Multiplexed Single Cell mRNA Sequencing Analysis of Mouse Embryonic Cells
Summary
Here we presented a multiplexed single cell mRNA sequencing method to profile gene expression in mouse embryonic tissues. The droplet-based single cell mRNA sequencing (scRNA-Seq) method in combination with multiplexing strategies can profile single cells from multiple samples simultaneously, which significantly reduces reagent costs and minimizes experimental batch effects.
Abstract
Single cell mRNA sequencing has made significant progress in the last several years and has become an important tool in the field of developmental biology. It has been successfully used to identify rare cell populations, discover novel marker genes, and decode spatial and temporal developmental information. The single cell method has also evolved from the microfluidic based Fluidigm C1 technology to the droplet-based solutions in the last two to three years. Here we used the heart as an example to demonstrate how to profile the mouse embryonic tissue cells using the droplet based scRNA-Seq method. In addition, we have integrated two strategies into the workflow to profile multiple samples in a single experiment. Using one of the integrated methods, we have simultaneously profiled more than 9,000 cells from eight heart samples. These methods will be valuable to the developmental biology field by providing a cost-effective way to simultaneously profile single cells from different genetic backgrounds, developmental stages, or anatomical locations.
Introduction
The transcriptional profile of each single cell varies among cell populations during embryonic development. Although single molecular in situ hybridization can be used to visualize the expression of a small number of genes1, single cell mRNA sequencing (scRNA-Seq) provides an unbiased approach to illustrate genome-wide expression patterns of genes in single cells. After it was first published in 20092, scRNA-Seq has been applied to study multiple tissues at multiple developmental stages in the recent years3,4,5. Also, as the human cell atlas has launched its developmental-focused projects recently, more single cell data from human embryonic tissues are expected to be generated in the near future.
The heart as the first organ to develop plays a critical role in embryonic development. The heart consists of multiple cell types and the development of each cell type is tightly regulated temporally and spatially. Over the past few years, the origin and cell lineage of cardiac cells at early developmental stages have been characterized6, which provide a tremendous useful navigation tool for understanding congenital heart disease pathogenesis, as well as for developing more technologically advanced methods to stimulate cardiomyocyte regeneration7.
The scRNA-Seq has undergone a rapid expansion in recent years8,9,10. With the newly developed methods, design and analysis of single cell experiments has become more achievable11,12,13,14. The method presented here is a commercial procedure based on the droplet solutions (see Table of Materials)15,16. This method features capturing cells and sets of uniquely barcoded beads in an oil-water emulsion droplet under control of a microfluidic controller system. The rate of cell loading into the droplets is extremely low so that the majority of droplet emulsions contain only one cell17. The procedure's ingenious design comes from single cell separation into droplet emulsions occurring simultaneously with barcoding, which enables the parallel analysis of individual cells using RNA-Seq on a heterogeneous population.
The incorporation of multiplexing strategies is one of the important additions to the traditional single cell workflow13,14. This addition is very useful in discarding cell doublets, reducing experimental costs, and eliminating batch effects18,19. A lipid based barcoding strategy and an antibody based barcoding strategy (see Table of Materials) are the two mostly used multiplexing methods. Specific barcodes are used to label each sample in both methods, and the labeled samples are then mixed for single cell capturing, library preparation, and sequencing. Afterwards, the pooled sequencing data can be separated by analyzing the barcode sequences (Figure 1)19. However, significant differences exist between the two methods. The lipid based barcoding strategy is based on lipid-modified oligonucleotides, which has not been found to have any cell type preferences. While the antibody based barcoding strategy can only detect the cells expressing the antigen proteins19,20. In addition, it takes about 10 min to stain the lipids but 40 min to stain the antibodies (Figure 1). Furthermore, the lipid-modified oligonucleotides are cheaper than antibody-conjugated oligonucleotides but not commercially available at the time of writing this article. Finally, the lipid-based strategy can multiplex 96 samples in one experiment, but the antibody-based strategy currently can only multiplex 12 samples.
The recommended cell number to multiplex in a single experiment should be lower than 2.5 x 104, otherwise, it will lead to a high percentage of cell doublets and potential ambient mRNA contamination. Through the multiplexing strategies, the cost of single cell capturing, cDNA generation, and library preparation for multiple samples will be reduced to the cost of one sample but the sequencing cost will remain the same.
Protocol
The animal procedure is in accordance with the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC).
1. Mouse Embryonic Heart Dissection and Single Cell Suspension Preparation
NOTE: This step could take a few hours depending on the numbers of embryos to dissect.
- To acquire E18.5 embryonic hearts, euthanize a pregnant CD1 mouse by CO2 administration. Use a razor to remove the unwanted hair in the abdominal area and disinfect the skin with 70% ethanol.
- Cut the skin of the abdomen using sterilized scissors and carefully dissect the embryos out and quickly put them into cold phosphate-buffered saline (PBS) on ice.
- Isolate hearts from each individual embryo carefully in a 10 cm dish filled with cold PBS under a stereoscopic microscope using forceps and scissors.
NOTE: Keep the four chambers intact by dissecting the lung with heart together and DO NOT directly catch/pull the heart with surgical instruments. - Transfer ~10 hearts into a new 10 cm dish filled with cold PBS and micro-dissect the hearts into left atrium, right atrium, left ventricle, and right ventricle.
NOTE: This is assumed to yield more than 1 x 106 cells from each sample. We recommend to start with at least 1 x 105 cells per sample. - Transfer each of the 4 chamber tissues to a 1.5 mL tube and chop them into pieces using scissors. Centrifuge at 300 x g for 3 min to collect the tissues.
- After aspirating the supernatant, add 1 mL of 0.25% Trypsin/EDTA to each tube and incubate in a 37 °C water bath for 10 min. Pipet up and down gently 7-8 times using a P1000 pipette.
- If the embryonic stage is older than E11.5, add 1 mL of 10 mg/mL collagenase A/B mixture and incubate at 37 °C for 10-20 min. Gently pipet up and down until most cells are dissociated.
