS stencils are water-tight and thus prevent water access between the gel top surface and PDMS stencil bottom surface. Also, the hydrogel stiffness does not need to be adjusted with respect to the PDMS stiffness. - Cover the hydrogel surface with sulfosuccinimidyl-6-(4-azido-2-nitrophenylamino) hexanoate (sulfo-SANPAH) dissolved in a 0.1 M HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer solution at a concentration of 1 : 1000 and place under a UV lamp (power 36 W) for 8 min.
- Aspirate the excess sulfo-SANPAH and HEPES solution and rinse the hydrogel twice with 0.1 M HEPES followed by an additional two rinses with ultra-pure water.
- Aspirate excess ultra-pure water and coat hydrogels with 0.1 mg/mL collagen-I overnight at 4 °C.
- Cover the dishes and protect fluorescent beads from photobleaching.
NOTE: PDMS stencils are used to create micropatterned monolayers. Micropatterned monolayers are utilized as they allow for multiple monolayers of the same geometry and dimensions to be observed simultaneously during each experiment. However, should micropatterns not be desired the above steps can be followed with the exception of step 4.2.
5. Creating HUVEC monolayers on hydrogels
- Use 1x trypsin to detach cells from tissue culture flasks for 3-5 min in the incubator.
- After trypsinization, add cell culture media to the trypsin solution and add to the 15 mL centrifuge tube.
- Centrifuge the cell solution for 3 min at 1710 x g. A small, white pellet of cells should be visible at the bottom of the centrifuge tube.
- Aspirate the supernatant and resuspend cells in media to a concentration of 50 x 104 cells/ mL.
- Remove collagen-I from the hydrogel and rinse 1x with PBS.
- Add 75 x 103 cells to the top of the PDMS stencil and allow cells to attach to the hydrogel surface for at least 1 h in the incubator at 37 °C and 5% CO2.
- Remove the PDMS stencil and add at least 2 mL of media to the Petri dish. Submerge the PDMS stencil in 10x trypsin to remove any attached cells and sterilize by spraying with 70% ethanol and then placing under the UV light for 5 min.
- Place the Petri dish in the incubator and wait at least 36 h or until a confluent monolayer is observed.
6. 2,5 dihydroxychalcone treatment for Cx43 disruption
- Dissolve 2,5 dihydroxychalcone (chalcone) in dimethylsulfoxide (DMSO) to make a 0.1875 mg/mL stock solution.
- Dilute the stock solution with cell culture media to make a low chalcone concentration (0.2 µg/mL) aliquot and a high chalcone concentration (2 µg/mL) aliquot.
7. Data acquisition
- Locate cell islands with a microscope.
- Acquire phase contrast and bead images to image cell morphology and hydrogel displacements, respectively.
NOTE: This protocol used a 10x objective for data acquisition. - At the end of the experiment, detach cells with 10x trypsin and acquire an image of the gel surface without cells (reference image).
8. Immunostaining
- Fix monolayers with 4% formaldehyde and incubate at 37 °C for 15 min.
- Remove 4% formaldehyde and add 0.2% Triton X-100 for 5 min at 37 °C to permeabilize cells.
- Remove 0.2% Triton X-100 and rinse monolayers with PBS 2x-3x.
- Cover monolayer with 2% bovine serum albumin (BSA) solution for 45 min at 37 °C.
- Remove the 2% BSA solution and rinse monolayers with PBS 2x-3x.
- Add primary Cx43 antibody at a concentration of 1:400 to the sample and incubate overnight at 4 °C.
- Remove primary antibody and rinse sample with PBS 2x-3x.
- Add secondary antibody at a concentration of 3:200 and incubate for 2 h at 37 °C.
NOTE: Samples should be covered to prevent photobleaching. - Remove secondary antibody and rinse with PBS 2x-3x.
- Cover sample with mounting medium (Fluromount-G DAPI) and seal with an 18 mm cover slip.
