Generation of Chimeric Axolotls with Mutant Haploid Limbs Through Embryonic Grafting
Summary
This goal of this protocol is to produce chimeric axolotls with haploid forelimbs derived from Cas9-mutagenized donor tissue using embryonic tissue grafting techniques.
Abstract
A growing set of genetic techniques and resources enable researchers to probe the molecular origins of the ability of some species of salamanders, such as axolotls, to regenerate entire limbs as adults. Here, we outline techniques used to generate chimeric axolotls with Cas9-mutagenized haploid forelimbs that can be used for exploring gene function and the fidelity of limb regeneration. We combine several embryological and genetic techniques, including haploid generation via in vitro activation, CRISPR/Cas9 mutagenesis, and tissue grafting into one protocol to produce a unique system for haploid genetic screening in a model organism of regeneration. This strategy reduces the number of animals, space, and time required for the functional analysis of genes in limb regeneration. This also permits the investigation of regeneration-specific functions of genes that may be required for other essential processes, such as organogenesis, tissue morphogenesis, and other essential embryonic processes. The method described here is a unique platform for conducting haploid genetic screening in a vertebrate model system.
Introduction
Historically, embryonic tissue grafting in amphibians has been an important technique for exploring fundamental mechanisms of developmental biology and regeneration. The axolotl, a species of salamander, possesses an impressive ability to regenerate tissues and complex structures such as limbs and organs after injury or amputation. Similarly impressively, they can receive, without rejection, tissue grafts from other individuals at embryonic, juvenile, and adult stages1,2,3. Regions of embryos that produce whole structures such as limbs, tails, eyes, and heads, and more specific tissues, such as neuroectoderm and somites, can be grafted between embryos to produce chimeric animals1,2,4,5,6. For nearly a century, studies of such chimeric animals have provided crucial insights into regeneration, tissue differentiation, size control, and patterning1,7,8.
In the last the decade, numerous transcriptional studies of regenerating tissues have produced insights into the genetic programs underlying salamander regeneration9,10,11,12,13. These studies have added to an expanding list of candidate genes that, to date, are largely uncharacterized in the context of regeneration. Targeted mutagenesis techniques, such as CRISPR/Cas, now permit the investigation of such genes, and such genetic approaches are greatly facilitated by the recent sequencing and assembly of the large axolotl genome14,15,16.
We sought to develop techniques that coupled classic developmental biology with new genetic technology for the purpose of dissecting the mechanisms of regeneration. Methods for generating haploid embryos of axolotls and other salamanders have been established for decades17. While these techniques have long been noted to be advantages of salamanders as genetic model organisms18, few subsequent genetic studies have incorporated haploid animals. We use in vitro activation in the axolotl to produce haploid embryos that serve as tissue donors for grafting19. Using embryos carrying fluorescent genetic markers, we have devised reliable methods for generating limbs derived almost entirely from donor tissues (Figure 1A). By combining these two techniques, we have bypassed the late embryonic lethality associated with haploidy, allowing for the production of fully developed, grafted haploid limbs (Figure 1B, Figure 1B', and Figure 2).
By conducting CRISPR/Cas-mediated mutagenesis in haploid embryos prior to grafting to create chimeric axolotls with mutant haploid limbs, we may investigate gene function specifically within the context of limb development and regeneration. This allows the rescue of limbs from potentially embryonic-lethal mutant phenotypes. While CRISPR/Cas microinjection can generate animals that are highly mutant, such animals are typically highly mosaic, with some degree of retention of wildtype alleles and a variety of distinct mutations at targeted sites14,20. CRISPR-based mutagenesis in haploid cells increases the penetrance of single allele loss-of-function mutations, as they cannot be masked by retained wildtype alleles. For this reason, CRISPR-based screening in haploid cell lines is increasingly used to investigate the genetic basis of many cellular processes21,22,23. By combining CRISPR-based lineage tracing with our haploid limb bud grafting protocols, the approach described here can serve as a platform for haploid genetic screens in living animals20.
Protocol
Experimental procedures used in this protocol were approved by the Yale University Institutional Animal Care and Use Committee (IACUC, 2017–10557) and were in accordance with all federal policies and guidelines governing the use of vertebrate animals. All animal experiments were carried out on Ambystoma mexicanum (axolotls) in facilities at Yale University.
1. Diploid Embryo Generation
- Obtain GFP+ diploid embryos to serve as graft hosts through natural mating using one or two gfp parents24.
- Collect freshly laid diploid eggs and place them in a metal sieve.
- Rinse the eggs thoroughly with 40% Holtfreter's solution (20 mM NaCl, 0.2 mM KCl, 0.8 mM NaHCO3, 0.2 mM CaCl2, 4 mM MgSO4, pH to 7.4).
- Place the eggs in fresh 40% Holtfreter's solution and store at 12 °C.
NOTE: Diploid embryos should always be obtained before moving on to haploid embryo generation.
2. Haploid Embryo Generation
- Female gamete donor preparation
- 48 h before conducting in vitro activation, anesthetize a sexually mature white or white/RFP female axolotl by immersion in 1 g/L HEPES-buffered MS-222 in 40% Holtfreter's solution25.
- Ensure that the animal is fully anesthetized after approximately 30 min of immersion by firmly pinching its tail between the thumb and forefinger. Fully anesthetized animals will not physically respond to any pinch.
- Prepare a solution of human chorionic gonadotropin (HCG) containing 10,000 U/mL in sterile saline.
- Using a 30 G insulin syringe, inject 0.15 CC of HCG (1,500 units) into the musculature dorsal to the hind limb of the anesthetized female at 45° angle to the midline to avoid contact with the spinal cord.
- Return the female to fresh 40% Holtfreter's solution and place in an 8−12 °C refrigerator.
- After 48 h, return the female to room temperature. Place a few stones or plastic plants in her container as surfaces for egg laying.
NOTE: Females injected with HCG will often deposit empty jelly cases for several hours before laying eggs. - Remove the stones or plastic plants after the female has begun consistently laying eggs.