- Transfer the cells to a 15 mL tube and add 8 mL Hank's balanced salt solution (HBSS) to dilute the enzymes. Spin down the cells at 300 x g for 5 min. Suspend the cells in 1 mL of PBS and transfer them to a 1.5 mL tube. Filter the cells through a 40 µm cell strainer.
- Take 15 µL of volume from each sample and mix with the same amount of 0.04% trypan blue. Load this on a cell counting chamber and count the cells in a cell counter.
NOTE: To generate high quality results, cell viability is recommended to be higher than 95%.
2. Single Cell Multiplexing Barcoding
NOTE: This step takes at least 40 min which varies based on the number of samples processed. A clean bench area treated with RNase decontamination solution is required for pre-amplification steps (step 2.11 to 3.11), and a separate clean bench area is required for the post-amplification steps (the steps after 3.11).
- Lipid based barcoding procedure (optional procedure 1)
- Based on cell concentration, keep less than 5 x 105 cells per sample. Make sure the cell suspension is free of debris and cell aggregates.
- Prepare 2 µM anchor/barcode stock solution and 2 µM co-anchor stock solution for each sample (Table 1).
NOTE: The anchor and co-anchor were kindly gifted by Dr. Zev J. Gartner lab. To synthesize these lipid-modified oligonucleotides, DNA sequences were conjugated with fatty acid on a solid support and purified by reversed-phase high-performance liquid chromatography (HPLC)18,19. - Wash the cells twice with PBS and collect the cells at 300 x g for 5 min. Suspend cells in 180 µL of PBS.
- Add 20 µL of anchor/barcode stock solution and pipette up and down gently to mix. Incubate on ice for 5 min.
- Add 20 µL of co-anchor stock solution and pipette up and down gently to mix, then incubate on ice for another 5 min.
- Add 1 mL of cold PBS with 1% BSA and centrifuge at 300 x g for 5 min at 4 °C. Wash at least 2 more times with ice cold 1% BSA in PBS.
- Combine all samples together and filter through 40 µm cell strainers. Count the cells and keep the cell suspension on ice to use in section 3.
- Antibody-based barcoding procedure (optional procedure 2)
- Centrifuge 1 x 106-2 x 106 cells for each sample (from step 1.8) at 300 x g for 5 min and suspend them in 100 µL of staining buffer (Table 1) in 1.5 mL low bind tubes.
- Add 10 µL Fc blocking reagent and incubate for 10 min at 4 °C.
- Prepare antibodies (see Table of Materials) by centrifuging at 14,000 x g for 10 min at 2-8 °C.
- Add 1 µg of each oligo-conjugated antibody to 50 µL of cell staining buffer to make antibody staining solution20. Add one antibody staining solution to each sample tube. Incubate for 30 min at 4 °C.
- Wash cells 3 times with 1 mL of PBS, spin for 5 min at 350 x g at 4 °C.
- Pool all samples at desired proportions in 1 mL of staining buffer, spin for 5 min at 350 x g at 4 °C.
- Resuspend cells in PBS at appropriate concentration (up to 1,500 cells/µL) and filter cells through a 40 µm cell strainer. Immediately proceed to the next step.
3. Droplet Generation and mRNA Reverse Transcription
NOTE: This step takes about 90 min for one multiplexed reaction.
- Equilibrate the gel beads (see Table of Materials) to room temperature for 30 min. Take out reagents from gel beads-in-emulsion (GEMs) kit (see Table of Materials) and keep them at their indicated temperature.
- Assemble the chip B into a chip holder (see Table of Materials).
- Dispense 75 µL of 50% glycerol solution into the unused wells in row 1; 40 µL in row 2; 280 µL in row 3. Do not add glycerol in any recovery wells on the top row of the chip.
- Prepare the master mix on ice according to Table 1. Add appropriate volume of cell suspension and nuclease-free water to master mix according to a cell suspension volume calculator table17 and gently pipette the mix. Dispense 75 µL of cell mixture into the bottom center of the sample well in row 1 without introducing bubbles.
- Vortex the gel beads for 30 s using a vortex adapter and slowly dispense 40 µL of gel beads into the bottom center of the gel bead well in row 2 without introducing bubbles.
NOTE: It is critical to wait for 30 s between adding cells and gel beads to avoid wetting failure. - For the partitioning oil well in row 3, dispense 280 µL of partitioning oil through the sidewall of the well.
NOTE: Loading less than 270 µL of partitioning oil will lead to abnormal GEMs generation. - Attach the gasket onto the chip, do not press down on the gasket and keep it horizontal to avoid wetting the gasket.
- Load the assembled chip with the gasket in the chromium controller and run the chromium single cell B program (see Table of Materials), immediately proceed to the next step when the program completes.
- Take the chip out and discard the gasket. Fold the lid back to expose wells at 45°, check the liquid level to make sure no clogs are present.
- Slowly aspirate 100 µL of GEMs from the lowest points of the recovery well and check the uniformity of the GEMs. Dispense GEMs into a new polymerase chain reaction (PCR) tube on ice with the pipette tips against the sidewall of the tube.
NOTE: If excess aqueous layer is observed, it is suggested to re-prepare the samples. Importantly, take a picture of the mixture when the GEMs are still in the pipette tips. This picture can tell if there is a wetting failure, partially emulsified GEMs, and reagent clogs. The photograph can also be used as evidence to get reimbursement from the reagent company with replacement reagents and chips. - Put the tube in a thermal cycler and perform the reverse transcription procedure (Table 2).
NOTE: Stop here or proceed to the next step. The PCR product can be stored at -20 °C for up to a week.
4. cDNA Amplification
NOTE: This step takes about 150 min.
- Post single cell reverse transcription cleanup
- Take out the cDNA amplification reagents from GEMs kit (see Table of Materials) and keep them at their indicated temperature.
- Add 125 µL of recovery agent to the sample at room temperature to acquire a biphasic mixture. No opaque liquid should be observed and avoid pipetting or vortexing the mixture.