9. Implementation of traction force microscopy (TFM) and monolayer stress microscopy (MSM)
- Hydrogel deformation calculation
NOTE: A step-by-step displacement calculation procedure using MATLAB is given below.- Open the main traction.m file in MATLAB (for all the MATLAB routines see Supplementary Materials).
NOTE: Follow instructions provided in the code to set the MATLAB directory. - Define the following variables: image format, pixel-to-micron conversion, Young's modulus, Poisson's ratio, and microscope objective.
- Locate the bead, trypsin image, and phase image using the OpenFiles subroutine.
NOTE: For large data sets, it is best to name files in sequential order. For example, files should be named "filename1.tif", "filename2.tif", etc. - Define a square ROI (region of interest) around the cell monolayer and execute the cell_cropper subroutine to crop the original image.
- Execute the displacement_finder subroutine to compute displacements.
- Execute the Dedrift subroutine to remove any additional non-cellular displacements that may be due to microscope stage drift.
- Open the main traction.m file in MATLAB (for all the MATLAB routines see Supplementary Materials).
- Computation of tractions
NOTE: Here, unconstrained tractions are calculated using our custom-written MATLAB routine. Step by step traction computation using the MATLAB routine previously mentioned is given below.- Define the following variables within the traction.m routine: boundary condition, gel thickness, gel height, and dedrift.
- Execute the traction_finder subroutine to calculate tractions and execute the plot_traction subroutine to plot tractions. All the tractions in x-direction (Tx) and y-direction (Ty) along with their corresponding pixel locations can be found in a traction.dat file that will be generated by the code.
- Computation of intercellular stresses
NOTE: Step by step instructions for intercellular stress computation using the MATLAB routine previously mentioned are given below.- Execute the mark_circular_domain subroutine to specify the monolayer boundary. This routine will prompt all cropped phase images sequentially for the user to manually draw a boundary around the monolayer. Keep a note of the nXPts generated in command window and use them later as grid parameters in both X axis and Y axis for FEM analysis (see step 9.3.2).
- Execute Run_StressCode to compute intercellular stresses. This subroutine directly reads parameters from "model.in" file and executes "island.exe" to perform FEM analysis. Before running this routine, make sure all parameters in the model.in file, i.e. grid parameters in X and Y, pixel to micron conversion, Young's modulus of the gel, Poisson's ratio, monolayer height, and monolayer pattern (strip or hole), are edited correctly.
- Plot all the FEM results using the plot_FEM_results subroutine.
NOTE: All results generated will automatically be stored in the Resultados folder in the MATLAB directory.
Representative Results
Phase contrast images of control, 0.2 µg/mL, and 2 µg/mL chalcone treated monolayers were taken 30 minutes before chalcone treatment (Figure 1A-C) and 2 hours after chalcone treatment (Figure 1D-F). Cell-induced bead displacements (µm) were observed to decrease in both low dose chalcone and high dose chalcone conditions (Figure 2E,F) when compared to control HUVEC monolayers (Figure 2D). Prior to chalcone treatment, rms tractions were around 51 ± 8 Pa (Figure 3A-C) for all conditions. After chalcone treatment, there was a small increase in rms tractions to 59 ± 11 Pa in a low dose chalcone treated monolayers (Figure 3E) and an almost 2-fold decrease in rms tractions to 18 ± 2 in high dose chalcone treated monolayers (Figure 3F) compared to the control (Figure 3D). Prior to chalcone treatment, average normal intercellular stresses were around 220 ± 66 Pa (Figure 4A-C). After chalcone treatment, there was an increase in average normal intercellular stress magnitude to 285 ± 75 Pa with low dose chalcone treatment (Figure 4E) but a significant decrease in average normal intercellular stress magnitude to 106 ± 4 Pa with high dose chalcone treatment (Figure 4F) when compared to control average normal intercellular stresses (235 ± 18 Pa, Figure 4D). Maximum shear intercellular stresses were around 241 ± 30 Pa (Figure 5A-C) before chalcone treatment, but after the chalcone treatment, there was a decrease to 227 ± 20 Pa at low chalcone concentration (Figure 5E) and a further decrease in maximum shear intercellular stress magnitude to 91 ± 6 Pa at high chalcone concentration treatment (Figure 5F) when compared to control maximum shear intercellular stresses (270 ± 30 Pa, Figure 5D). The analysis of tractions and intercellular stresses are presented in Figure 6A-C. All plotted results were tested for statistical significance (t-test and single factor ANOVA test), and in both tests results were found to be statistically significant (p < 0.05) when independently comparing 0.2 µg/mL chalcone concentration and 2 µg/mL chalcone to control conditions (without chalcone).