- Allow the female to sit in the empty tank for 30 min to 1 h.
NOTE: Withholding materials for the animal to lay eggs on will allow tighter control of oocyte collection.
- Male gamete collection
- While the female's egg laying is stalled, anesthetize a sexually mature GFP+ male axolotl to serve as a sperm donor, as in step 2.1.1.
- Place the fully anesthetized male on his back on damp paper towels under a dissecting microscope.
- If the experimenter is righthanded, position the tip of a P1000 pipette at the base of the cloaca with the right hand.
- Place the left forefinger and thumb of the left hand 2−3 cm rostral to the pelvis. Gently squeeze the animal while moving the fingers towards the hind legs to flush out spermic urine samples.
- Collect each that is flushed out of the cloaca into an individual microcentrifuge tube. Repeat this process to collect 6 to 10 samples.
- Pipette 5.0 µL from each sample onto a petri dish to inspect the quality of the sperm using an inverted microscope.
NOTE: Concentrated sperm samples are a milky white color, typically range from 5 to 20 µL, and are usually retrieved after several, higher volume spermic urine samples are extracted. Sperm will collect at the bottom of the tubes in higher volume samples when left undisturbed. Concentrated samples of healthy sperm are highly motile and lose activity as their concentration decreases. Healthy males can produce up to 50 µL of concentrated sperm.
- Female gamete collection
- After obtaining and confirming a healthy sperm sample, anesthetize the HCG-injected female axolotl as in step 2.1.1.
- Place the fully anesthetized female on damp paper towels on her back.
- Extract unfertilized eggs from the female using a hand motion similar to that in step 2.2.4.
- Collect the eggs using wet forceps and transfer them to a 10 cm petri dish. Treat eggs with irradiated sperm within 15 min of collection.
- Male gamete preparation and in vitro activation
- Use a P10 or P20 pipette to pipette the sperm up and down, breaking apart the clumps to form a homogenous suspension.
- Approximate the percentage of motile sperm by placing a 0.5 µL drop of undiluted sperm suspension on a petri dish lid or glass slide and examining with an inverted microscope.
- Add 9.5 µL of 0.1x Marc's modified Ringer's solution (MMR; Table of Materials) to this sample to make a 20x sperm dilution. Gently pipette up and down to mix.
- With an inverted microscope, count the sperm in three 1.0 µL drops of the 20x diluted aliquot on a petri dish cover or hemocytometer to estimate the sperm concentration of the undiluted suspension.
- Prepare the sperm for irradiation by diluting an aliquot of the original sperm sample to about 80,000 motile cells/mL in sterile 0.1x MMR.
- Count the number of eggs obtained within the past 15 min. Add 0.5 µL of the freshly diluted sperm from step 2.4.5 per egg to a petri dish. Use the pipette tip to spread this suspension into a 1 mm thick layer.
- Using a plastic lift, place the sample 4 cm from the bulbs of a 254 nm crosslinker. Genetically inactivate the sperm by irradiating the sample with 800,000 uJ/mm2.
- Using a P10 pipette, pipette the suspension onto the unfertilized eggs, coating each egg with 0.25−0.5 µL of irradiated sperm. Allow the eggs to sit at room temperature for 30 min.
- After 30 min, flood the eggs with 0.1x MMR. Immediately place the eggs in an 8−10 °C incubator for injections the next day or at 18 °C for injections within 7 hours post activation (hpa).
- Dejelly haploids using sharp forceps 30 min after hydration. Immediately place the eggs in an 8−10 °C incubator for injections the next day or at 18 °C for injections the same day.
3. Haploid Mutagenesis and Maintenance
- CRISPR/Cas9 microinjections
- Design sgRNAs using CRISPRscan and synthesize26,27.
- For multiplex mutagenesis, prepare a stock of 5 sgRNAs (10 ng/µL for each sgRNA) and Cas9 protein (1 µg/µL). Prepare a 100-fold dilution of this stock (0.1 ng/µL per sgRNA, 10 ng/µL Cas9) for injection. For single gene, high mutation frequency mutagenesis, follow the protocol outlined previously28.
- Inject haploid embryos at the single cell stage 7 hpa if stored at 18 °C or inject embryos the next day at the 2−8 cell stage if they are stored at 8−10 °C.
- Transfer the embryos to 1.0x MMR with 20% polysucrose 400.
- If single cell, inject each embryo with a 5 nL drop of the injection solution (approximately ¼ radius of the egg) containing a total mass of 0.5 pg/sgRNA and 50 pg Cas9 protein. If multicellular, distribute this mass by injecting smaller volumes into multiple cells.
- Allow the embryos to heal in the polysucrose 400 for a minimum of 4 h and a maximum of 18 h at 18 °C.
- Haploid embryo housing
- Transfer the embryos to 0.1x MMR with antibiotic-antimycotic.
- House each embryo in an individual well of a 24-well plate, as some will die or develop abnormally. Maintain at 16−18 °C. Lower temperatures can be used to prolong development, if necessary.
- Replace the media with fresh 0.1x MMR with antibiotic-antimycotic every other day.
4. Diploid Host Embryo Preparation
- Maintain embryos in the jelly coating at 12 to 16 °C until they reach stage 22 to 26.
- Collect the embryos that are ready for grafting, place them within a sieve (4 mm mesh size), and gently rinse them with 40% Holtfreter's solution.
- Transfer the embryos into filter-sterilized 0.1x MMR containing 1.5% bleach for up to 2 min. Completely submerge the embryos in the bleach solution and swirl them gently to ensure that the jelly coating makes full contact with the bleach solution to kill the microbes present on the embryos.
- After 2 min, dilute the bleach solution containing the embryos with an equal volume of filter-sterilized 0.1x MMR.
- Gently pour the embryos into a bleach-sanitized sieve (4 mm mesh size) and rinse the embryos five times with filter-sterilized 0.1x MMR.