- After waiting for 60 s, slowly remove 125 µL of recovery agent from the bottom of the tube.
- Vortex the magnetic beads (see Table of Materials) thoroughly for 30 s and immediately use it to prepare beads cleanup mixture (Table 1). Reagents should be added sequentially as listed.
- Vortex the beads cleanup mixture and add 200 µL to the sample. Pipette the mixture 10 times then incubate it for 10 min at room temperature.
- Add the reagents sequentially as listed in Table 1 to prepare the elution solution and elute the cDNA as follows.
- Place the samples on the magnet (high position) (see Table of Materials) until the solution clears, then remove the supernatant.
- Add 200 µL of 80% ethanol to the pellet. Wait for 30 s, then remove the ethanol.
- Repeat step 4.1.6.2 for another 2 times. Centrifuge briefly and place on the magnet (low position). Carefully remove the remaining ethanol and air dry for less than 2 min.
NOTE: DO NOT exceed air drying past 2 min, otherwise the elution efficiency will decrease. - Remove the sample from the magnet. Add 35.5 µL of elution solution and pipette to mix 15 times. Incubate 2 min at room temperature.
- Place the sample on the magnet (high position) until the solution clears. Transfer 35 µL of the sample to a new tube strip.
NOTE: This purification procedure is also used in steps 4.2.1.6, 4.2.2.3, 5.8, 6.1.10, and 6.2.6. Pay attention to the concentration of magnetic beads and the volume of EB buffer/ ultrapure water used to elute the samples at each step.
- Quantify the size, concentration and integrity of eluted cDNA using an automated electrophoresis instrument21 (see Table of Materials) (Figure 2).
- Amplification of cDNA
- cDNA amplification using the lipid-based barcoding strategy (optional procedure 1)
- Prepare amplification reaction mixture (Table 1) on ice.
- Add the amplification reaction mixture to 35 µL of cDNA samples (from step 4.1.6.7). Pipette the mix, and centrifuge briefly. Incubate the mixture in a thermal cycler following the cDNA amplification procedure (Table 2).
- After vortexing thoroughly, add 120 µL of select reagent and 100 µL of ultrapure water to 100 µL of sample to acquire a 0.6x concentration of select reagent (see Table of Materials). Pipette the mixture for 15 times.
- Incubate for 5 min at room temperature and then place the sample on magnet until the solutions become clear.
NOTE: Endogenous cDNA in the beads fraction and multiplexing barcoded cDNA is in the supernatant. - Transfer the supernatant into a 1.5 mL low bind tube for multiplexing barcoded cDNA library construction at step 6.1.
- Clean the endogenous cDNA by following the steps in 4.1.6 and elute them with 40 µL of EB buffer.
- Run 1 µL of purified cDNA sample on an automated electrophoresis instrument (see Table of Materials) to analyze/quantify the cDNA.
- Aliquot 10 µL of cDNA into a new PCR tube for endogenous library construction.
NOTE: Stop here or proceed to the next step. The remaining sample can be stored in -20 °C for up to 4 weeks to generate additional libraries if needed.
- cDNA amplification in the antibody based barcoding strategy (optional procedure 2)
- Add 2 pmol of HTO and ADT additive primer and 15 µL of cDNA primers to 50 µL of amplification reaction mixture (Table 1) and perform cDNA amplification with the cDNA amplification procedure (Table 2).
- Use 0.6x select reagent to separate endogenous cDNA (beads fraction) and multiplexing barcode cDNA (in supernatant). Remember to save the supernatant to perform barcoding library construction in step 6.2.
- Purify and elute the endogenous transcript cDNA by following the steps in 4.1.6, perform quality control (QC) of the libraries and aliquot 10 µL into a new PCR tube for endogenous library construction.
- cDNA amplification using the lipid-based barcoding strategy (optional procedure 1)
5. Endogenous Transcript Library Preparation
NOTE: This step takes about 120 min.
- Keep gene expression library construction reagents from the library kit (see Table of Materials) at their indicated temperature, respectively.
- Prepare the fragmentation mixture (Table 1) on ice, pipette to mix and centrifuge briefly.
- Add 25 µL EB buffer to the 10 µL purified cDNA sample (from step 4.2.1.8 or 4.2.2.3) and then add the newly prepared 15 µL fragmentation mixture to the sample, pipette the mixture 15 times on ice and centrifuge briefly.
- Transfer the sample into a pre-cooled thermal cycler and initiate the PCR program for fragmentation, end repair and A-tailing (Table 2).
- Vortex the select reagent to suspend magnetic beads and successively use 0.6x and 0.8x select reagents to make a double-sided size selection according to the user guide17,22. Use 50 µL of EB buffer to elute the DNA.
- Prepare adaptor ligation mixture (Table 1), then pipette the mixture thoroughly and centrifuge briefly.
- Add 50 µL of adaptor ligation mixture to 50 µL of sample, pipette mix again for 15 times and centrifuge briefly. Perform the adaptor ligation as per the protocol in a thermal cycler (Table 2).
- Use 0.8x select reagent to purify the ligation product and elute the purified sample with 30 µL of EB buffer (see step 4.1.6).
- Prepare the sample index PCR mixture (Table 1) and add 60 µL to the purified sample. Add 10 µL of sample index to the sample, pipette mix up and down for 5 times and centrifuge briefly, incubate in a thermal cycler following the sample index protocol (Table 2).
NOTE: Stop here or proceed to the next step. If more than one well is used, choose one specific sample index (see Table of Materials) for each well. Remember to record the index ID used for each well and ensure no overlap in a multiplexed sequencing run. - Successively use 0.6x and 0.8x select reagents to make a double-sided size selection to acquire 35 µL of purified endogenous cellular library DNA17.
- QC the endogenous cellular library before sequencing (Figure 2).
6. Preparation of Multiplexing Sample Barcode cDNA Libraries
NOTE: This step takes at least 120 min.