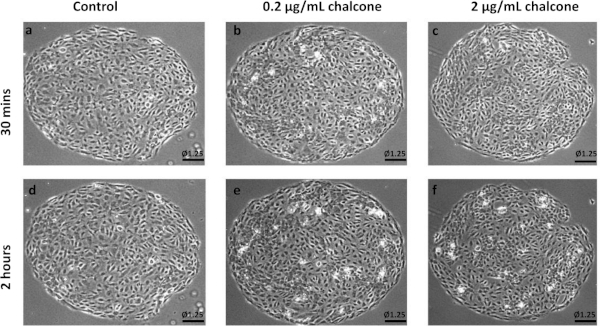
Figure 1: Representative phase contrast images of HUVEC monolayers. Example phase contrast images of control HUVECs at 30 min (A) and 2 h (D), phase contrast images of HUVECs treated with 0.2 µg/mL chalcone at 30 min (B) and 2 h (E), and phase contrast images of HUVECs treated with 2 µg/mL chalcone at 30 min (C) and 2 h (F) of experiment onset. Scale bar represents the monolayer diameter of 1.25 mm. Please click here to view a larger version of this figure.
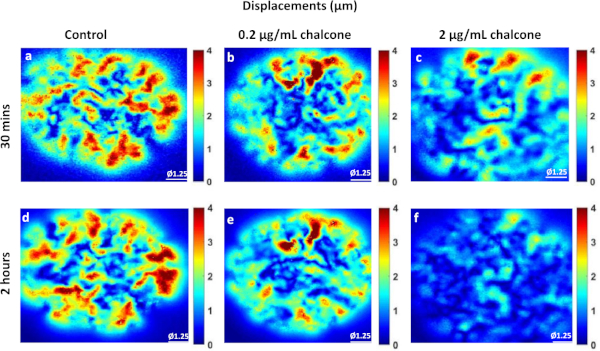
Figure 2: Illustration of displacement field produced by HUVEC monolayers. Representative displacements (µm) of control HUVECs at 30 min (A) and 2 h (D), HUVECs treated with 0.2µg/mL chalcone at 30 min (B) and 2 h (E), and HUVECs treated with 2 µg/mL chalcone at 30 min (C) and 2 h (F) of experiment onset. Scale bar represents the monolayer diameter of 1.25 mm. Color bar represents displacements in µm. Please click here to view a larger version of this figure.
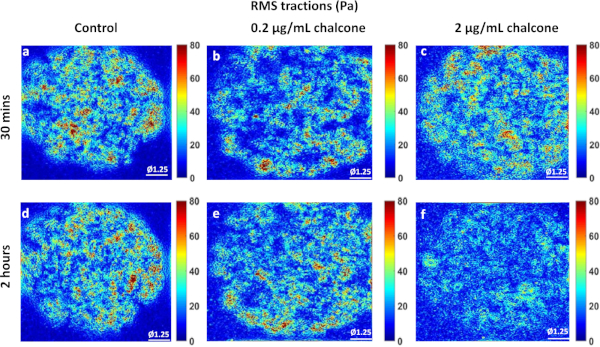
Figure 3: RMS traction distribution in HUVEC monolayers. Example of rms tractions (Pa) of control, 0.2 µg/mL chalcone, and 2 µg/mL chalcone HUVECs before chalcone treatment (A-C) and after an hour of chalcone treatment (D-F), respectively. Scale bar represents the monolayer diameter of 1.25 mm. Color bar represents RMS tractions in Pa. Please click here to view a larger version of this figure.