- Place the embryos into sterile 10 cm petri dishes with 0.1x MMR with antibiotics for dejellying (penicillin 100 units/mL, streptomycin 100 µg/µL, 0.25 µg/mL, gentamicin 25 µg/mL).
- Under a fluorescent stereomicroscope, remove the jelly coats and vitelline membranes from GFP+ embryos using sharp forceps (tip dimensions 0.05 x 0.01 mm2).
- Transfer the GFP+ embryos to a new petri dish. Minimize the amount of liquid transferred from the petri dish where they were dejellied.
- Rinse the GFP+ host embryos with sterile 0.1x MMR with antibiotics four to six times in order to remove contaminants.
- Place the clean embryos at 4 °C overnight before grafting.
NOTE: Cooling the embryos makes them more rigid and clean separation of the mesoderm from the endoderm feasible.
5. Haploid-diploid Chimera Generation
- Surgical dish and media preparation
- Prepare sterile surgical operating dishes by pouring autoclaved 2% agarose in 0.1x MMR into sterile 35 mm easy-grip petri dishes. Fill the petri dishes halfway with agarose.
- After the agarose cools, use a sterile scalpel to cut a 25 mm long, slanted trough in the agarose to hold the embryos in place.
- Fill the dish with sterile surgical media (0.1x MMR with anti-mycoplasma 2.5 µg/mL, amphotericin B 0.25 µg/mL, and ciprofloxacin 10.0 µg/mL) and refrigerate at 4 °C.
- Embryo grafting procedure
- Place one healthy haploid donor with one or two stage-matched GFP+ diploid host embryos inside the trough of the prechilled operating dish containing surgical media (Figure 3).
NOTE: Perform the procedure on a cooling stage (10 °C or lower) if possible. - Use two ultra-fine, autoclaved forceps (straight tip, tip dimensions 0.05 x 0.02 mm2) to remove the ectoderm and mesoderm layers from the host with the limb bud near the center (Figure 4).
NOTE: The rectangular tissue graft region encompasses the limb bud and extend from the ninth somite to the posterior half of the gill bulge, about 2 mm, along the anterior posterior axis. Along the dorsoventral axis, the grafted region spans approximately 1.5 mm, including the somites to just beyond the ventral edge of the gill bulge. The grafted region includes all lateral plate mesoderm and the lateral halves of the somites, without disturbing underlying endoderm. See Figure 4 and the accompanying video for details. - Set aside the host tissue and remove an equivalently sized tissue sheet from the haploid donor using the same methods.
- Place the haploid donor tissue sheet onto the corresponding region of the donor embryo.
- Secure the tissue by covering it with an autoclaved, rectangular glass shard from a crushed microscope cover glass and gently pressing it into the host embryo body.
- Flip the haploid donor embryo onto its other side to harvest the limb bud for the second host embryo. Repeat steps 5.2.2 through 5.2.5.
- Carefully remove the remaining excess host tissues and the donor embryo from the dish.
- Leave the grafts with the glass shard anchors in place for 60−75 min, checking every 20 min to ensure that the glass has not slipped off.
- After the tissue grafts fully adhere, use the forceps to slowly peel off the glass shard anchors.
- Place one healthy haploid donor with one or two stage-matched GFP+ diploid host embryos inside the trough of the prechilled operating dish containing surgical media (Figure 3).
- Chimera maintenance
- Transfer the engrafted embryos to fresh surgical media and maintain them at 8−12 °C overnight to heal. Individually house engrafted embryos in 12 or 24-well plates.
- After 36−48 h, transfer the engrafted embryos to sterile 0.1x MMR antibiotic-antimycotic. The strong antibiotics in the surgical media cause toxicity in engrafted embryos after 3 days.
- Replace the media with fresh 0.1x MMR and antibiotics every 2−3 days.
- Maintain the engrafted embryos at 18 °C until they are able to feed28.
- After 1 to 2 months of development and care, haploid limbs can be scored for purity of the graft using a fluorescent dissection microscope.
NOTE: The presence of non-neural or non-blood host-derived GFP-tissue in limbs is an indicator of impure grafting and is often associated with abnormal limb development. These animals should be excluded from further analysis.
Representative Results
Developing haploid embryos can be distinguished from diploid embryos by their 'haploid syndrome' phenotype29. At graft-stage, haploid embryos exhibit reduced curvature along the anterior-posterior axis and incomplete enclosure of the yolk plug (Figure 3A). A fluorescent microscope can be used to ensure that haploid embryos are free of paternally derived GFP expression (Figure 3B).
When grafts are clean, GFP expression should be limited primarily to the brachial plexus, the neural network derived from the host's spinal cord, as seen in Figure 2. Punctate GFP expression will also be present in single cells that appear to be sensory neurons and blood-derived host cells, which migrate into the developing limb. When RFP+ donors are used, haploid graft limbs will show universal expression of RFP (Figure 1B'). Throughout development, non-mutagenized haploid limbs are significantly shorter than the opposing diploid forelimbs in size-matched chimeric animals (Figure 1B, Figure 1B', and Figure 5, n = 16 limb pairs, haploid mean = 0.522 cm, SD = ± 0.087 cm, diploid mean = 0.667 cm, SD = ± 0.069 m, paired T-test p-value < 0.0001, mean ratio = 0.784, SD = ± 0.113). Non-mutagenized haploid limbs also fully regenerate (4/4 haploid and 4/4 diploid complete regeneration, Figure 6), though they show a slight delay in reaching the digital outgrowth stage compared to diploids (n = 4 haploid, n = 4 diploid; 23 days after amputation: haploids = 3 palette and 1 digital outgrowth stage limbs; diploids = 4 digital outgrowth stage limbs).
Successful grafting requires practice, and consistency will vary depending on the technician's micromanipulation skills and sterile technique. Failed and impure grafts produce a variety of phenotypes, as seen in Figure 7 and Figure 8. In our hands, approximately 38.7% (SD = ± 8.78%) of oocytes develop into normally developed haploid embryos and 11.1% (SD = ± 5.46%) of all haploid-diploid grafts produce viable animals with normally developed, cleanly grafted haploid limbs (Figure 9).