- Sample barcode library generation for the lipid based multiplexing strategy (optional procedure 1)
- Add 520 µL of select reagent and 360 µL of isopropanol to sample barcode cDNA from step 4.2.1.5 to get a 3.2x select reagent concentration. Pipette the mixture 10 times and incubate at room temperature for 5 min.
- Place the tube on a magnetic rack and wait for the solution to clear. Then discard the supernatant.
- Use 500 µL of 80% ethanol to wash beads twice on a magnet and wait for 30 s after each wash.
- Briefly centrifuge the beads and place on a magnet. Remove the remaining ethanol with a P10 micropipette and leave beads for 2 min.
- Remove the tube from the magnet rack and resuspend beads in 50 µL of EB buffer. Pipet up and down to mix thoroughly. Incubate at room temperature for 2 min.
- Return the tube to a magnet and wait for the solution to clear. Transfer the supernatant (sample barcode cDNA) to a new PCR tube. Be careful not to transfer any beads.
- Quantify the concentration of sample barcode cDNA23.
- Prepare lipid barcode library mixture (Table 1), add 3.5 ng of purified barcoded cDNA (from step 6.1.6) and nuclease-free water for a total volume of 50 µl.
- Keep it in a thermal cycler following the lipid-based barcode library PCR (Table 2).
- Use 1.6x select reagent to purify the PCR product and elute the DNA with 25 µL of EB buffer (see step 4.1.6).
- Quantify the library concentration using a high sensitivity DNA analysis method24 from the initial dilution of 1:5 (Figure 2).
- Sample barcode library generation for the antibody based multiplexing strategy (optional procedure 2)
- Add additional 1.4x reaction volume of select reagent to the supernatant containing sample barcodes acquired from step 4.2.2.3 to get a 2x select reagent ratio.
- Wash the beads with 80% ethanol by following the steps in 4.1.6 and elute the barcoded cDNA with ultrapure water.
- Perform the selection protocol with 2x select reagent for a second time and elute using ultrapure water.
- Prepare antibody barcode library mixture (Table 1), and add 45 µL of purified barcoded cDNA from the last step.
- Incubate in a thermal cycler following the antibody barcode library PCR (Table 2)20.
- Use 1.6x select reagent to purify the PCR product and elute the purified sample with 30 µL of ultrapure water (see step 4.1.6).
7. Library Sequencing
NOTE: Multiple next generation sequencing platforms such as HiSeq 4000 and NovaSeq can be used to sequence the endogenous transcript libraries and multiplexing barcode libraries.
- Use a next generation sequencing platform of choice to sequence the endogenous transcript libraries and multiplexing barcode libraries.
- Dilute the libraries according to the recommendations from an expert at a sequencing company or sequencing facility. Minimum 20,000 reads per cell is recommended for the endogenous transcript library and 3000 reads for the barcode libraries.
8. Data Analysis
NOTE: De-multiplex the sequencing data using the cloud-based resource BaseSpace or by running the bcl2fastq package on a UNIX server.
- Endogenous transcriptome data analysis
- With the fastq data generated from demultiplexing software, run "mkfastq" on the commercially available data analysis pipeline (see Table of Materials) to further demultiplex each GEMs barcode.
- Run "count" to perform the alignment, filtering, barcode counting, and UMI counting.
- Optionally, run "aggr" to aggregate multiple sequencing lanes from a single experiment.
- Use "cell browser" (see Table of Materials) to visualize data, cluster cells, identify differentially expressed genes, and generate tSNE or gene expression heatmap plots.
- Optionally, use a well-maintained R-based platform25 (see Table of Materials) to normalize and scale data, identify differentially expressed genes, and generate tSNE/UMAP plots and gene expression heatmaps (Figure 3).
- Multiplexing barcode data analysis
- Analysis of the data from lipid based barcoding strategy
- Use the commercially available data analysis pipeline or deMULTIplex R package (https://github.com/chris-mcginnis-ucsf/MULTI-seq) to convert the sample barcode FASTQ files into a sample barcode UMI count matrix.
- Load the barcode UMI count matrix together with endogenous transcriptome data to an R-based platform (see Table of Materials) for integration analysis (Figure 3).
- Analysis of the data from antibody-based barcoding strategy
- Use "count" from the commercially available data analysis pipeline to map the barcodes by providing the library CSV file and hashtag feature reference CSV file.
- Load the output unified feature-barcode matrix, which contains gene expression counts alongside feature barcode counts for each cell barcode, to an R-based platform for downstream analysis.
- Analysis of the data from lipid based barcoding strategy
Representative Results
In this study, we used mouse embryonic heart as an example to exhibit how multiplexed single cell mRNA sequencing was performed to process the different samples from separate parts of an organ simultaneously. E18.5 CD1 mouse hearts were isolated and dissected into left atrium (LA), right atrium (RA), left ventricle (LV) and right ventricle (RV). The atrial and ventricular cells were then barcoded independently using a lipid-based barcoding procedure and mixed together before GEMs generation and reverse-transcription. The schematic overview is shown in Figure 1. We quantified the cDNA concentration before library construction (Figure 2A). One of the distinctions in performing multiplexed scRNA-Seq from the standard scRNA-Seq is that the endogenous cDNA library and the sample barcode cDNA library were acquired separately after cDNA amplification and purification (Step 4.2.1 and 4.2.2.2). The two libraries were also qualified in our experiment (Figure 2B,C). Next generation sequencing and data analysis were performed followed by library construction and QC.
We used HiSeqX platform to sequence both libraries in the same sequencing lane. With the sequencing data, we first separated the endogenous transcript data and barcode data using the BaseSpace program. Then we analyzed barcode expression in each single cell and found 8 groups of single cells that uniquely express one type of barcode, representing cells from 8 different samples (Figure 3A). In addition, we also found that some cells do not express any barcode, which we defined as negative cells, and some cells express two different barcodes, which represent doublets (Figure 3B). In summary, we found that around 70% of cells are singlets, 25% of cells are negative and 5% of the cells are doublets.