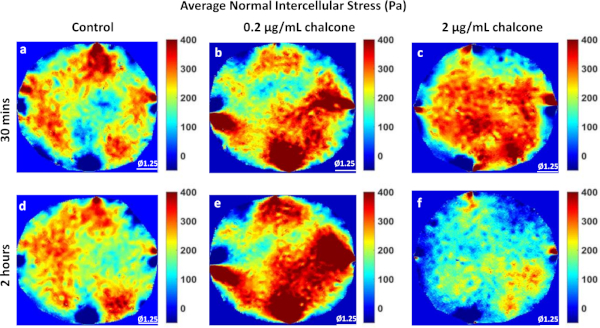
Figure 4: Average Normal Intercellular Stress distribution in HUVEC monolayers. Average normal intercellular stress (Pa) distribution of control HUVECs at 30 min (A) and 2 h (D), HUVECs treated with 0.2 µg/mL chalcone at 30 min (B) and 2 h (E), and HUVECs treated with 2 µg/mL chalcone at 30 min (C) and 2 h (F). Scale bar represents the monolayer diameter of 1.25 mm. Color bar represents stresses in Pa. Please click here to view a larger version of this figure.
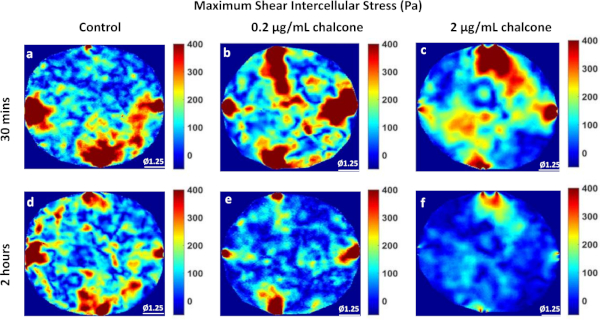
Figure 5: Maximum Shear Intercellular Stress distribution in HUVEC monolayers. Maximum shear intercellular stress (Pa) distribution of control HUVECs at 30 min (A) and 2 h (D), HUVECs treated with 0.2µg/mL chalcone at 30 min (B) and 2 h (E), and HUVECs treated with 2 µg/mL chalcone at 30 min (C) and 2 h (F). Scale bar represents the monolayer diameter of 1.25 mm. Color bar represents stresses in Pa. Please click here to view a larger version of this figure.
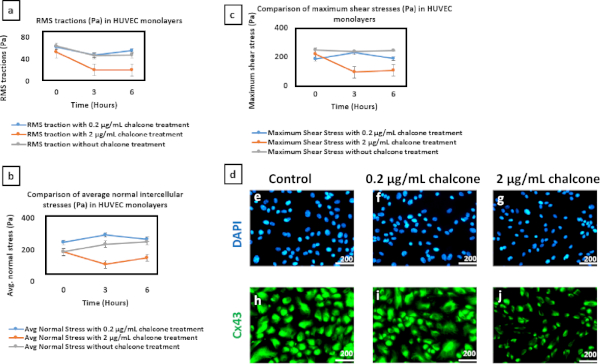
Figure 6: Comparison of RMS tractions and Intercellular Stresses in HUVEC monolayers and impact of chalcone treatment on HUVEC gap junction Cx43 structures. Plots of average normal intercellular stress (A), maximum shear intercellular stress (B) and RMS tractions (C) show the impact of chalcone doses (0.2 µg/mL and 2 µg/mL) on HUVEC monolayers compared to control. Error bars show standard error. Results were found to be statistically significant (sample size = 6 islands, with a confidence level of 95%) using both t-tests compared to control (p < 0.05) and single factor ANOVA (p < 0.05). In separate dishes, immunostaining was performed 5 h post-addition of the drug for islands of cells residing on soft 1.2 kPa hydrogels. The green color represents Cx43 and blue represents DAPI (nucleus). Panel D depicts the following: control (E,H), 0.2 µg/mL chalcone treated cells (F,I) and 2µg/mL chalcone treated cells (G,J). Scale bar = 200 µm, objective = 20x. Please click here to view a larger version of this figure.