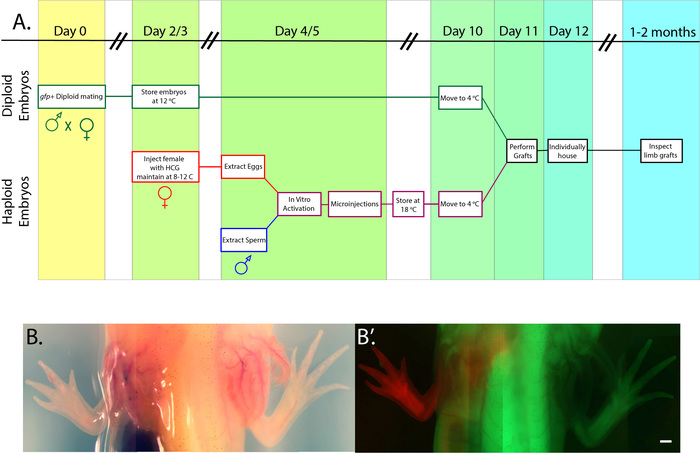
Figure 1: Overview of protocol and an example of a chimeric axolotl. (A) Schematic outlining the relative timing of steps taken to obtain diploid host embryos, sperm, and eggs in order to generate haploids and chimeric embryos. (B) Composite bright image of a juvenile axolotl produced by embryonic limb bud grafting from an RFP+ haploid embryo to a GFP+ diploid host (B') Overlay of green and red fluorescent images of the same 8 cm juvenile axolotl. The haploid limb is grossly normal, but shorter than the opposing diploid limb. Scale bar = 1 mm. Please click here to view a larger version of this figure.
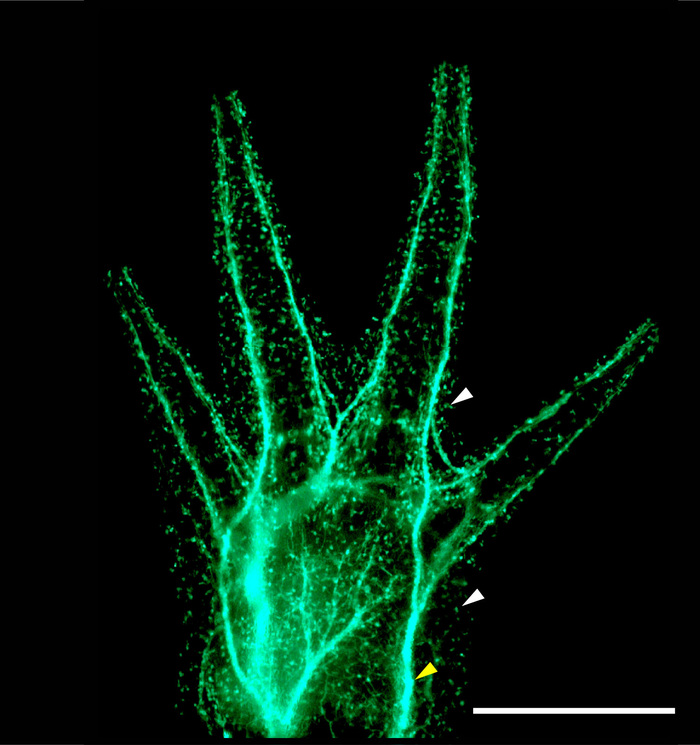
Figure 2: Fluorescent image of a normally developed and cleanly grafted haploid limb. GFP- haploid limb grafted to a GFP+ diploid host shows GFP expression pattern that appears to be restricted to spinal nerves innervating the limb (yellow arrow) and individual sensory neurons and blood-derived cells (white arrows). The limb pictured belonged to a 4 cm juvenile. Scale bar = 1 mm. Please click here to view a larger version of this figure.
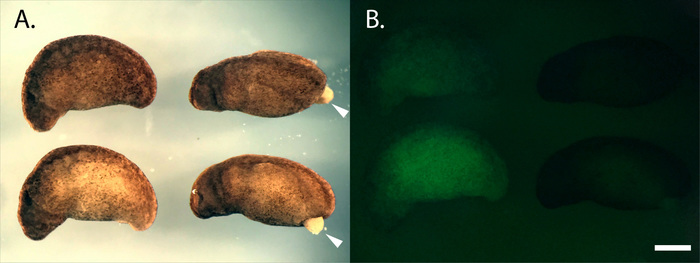
Figure 3: Comparison of diploid and haploid embryos. (A) Light image of diploids (left) and haploids (right). Note the reduced AP-axis curvature and the protruding endoderm of the haploid embryos (white arrows). (B) Green fluorescent image of the same embryos. GFP- haploids were generated using eggs from a GFP- female and irradiated sperm from a GFP+. Diploids are GFP+. Embryos pictured are approximately stage 2530. Staging series. Scale bar = 1 mm. Please click here to view a larger version of this figure.
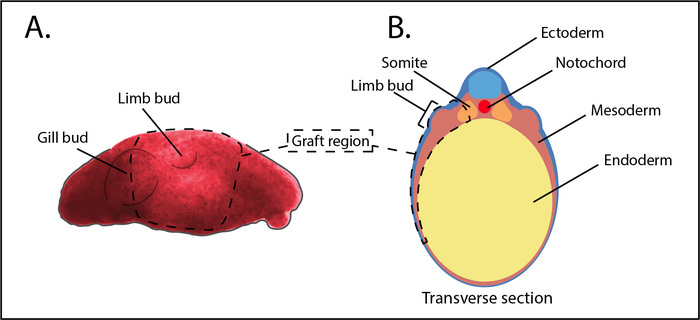
Figure 4: Schematic outlining the extent of a haploid limb bud graft. (A) Lateral view of a stage 25 haploid embryo. The dotted lines show the approximate area that should be transplanted, relative to the gill bulge and limb bud. (B) Transverse schematic of stage 25. The dotted lines show the approximate area and tissue types that should be removed from the host and replaced with the corresponding haploid donor tissue. Please click here to view a larger version of this figure.