With the singlet cells, we can perform further downstream analyses to understand the cellular heterogeneity and molecular regulations. The potential analyses can be cell type annotation (Figure 4A), novel/rare cell type identification (Figure 4B), anatomical zone comparative analysis (Figure 4C), and gene ontology pathway analysis such as cell cycle phase separations (Figure 4D).
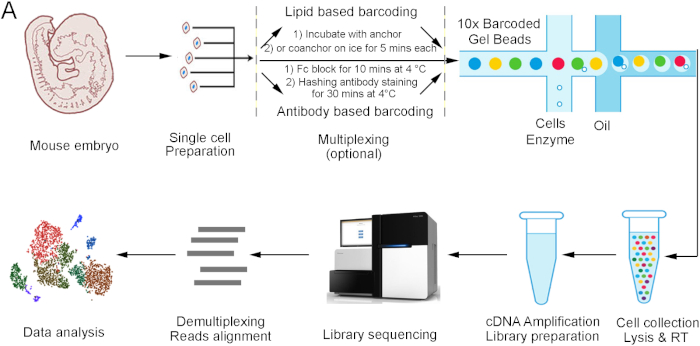
Figure 1: Multiplexed single cell mRNA sequencing workflow. Embryonic day 18.5 stage hearts were analyzed using a multiplexed droplet-based single cell sequencing procedure. RT = reverse transcription. Please click here to view a larger version of this figure.
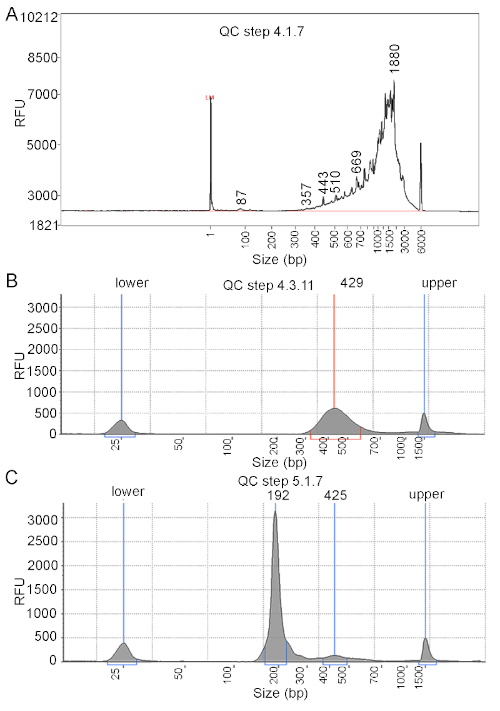
Figure 2: Representative QC results at different steps. (A) QC analysis of cDNA from step 4.1.7. The target fragment size is 200 to 9000 bp. (B) Endogenous library and (C) barcode library were analyzed with an automated electrophoresis instrument. The target fragment size for the endogenous library is 300-600 bp, and the barcode library DNA size is around 172 bp. Please click here to view a larger version of this figure.
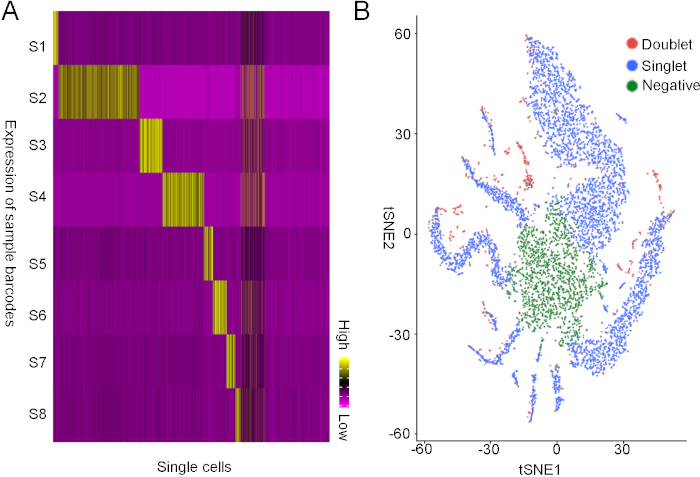
Figure 3: Demultiplexing the sequencing data from the lipid based barcoding strategy. (A) Unsupervised analysis of the barcode expression. X-axis represents single cells, and y-axis represents barcodes. Each of the 8 single cell populations were identified to uniquely express one of the 8 barcodes. Note some cells express more than one barcode, and some cells do not express any barcodes. (B) t-SNE plot of the singlet cells, doublet cells, and negative cells. Please click here to view a larger version of this figure.
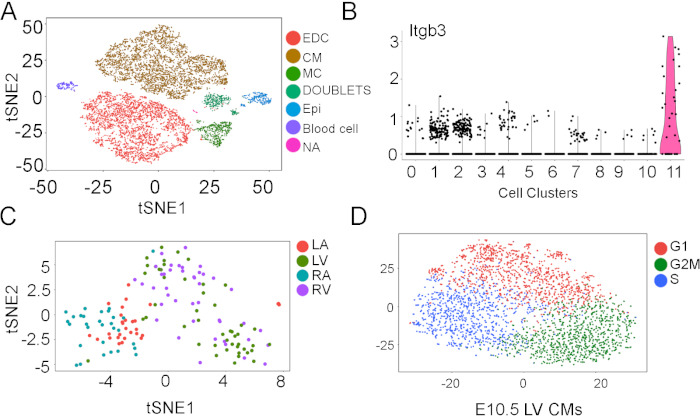
Figure 4: Advanced analysis of single cell transcriptional data. (A-D) Single cell data can be analyzed in different ways to understand the cellular heterogeneity and molecular pathways. We have listed several applications here as examples. Single cells were loaded into an R package to identify cell types (A), rare cell populations (B), cell anatomical zones (C), and cell cycle phases (D). Please click here to view a larger version of this figure.