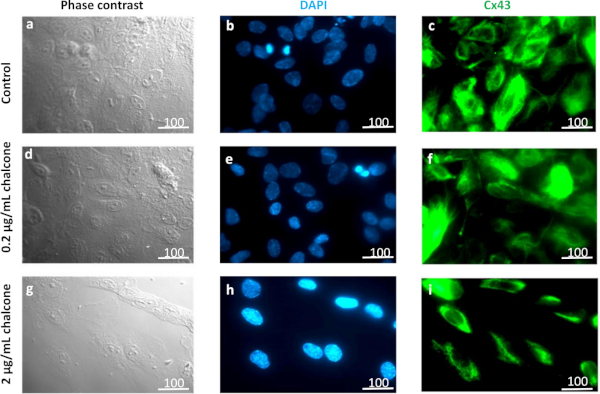
Supplementary Figure 1: High resolution Cx43 staining in HUVEC monolayers. Fixed HUVEC monolayers were stained for Cx43 (green) and nucleus (DAPI, blue) to observe the dose-dependent effect of chalcone. Higher magnification images (63x objective) reveal Cx43 localization mostly around the nucleus (evident from green fluorescence intensity). Shown are control (A-C), 0.2 µg/mL chalcone treated cells (D-F) and 2 µg/mL chalcone treated cells (G-I). Scale bar = 100 µm, objective = 63x oil immersion. Please click here to view a larger version of this figure.
| 1200 Pa | 870 Pa | 1 kPa | 4 kPa | 6.3 kPa | 11 kPa | 90 kPa | 150 kPa | |
| Solution composition | 0.05% BIS | 0.1% BIS | 0.03% BIS | 0.1% BIS | 0.03% BIS | 0.07% BIS | 0.3% BIS | 0.6% BIS |
| 5.5% Acryl | 2% Acryl | 5% Acryl | 5% Acryl | 10% Acryl | 10% Acryl | 12% Acryl | 12% Acryl | |
| Ultra pure water | 12.49 mL | 13.38 mL | 12.78 mL | 12.255 mL | 10.905 mL | 10.63 mL | 8.30 mL | 5.9 mL |
| 40% Acrylamide | 2.062 mL | 750 µL | 1.875 mL | 1.875 mL | 3.75 ml | 3.75 mL | 4.5 mL | 4.5 mL |
| 2% BIS Acrylamide | 375 µL | 750 µL | 225 µL | 750 µL | 225 µL | 525 µL | 2.12 mL | 4.5 mL |
| Fluroscent beads (0.2 µm or 0.5 µm) | 80 µL | 80 µL | 80 µL | 80 µL | 80 µL | 80 µL | 80 µL | Tractions are not measurable |
Table 1: Polyacrylamide gel making formulations for different Young's moduli.
| Volume | Thickness | Coverslip | ||
| 20-mm sl well | Thick | 500 µL | ~1 mm | 25 mm |
| Thin | 24 µL | ~100 µm | 18 mm | |
| 14-mm sl well | Thick | 175 µL | ~700 µm | 18 mm |
| Thin | 10.3 µL | ~100 µm | 12 mm | |
| 14 mm 6-well | Thick | 280 µL | ~1.5 mm | 18 mm |
Table 2: Gel volume and thickness.
Supplemental File: MATLAB Routines. Please click here to view this file (Right click to download).