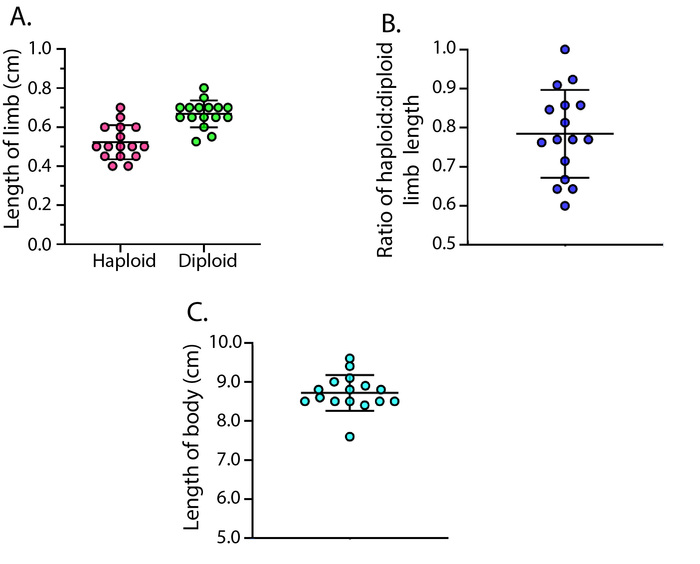
Figure 5: Comparison of haploid and diploid limb lengths. (A) Scatter plot of the lengths of haploid limbs (pink) and opposing diploid limbs (green) of 16 similarly sized chimeric animals (haploid mean = 0.522 cm, SD = ± 0.087 cm, diploid mean = 0.667 cm, SD = ± 0.069 cm). Limbs were measured from the base of the zeugopod to the junction of digits 2 and 3. (B) Scatter plot of the haploid limb length to diploid limb length ratios of the 16 animals (mean ratio = 0.784, SD = ± 0.113 cm). (C) Scatter plot of the body lengths (cm) of the 16 measured chimeras (mean animal length = 8.72 cm ± 0.456 cm). Animals were measured from the tip of the snout to the tail tip. Please click here to view a larger version of this figure.
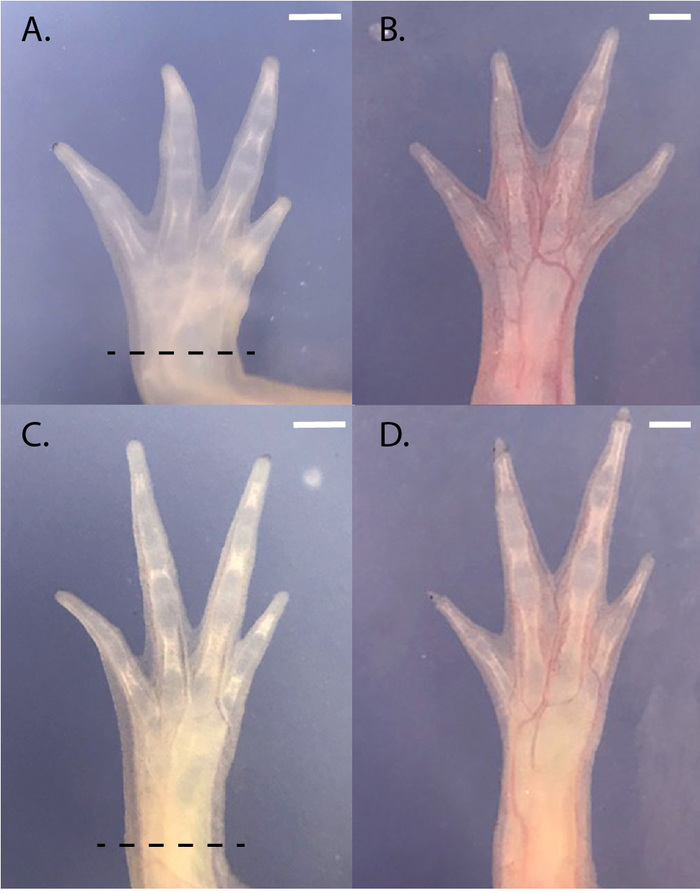
Figure 6: Comparison of regeneration in haploid and diploid limbs. (A) Haploid limb pre-amputation. (B) The same haploid limb after regeneration. (C) Diploid limb pre-amputation. (D) The same diploid limb after regeneration. 4/4 haploid limbs and 4/4 diploid limbs regenerated with normal morphology. Black dotted lines indicate the amputation plane. Scale bars = 1 mm. Amputations were performed on size-matched juvenile animals (8.5 cm). Please click here to view a larger version of this figure.
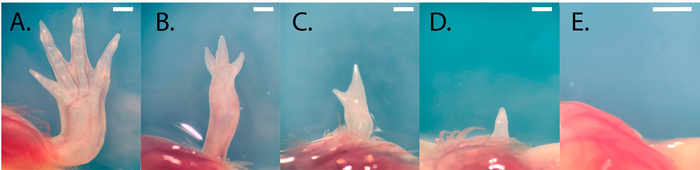
Figure 7: Examples of normal and abnormal development of haploid limbs. (A) A haploid limb with grossly normal morphology. (B-E) Examples of grafts that failed to support complete development of haploid. (B) Oligodactyly. (C) More severe oligodactyly. (D) No distinct digital structures are present. (E) No limb. The limbs pictured belong to similarly sized juveniles (8.0 to 10 cm). Scale bars = 1 mm. Please click here to view a larger version of this figure.