| Mixture Name | Composition |
| Collagenase mixture | 10 mg/mL collagenase A and 10 mg/mL collagenase B, dissolved in HBSS++ with 40% FBS. |
| 2 μM Anchor/Barcode stock solution | Mix 50 μM anchor and 10 μM barcode strand in 1:1 molar ratio in PBS (without FBS or BSA) for a total volume of 25 μL. |
| 2 μM Co-Anchor stock solution | Dilute 1 μL 50 μM Co-Anchor with 24 μL PBS (without FBS or BSA). |
| Staining buffer | PBS containing 2% BSA, 0.01% Tween 20 |
| Master Mixture | 20 μL RT Reagent, 3.1 μL Oligo, 2 μL Reducing Agent B, 8.3 μL RT Enzyme C. |
| Beads Cleanup Mixture | 182 μL Cleanup Buffer, 8 μL Selection Reagent, 5 μL Reducing Agent B, 5 μL Nuclease-free Water. |
| Amplification Reaction Mixture | 1 μL of 10 μM Lipid-tagged additive primer, 15 μL cDNA primer, 50 μL Amp Mix |
| Elution Solution | 98 μL Buffer EB, 1 μL 10% Tween 20, 1 μL Reducing Agent B. |
| Fragmentation Mixture | 5 μL Fragmentation Buffer, 10 μL Fragmentation Enzyme. |
| Adaptor Ligation mixture | 20 μL Ligation Buffer, 10 μL DNA Ligase, 20 μL Adaptor Oligos. |
| Sample Index PCR Mixture | 50 μL Amp Mix, 10 μL SI Primer |
| Lipid barcode library mixture | 26.25 μL of 2× Hot Start master mix, 2.5 μL of 10 μM RPIX primer, 2.5 μL of 10 μM TruSeq Universal Adapter primer (see table of materials) |
| Antibody barcode library mixture | 50 μL of 2× Hot Start master mix, 2.5 μL of 10 μM RPIX primer, 2.5 μL of 10 μM P5-smart-pcr hybrid oligo |
Table 1: The reagent mixtures used in the protocol.
| Incubating Procedure | Temperature(1) | Time |
| GEM-RT Incubation | Lid Temperature 53 °C | |
| Step 1 | 53 °C | 45 min |
| Step 2 | 85 °C | 5 min |
| Step 3 | 4 °C | Hold |
| 10x Genomics cDNA Amplification | Lid Temperature 105 °C | |
| Step 1 | 98 °C | 3 min |
| Step 2 | 98 °C | 15 s |
| Step 3 | 63 °C | 20 s |
| Step 4 | 72 °C | 1 min |
| Step 5 | Repeat steps 2 to 4 for 12 cycles in total (2) | |
| Step 6 | 72 °C | 1 min |
| Step 7 | 4 °C | Hold |
| Library construction | Lid Temperature 65 °C | |
| Pre-cool block | 4 °C | Hold |
| Fragmentation | 32 °C | 5 min |
| End Repair and A-tailing | 65 °C | 30 min |
| Hold | 4 °C | Hold |
| Adaptor ligation | Lid Temperature 30 °C | |
| Step 1 | 20 °C | 15 min |
| Step 2 | 4 °C | Hold |
| Sample index PCR | Lid Temperature 105 °C | |
| Step 1 | 98 °C | 45 s |
| Step 2 | 98 °C | 20 s |
| Step 3 | 54 °C | 30 s |
| Step 4 | 72 °C | 20 s |
| Step 5 | Repeat steps 2 to 4 for 12 cycles in total (3) | |
| Step 6 | 72 °C | 1 min |
| Step 7 | 4 °C | Hold |
| Lipid barcode library PCR | ||
| Step 1 | 95 °C | 5 min |
| Step 2 | 98 °C | 15 s |
| Step 3 | 60 °C | 30 s |
| Step 4 | 72 °C | 30 s |
| Step 5 | Repeat steps 2 to 4 for 10 cycles in total (4) | |
| Step 6 | 72 °C | 1 min |
| Step 7 | 4 °C | Hold |
| Antibody barcode library PCR | ||
| Step 1 | 95 °C | 3 min |
| Step 2 | 95 °C | 20 s |
| Step 3 | 60 °C | 30 s |
| Step 4 | 72 °C | 20 s |
| Step 5 | Repeat steps 2 to 4 for 8 cycles in total (5) | |
| Step 6 | 72 °C | 5 min |
| Step 7 | 4 °C | Hold |
Table 2: The incubating procedure used in the protocol. (1) Pay attention to the different lid temperature used in every Procedure. (2) Set total cycle numbers according to the cell load: 13 cycles for <500 cell load; 12 cycles for 500-6,000 cell load; 11 cycles for >6,000 cell load. (3) Set total cycle numbers according to the cDNA input: 14-16 cycles for 1-25 ng cDNA; 12-14 cycles for 25-150 ng cDNA; 10-12 cycles for 150-500 ng cDNA; 8-10 cycles for 500-1,000 ng cDNA; 6-8 cycles for 1000-1500 ng cDNA. (4) Set total cycle numbers according to the cDNA input: 8-12 cycles. (5) Set total cycle numbers according to the cDNA input: 6-10 cycles.
| Lipid based barcoding Oligonucleotides | |
| Anchor LMO | 5'-TGGAATTCTCGGGTGCCAAGGGTAACGATCCAGCTGTCACT-Lipid-3' |
| Co-Anchor LMO | 5'-Lipid-AGTGACAGCTGGATCGTTAC-3' |
| Barcode Oligo | 5'-CCTTGGCACCCGAGAATTCCANNNNNNNNA30-3' |
| Lipid barcoding Additive Primer | 5'-CTTGGCACCCGAGAATTCC-3' |
| RPIX Primer | 5'-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCC TTGGCACCCGAGAATTCCA-3' |
| Universal Adapter Primer | 5'-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGAC GCTCTTCCGATCT-3' |
| Antibody based barcoding Oligonucleotides | |
| Antibody barcoding oligo | 5'-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNNN NNBAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA*A*A-3' |
| HTO additive Primer | 5'-GTGACTGGAGTTCAGACGTGTGCTC-3' |
| ADT additive Primer | 5'-CCTTGGCACCCGAGAATTCC-3' |
| P5-smart-pcr hybrid oligo | 5'-AATGATACGGCGACCACCGAGATCTACACGCCTGTCCGCGGAA GCAGTGGTATCAACGCAGAGT*A*C-3' |
Table 3: Oligonucleotide sequences used in this protocol. N = Barcode or index sequence; * = Phosphorothioate bond
Discussion
In this study, we have demonstrated a protocol to analyze single cell transcriptional profiles. We have also provided two optional methods to multiplex samples in the scRNA-Seq workflow. Both methods have proved to be feasible at various labs and provided solutions to run a cost-effective and batch effect-free single cell experiment18,26.