Discussion
Our group, as well as others, has been successfully using TFM and MSM to probe the influence of cell-cell junctions in various pathological and physiological cellular processes in vitro7,15,18,27. For example, Hardin et al. presented a very insightful study that suggests intercellular stress transmission guides paracellular gap formation in endothelial cells15. While it possible to relate the Cx43-related changes we report here to the changes reported by Hardin et al., we do not specifically address paracellular gap formation in this protocol. Here, we presented a mechanics-based protocol to specifically target the gap junction Cx43 and investigate its influence on endothelial biomechanics.
To make this protocol successful, there were a few challenges that we had to overcome, some of which could occur should other researchers decide to adopt our protocol for similar studies. A major challenge was to find an optimum chalcone dose range where Cx43 expression could be inhibited, while simultaneously keeping our endothelial monolayers intact. The IC50 of chalcone for HUVECs has previously been reported to be 10.01 µg/mL28. However, when we exposed our HUVEC monolayers to multiple chalcone concentrations ranging from 0.2 µg/mL to 20 µg/mL, we found a chalcone concentration of 2 µg/mL to be the highest concentration our monolayers could withstand while still remaining confluent. A confluent monolayer was essential for this protocol, as a monolayer is required to measure intercellular stresses using MSM. Next, we performed an immunofluorescence assay to determine if the selected doses of chalcone successfully disrupted Cx43 structure or apparent expression. Our result revealed that while disruption of Cx43 structure with 0.2 µg/mL chalcone was visually difficult to distinguish from control, cells treated with 2 µg/mL of chalcone appeared to show an apparent difference in Cx43 structure and potentially expression (Figure 6D and Supplementary Figure 1). In addition, our results showed that Cx43 disruption indeed influences endothelial biomechanics by reducing tractions and intercellular stresses at its highest concentration. These findings are in agreement with Bazellieres et al. who showed silencing of Cx43 with siRNA to also reduce tractions and intercellular stresses, but in a sheet of epithelial cells27. Although our results are in agreement with others it should be noted that the molecule we used to disrupt Cx43 expression, chalcone, has also been suggested to influence activation of MAPK and NFkB in addition to Cx43 disruption26. Therefore, since we did not specifically look at the above-mentioned molecules or their associated pathways, we cannot rule out the influence a potential MAPK and NFkB perturbation can have on endothelial biomechanics as well.
Another key point worth mentioning is that the recovered tractions and intercellular stresses are 2D in nature and ignore out of plane (z-direction) tractions and intercellular stresses12,13. While there is a small error associated with ignoring out-of-plane stresses, this error is negligible13. In addition, the lateral dimension of the monolayer is sufficiently large (1.25 mm) relative to the monolayer height (~ 5 µm) such that we would not expect significant displacements in the z-direction. Furthermore, the MSM calculation offers error at the monolayer boundary12,13. However, Tambe et al. experimentally showed that errors are highest at the optical edges (i.e., the monolayer boundary) and decays quickly distal from boundary edges13. We perform micropatterning and then calculate intercellular stresses of the entire monolayer to avoid boundary errors that can occur during intercellular stress computation.
Monolayer stress microscopy is utilized in this protocol as the intercellular stress information yielded from this method is essential in order to have a more complete understanding of the role gap junction disruption has on endothelial biomechanics. In addition, intercellular stresses have been suggested to be important in endothelial barrier function, as suggested by Hardin et al.15 and Krishnan et al.17, for example. Furthermore, while intercellular stresses were correlated to tractions in the representative data presented here, this is not always the case depending on the stimulus. In the study of Steward et al., for example, endothelial intercellular stresses were shown to decrease under fluid shear, while tractions remained relatively unchanged7. Also, the protocol we present here does not allow for the simultaneous measurement of cell-derived mechanical forces and staining of junctions and focal adhesions, but such an addition would be complimentary to this protocol. In closing, the protocol we present here describes a mechanics-based method to investigate the influence Cx43 has solely on endothelial biomechanics, specifically endothelial cell-derived forces. We believe our mechanics-based protocol can be used in conjunction with currently existing biological-based protocols to provide truly groundbreaking work in the field.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by the University of Central Florida start-up funds and the National Heart, Lung, And Blood Institute of the National Institute of Health under award K25HL132098.