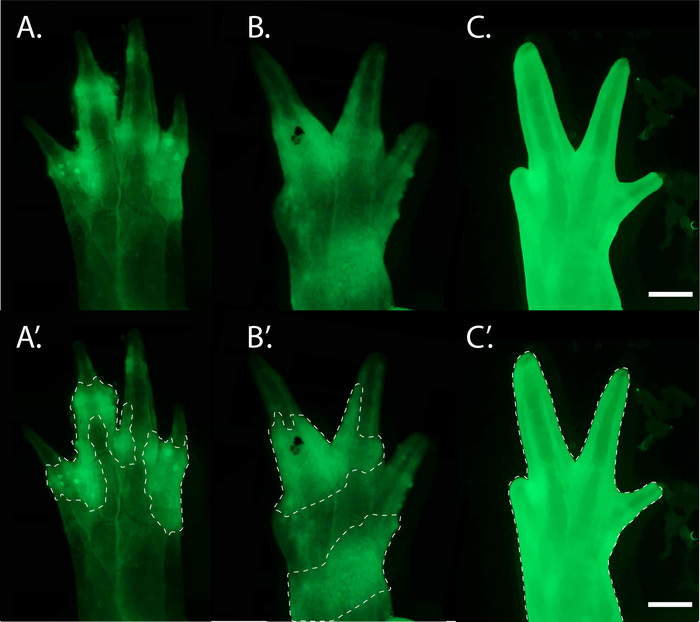
Figure 8: Examples of GFP+ cells in impure haploid limb grafts. (A,A') Morphologically normal haploid graft limb with GFP+ diploid cells in the skin and dermis (white dashed outline) revealed by fluorescent microscopy. (B,B') A grafted abnormal haploid limb with GFP+ diploid cells contributing extensively to the dermis and skin (white outline). (C,C') A grafted abnormal haploid limb in which the epidermis has been replaced with GFP+ diploid cells (white outline). The limbs pictured belong to similarly sized juveniles (8.0 to 10.0 cm). Scale bar = 1 mm. Please click here to view a larger version of this figure.
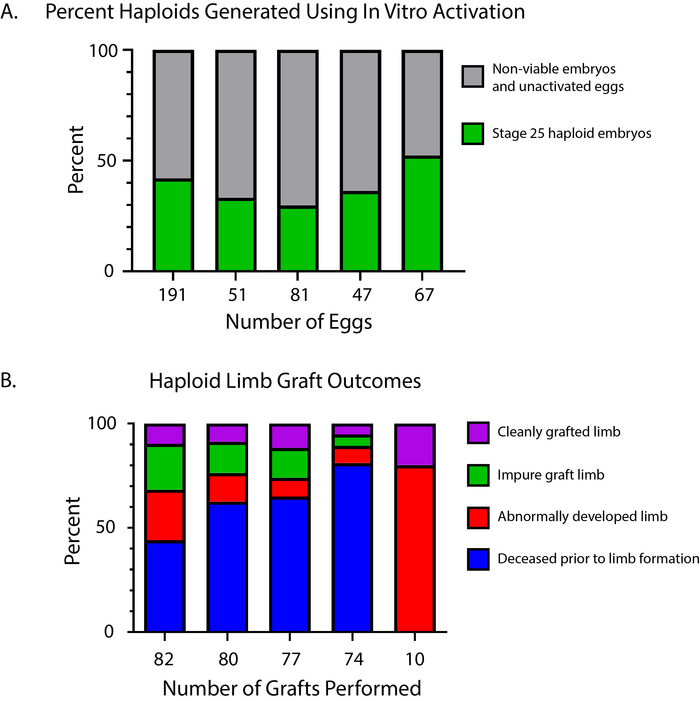
Figure 9: Success rates of haploid embryo and haploid limb generation. (A) Percent of viable haploid embryos generated using in vitro activation. Data were collected from five independent in vitro activation experiments. Green indicates the fraction of eggs that produced stage 25 haploid embryos that could be used for grafting (mean = 38.7%, SD = ± 8.78%). Grey indicates the fraction of eggs that did not show signs of cleavage, were non-viable, or were normal in development. (B) Percent of normally developed, cleanly grafted haploid limbs. Purple indicates the fraction of grafts that yielded clean, normally developed haploid limbs (mean = 11.1%, SD = ± 5.46%). Green indicates limbs that developed normally but had contaminating GFP+ host tissues (mean = 11.3%, SD = ± 8.64%). Red indicates graft limbs that did not develop normally (mean = 27.1%, SD = ± 30.3%). Blue indicates all grafted embryos and animals that did not survive to complete limb development (mean = 50.5%, SD = ± 31.2%). Loss of grafted animals was distributed across late embryonic and larval stages. Please click here to view a larger version of this figure.
Discussion
There are a few critical steps in our protocol for generating haploid-diploid chimeras that the operating technician should consider for consistent grafting results.
The most likely reason for haploid generation to fail is due to poor in vitro activation conditions. The proper quantities of motile sperm must be used to activate eggs. To prolong motility, sperm samples should always be maintained at 4 °C. Before applying any sperm sample to eggs, check the viability of the sperm using an inverted microscope. Completely non-motile sperm should never be used, and the concentration should be adjusted to 80,000 motile cells/mL. Too much sperm in the activation step can also prevent normal egg development. Sperm pits, which appear as small, dark indentations on the surface of the egg, are a physical indication that a sperm cell has penetrated the egg and typically appear 20 to 60 min after applying sperm. Eggs with ten or more sperm pits may not activate properly.
Unfertilized eggs can be activated if collected within 15 min after being laid under water, placed in a dry petri dish, and dried thoroughly with paper towels prior to sperm application. Alternatively, as shown in the video, eggs may be squeezed from a hormone-injected female. We find that these "dry" eggs give more consistent results, though we regularly employ both methods.