There are a few steps that should be followed carefully when going through the protocol. An ideal single cell suspension should have >90% of viable cells and the cell density should also be within a specific range27. It is critical to obtain a good quality of cells to minimize the presence of cellular aggregates, debris, and fibers. Cellular aggregates have negative impact on sample multiplexing and have a potential risk to clog the droplet generating machine17. Generally speaking, a 30-40 µm cell strainer is ideal for removing large clumps and debris while preserving the cell samples because most cells will shrink below 30 µm after dissociation. Single cell nuclei are recommended to use instead if the cell diameter is larger than 30 µm. At early embryonic stages, the cell size for all types of mouse cells should be smaller than 30 µm. However, at later stages, the cardiomyocytes in the heart, neurons in the brain, muscle cells in limbs, and some fat cells may have a cell size larger than 30 µm. Cell size should be measured for these types of cells before starting the single cell experiments.
The multiplexing strategies provide a way to simultaneously analyze a large number of samples in a cost-effective way. In addition, by profiling multiple samples together, we can significantly avoid the batch effects and identify cell doublets. These advantages will be very attractive to the single cell field. However, there are some factors that may limit their usage. As more cells are multiplexed in a single experiment, the cell doublet ratio will also increase. Although those doublets can be identified and removed by analyzing the multiplexing barcode data, it will lead to a large waste of sequencing reads. In addition, as more cells are pooled together, the cells are easier to break and cause an increase of the ambient mRNA, which will be captured into droplets with cells and interfere with the detection sensitivity. We are expecting that further optimization of the experimental workflow or bioinformatics analysis pipeline will resolve these two issues in the near future.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank David M. Patterson and Christopher S. McGinnis from Dr. Zev J. Gartner lab for their kind supply of the lipid based barcoding reagents and suggestions on the experimental steps and data analysis. This work was founded by the National Institutes of Health (HL13347202).
Materials
| 10% Tween-20 | Bio-Rad | 1610781 | |
| 10x Chip Holder | 10x Genomics | 120252 330019 | |
| 10x Chromium Controller | 10x Genomics | 120223 | |
| 10x Magnetic Separator | 10x Genomics | 120250 230003 | |
| 10x Vortex Adapter | 10x Genomics | 330002, 120251 | |
| 10x Vortex Clip | 10x Genomics | 120253 230002 | |
| 4200 TapeStation System | Agilent | G2991AA | |
| Agilent High Sensitivity DNA Kit | Agilent | 5067-4626 | University of Pittsburgh Health Sciences Sequencing Core |
| Barcode Oligo | Integrated DNA Technologies | Single-stranded DNA | 25 nmol |
| Buffer EB | Qiagen | 19086 | |
| CD1 mice | Chales River | Strain Code 022 | ordered pregnant mice |
| Centrifuge 5424R | Appendorf | 2231000214 | |
| Chromium Chip B Single Cell Kit, 48 rxns | 10x Genomics | 1000073 | Store at ambient temperature |
| Chromium i7 Multiplex Kit, 96 rxns | 10x Genomics | 120262 | Store at -20 °C |
| Chromium Single Cell 3' GEM Kit v3,4 rxns | 10x Genomics | 1000094 | Store at -20 °C |
| Chromium Single Cell 3' Library Kit v3 | 10x Genomics | 1000095 | Store at -20 °C |
| Chromium Single Cell 3' v3 Gel Beads | 10x Genomics | 2000059 | Store at -80 °C |
| Collagenase A | Sigma/Millipore | 10103578001 | Store powder at 4 °C, store at -20 °C after it dissolves |
| Collagenase B | Sigma/Millipore | 11088807001 | Store powder at 4 °C, store at -20 °C after it dissolves |
| D1000 ScreenTape | Agilent | 5067-5582 | University of Pittsburgh Health Sciences Sequencing Core |
| DNA LoBind Tube Microcentrifuge Tube, 1.5 mL | Eppendorf | 022431021 | |
| DNA LoBind Tube Microcentrifuge Tube, 2.0 mL | Eppendorf | 022431048 | |
| Dynabeads MyOne SILANE | 10x Genomics | 2000048 | Store at 4 °C, used in Beads Cleanup Mix (Table 1) |
| DynaMag-2 Magnet | Theromo Scientific | 12321D | |
| Ethanol, Pure (200 Proof, anhydrous) | Sigma | E7023-500mL | |
| Falcon 15mL High Clarity PP Centrifuge Tube | Corning Cellgro | 14-959-70C | |
| Falcon 50mL High Clarity PP Centrifuge Tube | Corning Cellgro | 14-959-49A | |
| Fetal Bovine Serum, qualified, United States | Fisher Scientific | 26140079 | Store at -20 °C |
| Finnpipette F1 Multichannel Pipettes, 10-100μl | Theromo Scientific | 4661020N | |
| Finnpipette F1 Multichannel Pipettes, 1-10μl | Theromo Scientific | 4661000N | |
| Flowmi Cell Strainer | Sigma | BAH136800040 | Porosity 40 μm, for 1000 uL Pipette Tips, pack of 50 each |
| Glycerin (Glycerol), 50% (v/v) | Ricca Chemical Company | 3290-32 | |
| HBSS, no calcium, no magnesium | Thermo Fisher Scientific | 14170112 | |
| Human TruStain FcX (Fc Receptor Blocking Solution) | BioLegend | 422301 | Add 5 µl of Human TruStain FcX per million cells in 100 µl staining volume |
| Isopropanol (IPA) | Fisher Scientific | A464-4 | |