Materials
| 18 mm coverslip | ThermoFisher | 18CIR-1 | Essential to flatten polyacrylamide gels |
| 2% bis-acrylamide | BIO-RAD | 1610143 | Component of polyacrylamide gel |
| 2′,5′-Dihydroxychalcone | SIGMA | IDF00046 | To disrupt Cx43 structure |
| 3-(Trimethoxysilyl)propyl methacrylate | SIGMA | 2530-85-0 | Stock solution to make bind silane mixture with acetic acid and ultra-pure water |
| 40% Acrylamide | BIO-RAD | 1610140 | Component of polyacrylamide gel |
| Acetic acid | Fisher-Sceintific | 64-19-7 | Essential to make bind saline solution |
| Alexa Fluro 488 goat anti-mouse IgG; | ThermoFisher | Catalog # A-11001 | Secondary antibody |
| Ammonium persulfate | BIO-RAD | 1610700 | Polyacrylamide gel polymerizing agent |
| Bovine Serum Albumin (BSA) | SIGMA | 9048-46-8 | To make blocking solution |
| Bovine Type I Atelo-Collagen Solution, 3 mg/mL, 100 mL | Advance Biomatrix | 5005-100ML | Use as a extracellular matrix |
| Corning Cell Culture Phosphate Buffered Saline (1x) | Fisher-Sceintific | 21040CV | Buffer Saline needed for cell culture |
| Dimethyl Sulfoxide, Fisher BioReagents | Fisher-Sceintific | 67-68-5 | To dissolve chalcone and make stock solution |
| Fluoromount-G with DAPI | ThermoFisher | 00-4959-52 | Mounting medium for immunostaing used to stain for DAPI |
| Fluroscent microsphere Carboxylate-modified beads | ThermoFisher | F8812 | 0.5 micron carboxylate-modified beads (red), 2% solids |
| HEPES buffer solution 1 M | SIGMA | 7365-45-9 | Essential to |
| LVES | ThermoFisher | A1460801 | Essential HUEVC media 200 supplement |
| Medium 200 | ThermoFisher | M200500 | Essential media for HUVEC cell culture |
| Mouse monoclonal Cx43 antibody (CX – 1B1) | ThermoFisher | Catalog #13-8300 | Primary antibody for Cx43 |
| Petri dish (35 mm dia) | CellVis | D35-20-1.5H | 35 mm petri dish with a 20 mm center well |
| Sulfo-SANPAH Crosslinker 100 mg | Proteochem | 102568-43-4 | Essential to functionalize polyacrylamide gel surface |
| SYLGARD 184 Silicone Elastomer Kit | DOW corning | 2646340 | Silicon elastomer with curing agent to make PDMS |
| TEMED | BIO-RAD | 1610801 | Polyacrylamide gel polymerizing agent |
| Triton-X 100 | SIGMA | 9002-93-1 | To permeabilize cells |
| Trypsin -EDTA | ThermoFisher | 25300054 | Used to detach cells |
Referências
- Mammoto, T., Mammoto, A., Ingber, D. E. Mechanobiology and developmental control. Annual Review of Cell and Developmental Biology. 29, 27-61 (2013).
- Schwarz, U. S., Soine, J. R. Traction force microscopy on soft elastic substrates: A guide to recent computational advances. Biochimica et Biophysica Acta. 1853 (11 Pt B), 3095-3104 (2015).
- Style, R. W., et al. Traction force microscopy in physics and biology. Soft Matter. 10 (23), 4047-4055 (2014).
- Colin-York, H., et al. Super-Resolved Traction Force Microscopy (STFM). Nano Letters. 16 (4), 2633-2638 (2016).
- Zimmermann, J., et al. Intercellular stress reconstitution from traction force data. Biophysical Journal. 107 (3), 548-554 (2014).
- Islam, M. M. Recent Advances in Experimental Methods of Cellular Force Sensing. Biomedical Journal of Science & Technical Research. 17 (3), (2019).