Successful grafting requires the proper coupling of diploid embryos with appropriately staged haploid limb bud tissue. This protocol contains a number of measures to ensure stage-matching. As natural mating does not always result in egg laying, HCG injection to obtain haploid oocytes should not be performed until the naturally mated female begins to lay diploid eggs. After this injection, the induced female should be housed at reduced temperature (8 to 12 °C), which will reduce the rate of egg laying. Housing the hormone-stimulated female at normal temperatures may result in most of the eggs being laid in a short period of time. The onset of egg laying in a chilled female indicates that the female is ready for anesthesia and oocyte extraction. Oocyte activation must occur within 15 min of oocyte extraction. Because activated haploid embryos will be several days behind the naturally produced diploid embryos in development, we recommend incubating diploid embryos at 12 °C while increasing the relative rate of development of the haploids by incubating them at 18 °C. Because there will be some variation in the interval between HCG-injection and egg laying of induced females, slight adjustments in the incubation temperatures of either haploids or diploids may be necessary so that the staging of host and donor embryos are paired at the time of grafting. Embryos should be chilled to 4 °C for several hours prior to grafting, and this nearly arrests development. Differing the time of onset of chilling haploid and diploid embryos prior to grafting can enable stage-matching. While haploid embryos differ morphologically from diploids, both haploids and diploids reach stage 21 after the neural folds close and the embryos lie one their sides. Like diploids, stage 25 haploid embryos have prominent gill and pronephric bulges. The large head protrudes at an angle relative to the body in haploids, though this is greatly reduced relative to stage 25 diploid embryos, whose heads protrude from the body at nearly a right angle30.
Sterile technique is absolutely critical for successful tissue grafting. Sterile conditions should be maintained through the entirety of the experiment from haploid generation through the chimeras' embryonic development. We recommend keeping all parent animals in clean, non-system water conditions during the egg laying period to minimize the amount of contaminating organic material at the start of the procedure. Consistent, daily removal of deceased or dying embryos through the entire process is important for maintaining the health of the other embryos. We recommend individually housing all haploid and chimera embryos to minimize cross contamination.
Embryonic microdissection and tissue grafting skills are acquired through practice. During the grafting process, it is important not to puncture or tear the limb bud itself. Tissues being removed from the host and donor embryos should extend beyond the limb bud, as pictured, to provide a buffering zone. If too small of an area of tissue is grafted, the haploid limbs will be infiltrated by GFP+ diploid skin and other tissues.
One of the limitations of this grafting technique is that the neural tissue and blood in the limbs are derived from the host body. In a few rare cases, we have been able to generate grafted haploid limbs which completely lack all signs of GFP+ expressing nerves. In these cases, we were able to replace enough of the neural tissue in the host animal with that of the donor. However, such extensive grafting is difficult, unreliable, and often produces developmental abnormalities outside of the limb.
Haploidy is embryonic lethal in the axolotl. Similarly, mutations in many genes potentially essential for limb development and regeneration are also early embryonic lethal. Our protocol bypasses the embryonic lethality of haploidy for the production of experimental limbs and could also be used in cases where mutation of a candidate regeneration gene produces an embryonic lethal phenotype. Our limb bud grafting technique provides an advantage for accomplishing this versus previously described methods of limb bud and contributing tissue grafting, as it is performed during early embryonic development and involves the transplantation of complete ectoderm and mesoderm layers1,2.
Haploid limb bud grafting has been previously described; however, in this earlier study, haploid limb buds were grafted ectopically31. Microscopic nuclear analysis of the ectopic limbs derived from these grafts revealed extensive contributions of diploid tissue. While these limbs developed abnormally, they were able to regenerate31. Rather than grafting ectopic limb buds, we replace the entire endogenous limb bud with equivalent haploid tissues. Through the use of fluorescent markers, we are able to visually identify haploid limbs that are free of diploid contributions, with the exception of nerve and blood cells, without sacrificing the haploid limb itself. We find that haploid limbs develop and regenerate normally. However, limbs in which GFP+ hosts contribute to tissues other than neural and blood-derived cells often display abnormalities. Thus, when investigating gene function in haploid limbs, those with excessive host contributions must be excluded from analysis before a genotype can be associated with any phenotype observed in mutant haploid limbs.
Recent innovations, like the assembly of the axolotl genome and recent CRISPR/Cas-based insertional methods15,16,32, have dramatically expanded the genetic malleability of the axolotl. Methods to restrict genetic manipulations temporally and spatially hold great promise, but their current applications are restricted by the resources needed to generate, house, and distribute animal lines. The protocol detailed here accelerates the process by which the consequences of genetic perturbations of genes may be investigated in limb development and regeneration. There has been little characterization of many of the genes found to be upregulated in regenerating limbs, and these genes may be involved in a variety of essential developmental and cellular processes. While we anticipate that many genes required for limb regeneration will also be required for limb development, this method may uncover whether any genes implicated in regeneration have loss-of-function phenotypes that are regeneration-specific. CRISPR/Cas mutagenesis in axolotls typically produces allelic mosaicism that includes retained wildtype alleles, and this mosaicism itself may be regarded as a quantifiable phenotype20. Using Next Generation Sequencing of amputated original and regenerated limbs, researchers may quantify whether haploid cells with mutations in a gene of interest are preferentially lost after regeneration. This approach may permit the investigation of regeneration phenotypes produced by perturbation of otherwise essential developmental genes.
Haploid loss-of-function genetic screens facilitate the investigation of the genetic origins of many biological processes without the need to establish biallelic mutant cell lines. By combining haplogenesis, CRISPR/Cas9 mutagenesis, and limb bud grafting, we provide a novel platform for a haploid genetic screen exploring limb development and regeneration in a living animal.
Declarações
The authors have nothing to disclose.
Acknowledgements
We would like to thank Katherine Roberts for her care of the axolotl colony. Funding for this work was provided by the Connecticut Innovations Regenerative Medicine Research Fund (15RMA-YALE-09 and 15-RMB-YALE-01) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Individual Postdoctoral Fellowship F32HD086942).
Materials
| #55 Dumont Forceps | Fine Science Tools | 11295-1 | Only use Dumostar material (can be autoclaved) |
| Amphotericin B | Sigma Aldrich | A2942-20ML | 20 mL |
| Antibiotic-Antimycotic 100x | Thermo Fisher | 15240062 | |
| Ciprofloxacin | Sigma Aldrich | 17850-5G-F | |
| Ficoll 400 (polysucrose 400) | bioworld | 40600032-3 | Ficoll 400 |
| Gentamicin | Sigma Aldrich | G1914-250MG | |
| Heating/Cooling Incubator | RevSci | RS-IF-233 | |
| Human Chorionic Gonadotropin | Merk | Chorulon | |
| Megascript T7 Transcription Kit | Thermo Fisher | AM1334 | 40 reactions |
| Miroscope Cooling Stage | Brook Industries | Custom | Custom |
| NLS Cas9 Protein | PNAbio | CP01-200 | 4 vials of 50 µg protein each |
| Plasmocin | Invivogen | ant-mpt-1 | Treatment level |
| Recipes | |||
| 1.0x Marc's modified Ringer's solution (MMR) | 0.1 M NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 0.1 mM EDTA, 5 mM HEPES (pH 7.8), ph 7.4 | ||
| 40% Holtfreter's solution | 20 mM NaCl, 0.2 mM KCl, 0.8 mM NaHCO3, 0.2 mM CaCl2, 4 mM MgSO4, pH to 7.4 |
Referências
- Kragl, M., et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 460 (7251), 60-65 (2009).
- Maden, M., Goodwin, B. C. Experiments on developing limb buds of the axolotl Ambystoma mexicanum. Journal of Embryology and Experimental Morphology. 57, 177-187 (1980).
- McCusker, C. D., Diaz-Castillo, C., Sosnik, J., Phan, A. Q., Gardiner, D. M. Cartilage and bone cells do not participate in skeletal regeneration in Ambystoma mexicanum limbs. Biologia do Desenvolvimento. 416 (1), 26-33 (2016).
- Brun, R. B. Experimental analysis of the eyeless mutant in the mexican axolotl (Ambystoma mexicanum). Integrative and Comparative Biology. 18 (2), 273-279 (1978).
- Lopez, D., et al. Mapping hematopoiesis in a fully regenerative vertebrate: the axolotl. Blood. 124 (8), 1232-1242 (2014).
- de Both, N. J. Transplantation of Axolotl Heads. Science. 162 (3852), 460-461 (1968).
- Harrison, R. G. Some Unexpected Results of the Heteroplastic Transplantation of Limbs. Proceedings of the National Academy of Sciences of the United States of America. 10 (2), 69-74 (2006).
- Fields, E., French, V., Bryant, P. J., Bryant, S. V Pattern regulation in epimorphic fields. Science. 193 (4257), 969-981 (2013).
- Gerber, T., et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 362 (6413), (2018).
- Knapp, D., et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PloS One. 8 (5), e61352 (2013).
- Campbell, L. J., et al. Gene expression profile of the regeneration epithelium during axolotl limb regeneration. Developmental Dynamics: an official publication of the American Association of Anatomists. 240 (7), 1826-1840 (2011).
- Bryant, D. M., et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Reports. 18 (3), 762-776 (2017).
- Gardiner, D. M., et al. Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression. Regeneration. 2 (3), 120-136 (2015).
- Flowers, G. P., Timberlake, A. T., McLean, K. C., Monaghan, J. R., Crews, C. M. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development. 141 (10), 2165-2171 (2014).
- Smith, J. J., et al. A Chromosome-Scale Assembly of the Enormous (32 Gb) Axolotl Genome. bioRxiv. , 373548 (2018).
- Nowoshilow, S., et al. The axolotl genome and the evolution of key tissue formation regulators. Nature. 559 (7712), 50-55 (2018).
- Fankhauser, B. Y. G. The Effects of Changes in Chromosome Number on Amphibian Development. The Quarterly Review of Biology. 20 (1), 20-78 (1945).
- Malacinski, G. M., Brothers, A. J. Mutant Genes in the Mexican Axolotl. Science. 184 (4142), 1142-1147 (1974).
- Armstrong, B. Gynogenesis in the mexican axolotl. Genética. 83 (4), 783-792 (1976).
- Flowers, G. P., Sanor, L. D., Crews, C. M. Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. eLife. 6, 1-15 (2017).
- Shalem, O., et al. Genome – scale CRISPR – Cas9 knockout screening in human cells. Science. 343 (6166), 84-87 (2014).
- Wang, T., Wei, J. J., Sabatini, D. M., Lander, E. S. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 343 (6166), 80-84 (2014).
- Yin, Z., Chen, L. Simple Meets Single: The Application of. CRISPR/Cas9 in Haploid Embryonic Stem Cells. Stem Cells International. 2017, 1-6 (2017).
- Khattak, S., et al. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nature Protocols. 9 (3), 529-540 (2014).
- Vachon, P., Zullian, C., Dodelet-Devillers, A., Roy, S. Evaluation of the anesthetic effects of MS222 in the adult Mexican axolotl (Ambystoma mexicanum). Veterinary Medicine: Research and Reports. 7, 1-7 (2016).
- Montague, T. G., et al. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS One. 9 (5), (2014).
- Moreno-Mateos, M. A., et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature Methods. 12 (10), 982-988 (2015).
- Kumar, A., Simon, A. . Salamanders in Regeneration Research: Methods and Protocols. , (2015).
- Hronowski, L., Gillespie, L. L., Armstrong, J. B. Development and Survival of Haploids of the Mexican Axolotl, Ambystoma mexicanum. Journal of Experimental Zoology. 209, 41-47 (1979).
- Schreckenberg, G. M., Jacobson, A. G. Normal stages of development of the axolotl, Ambystoma mexicanum. Biologia do Desenvolvimento. 42 (2), 391-399 (1975).
- Hertwig, G. Beitrage Zum Determinations- Und Regenerationsproblem Mittels Der Transplantation Haploidkerniger Zellen. Archiv f. Entwicklungsmechanik. 111, 292-316 (1927).
- Fei, J. -. F., et al. Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. Proceedings of the National Academy of Sciences of the United States of America. 114 (47), 12501-12506 (2017).