- Steward Jr, R., Tambe, D., Hardin, C. C., Krishnan, R., Fredberg, J. J. Fluid shear, intercellular stress, and endothelial cell alignment. American Journal of Physiology-Cell Physiology. 308 (8), C657-C664 (2015).
- Trepat, X., et al. Physical forces during collective cell migration. Nature Physics. 5, 426-430 (2009).
- Li, Z., et al. Cellular traction forces: a useful parameter in cancer research. Nanoscale. 9 (48), 19039-19044 (2017).
- Brugues, A., et al. Forces driving epithelial wound healing. Nature Physics. 10 (9), 683-690 (2014).
- Pasqualini, F. S., et al. Traction force microscopy of engineered cardiac tissues. PLoS One. 13 (3), e0194706 (2018).
- Tambe, D. T., et al. Collective cell guidance by cooperative intercellular forces. Nature Materials. 10 (6), 469-475 (2011).
- Tambe, D. T., et al. Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS One. 8 (2), e55172 (2013).
- Butler, J. P., Tolic-Norrelykke, I. M., Fabry, B., Fredberg, J. J. Traction fields, moments, and strain energy that cells exert on their surroundings. American Journal of Physiology-Cell Physiology. 282 (3), C595-C605 (2002).
- Hardin, C. C., et al. Long-range stress transmission guides endothelial gap formation. Biochemical and Biophysical Research Communications. 495 (1), 749-754 (2018).
- Cho, Y., Son, M., Jeong, H., Shin, J. H. Electric field-induced migration and intercellular stress alignment in a collective epithelial monolayer. Molecular Biology of the Cell. 29 (19), 2292-2302 (2018).
- Krishnan, R., et al. Substrate stiffening promotes endothelial monolayer disruption through enhanced physical forces. American Journal of Physiology-Cell Physiology. 300 (1), C146-C154 (2011).
- Islam, M. M., Steward, R. L. Probing Endothelial Cell Mechanics through Connexin 43 Disruption. Experimental Mechanics. 59, 327 (2019).
- Figueroa, X. F., Duling, B. R. Gap junctions in the control of vascular function. Antioxidants & Redox Signaling. 11 (2), 251-266 (2009).
- Nielsen, M. S., et al. Gap junctions. Comprehensive Physiology. 2 (3), 1981-2035 (2012).
- Sohl, G., Willecke, K. Gap junctions and the connexin protein family. Cardiovascular Research. 62 (2), 228-232 (2004).
- Haefliger, J. A., Nicod, P., Meda, P. Contribution of connexins to the function of the vascular wall. Cardiovascular Research. 62 (2), 345-356 (2004).
- Marquez-Rosado, L., Solan, J. L., Dunn, C. A., Norris, R. P., Lampe, P. D. Connexin43 phosphorylation in brain, cardiac, endothelial and epithelial tissues. Biochimica et Biophysica Acta. 1818 (8), 1985-1992 (2012).
- Liao, Y., Day, K. H., Damon, D. N., Duling, B. R. Endothelial cell-specific knockout of connexin 43 causes hypotension and bradycardia in mice. Proceeding of the National Academy of Sciences of the United States of America. 98 (17), 9989-9994 (2001).
- Walker, D. L., Vacha, S. J., Kirby, M. L., Lo, C. W. Connexin43 deficiency causes dysregulation of coronary vasculogenesis. Biologia do Desenvolvimento. 284 (2), 479-498 (2005).
- Lee, Y. N., et al. 2′,5′-Dihydroxychalcone down-regulates endothelial connexin43 gap junctions and affects MAP kinase activation. Toxicology. 179 (1-2), 51-60 (2002).
- Bazellieres, E., et al. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nature Cell Biology. 17 (4), 409-420 (2015).
- Nam, N. H., et al. Synthesis and cytotoxicity of 2,5-dihydroxychalcones and related compounds. Archives of Pharmacal Research. 27 (6), 581-588 (2004).