| Kapa HiFi HotStart ReadyMix (2X) | Fisher Scientific | NC0295239 | Store at -20 °C, used in Lipid-tagged barcode library mix (Table 1) |
| Lipid Barcode Primer (Multi-seq Primer) | Integrated DNA Technologies | Single-stranded DNA | 100 nmol |
| Low TE Buffer (10 mM Tris-HCl pH 8.0, 0.1 mM EDTA) | Thermo Fisher Scientific | 12090-015 | |
| MasterCycler Pro | Eppendorf | 950W | |
| Nuclease-Free Water (Ambion) | Thermo Fisher Scientific | AM9937 | |
| PCR Tubes 0.2 ml 8-tube strips | Eppendorf | 951010022 | |
| Phosphate-Buffered Saline (PBS) 1X without calcium & magnesium | Corning Cellgro | 21-040-CV | |
| Phosphate-Buffered Saline (PBS) with 10% Bovine Albumin (alternative to Thermo Fisher product) | Sigma-Aldrich | SRE0036 | |
| Pipet 4-pack (0.1–2.5μL, 0.5-10μL, 10–100μL, 100–1,000μL variable-volume pipettes | Fisher Scientific | 05-403-151 | |
| Selection reagent (SPRIselect Reagent Kit) | Beckman Coulter | B23318 (60ml) | |
| Template Switch Oligo | 10x Genomics | 3000228 | Store at -20 °C, used in Master Mix (Table 1) |
| The antibody based barcoding strategy is also known as Cell Hashing | |||
| The cell browser is Loup Cell Browser | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/visualization/latest/what-is-loupe-cell-browser | |
| The commercial available analysis pipline in step 8.1 is Cell Ranger | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger | |
| The lipid based barcoding strategy is also known as MULTI-seq | |||
| The well maintained R platform is Seurat V3 | satijalab | https://satijalab.org/seurat/ | |
| TipOne RPT 0.1-10/20 ul XL ultra low retention filter pipet tip | USA Scientific | 1180-3710 | |
| TipOne RPT 1000 ul XL ultra low retention filter pipet tip | USA Scientific | 1182-1730 | |
| TipOne RPT 200 ul ultra low retention filter pipet tip | USA Scientific | 1180-8710 | |
| TotalSeq-A0301 anti-mouse Hashtag 1 Antibody | BioLegend | 155801 | 0.1 – 1.0 µg of antibody in 100 µl of staining buffer for every 1 million cells |
| TotalSeq-A0302 anti-mouse Hashtag 2 Antibody | BioLegend | 155803 | 0.1 – 1.0 µg of antibody in 100 µl of staining buffer for every 1 million cells |
| TotalSeq-A0302 anti-mouse Hashtag 3 Antibody | BioLegend | 155805 | 0.1 – 1.0 µg of antibody in 100 µl of staining buffer for every 1 million cells |
| TrueSeq RPI primer | Integrated DNA Technologies | Single-stranded DNA | 100 nmol, used in Lipid-tagged barcode library mix (Table 1) |
| Trypan Blue Solution, 0.4% | Fisher Scientific | 15250061 | |
| Trypsin-EDTA (0.25%), phenol red | Fisher Scientific | 25200-056 | |
| Universal I5 | Integrated DNA Technologies | Single-stranded DNA | 100 nmol |
Referências
- Raj, A., Van Den Bogaard, P., Rifkin, S. A., Van Oudenaarden, A., Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 5 (10), 877 (2008).
- Tang, F., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nature Methods. 6 (5), 377 (2009).
- Li, G., Plonowska, K., Kuppusamy, R., Sturzu, A., Wu, S. M. Identification of cardiovascular lineage descendants at single-cell resolution. Development. 142 (5), 846-857 (2015).
- DeLaughter, D. M., et al. Single-cell resolution of temporal gene expression during heart development. Developmental Cell. 39 (4), 480-490 (2016).
- Li, G., et al. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development. 146 (12), dev173476 (2019).
- Meilhac, S. M., Buckingham, M. E. The deployment of cell lineages that form the mammalian heart. Nature Reviews Cardiology. 1, (2018).
- Liu, Z., et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 551 (7678), 100 (2017).
- Lafzi, A., Moutinho, C., Picelli, S., Heyn, H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nature Protocols. 1, (2018).
- The Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 562 (7727), 367 (2018).
- Gawad, C., Koh, W., Quake, S. R. Single-cell genome sequencing: current state of the science. Nature Reviews Genetics. 17 (3), 175 (2016).
- Grün, D., van Oudenaarden, A. Design and analysis of single-cell sequencing experiments. Cell. 163 (4), 799-810 (2015).
- Ziegenhain, C., et al. Comparative analysis of single-cell RNA sequencing methods. Molecular cell. 65 (4), 631-643 (2017).
- Hashimshony, T., et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biology. 17 (1), 77 (2016).
- Islam, S., et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Research. 21 (7), 1160-1167 (2011).
- Macosko, E. Z., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161 (5), 1202-1214 (2015).
- Klein, A. M., et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 161 (5), 1187-1201 (2015).
- . Library Prep -Single Cell Gene Expression -Official 10x Genomics Support Available from: https://support.10xgenomics.com/single-cell-gene-expression/library-prep (2018)
- McGinnis, C. S., et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nature Methods. 1, (2019).
- Stoeckius, M., et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biology. 19 (1), 224 (2018).
- Chan, M. M., et al. Molecular recording of mammalian embryogenesis. Nature. 570 (7759), 77-82 (2019).
- . Agilent 4200 TapeStation System Available from: https://www.agilent.com/cs/library/datasheets/public/5991-6029EN.pdf (2019)
- . SPRIselect User Guide Available from: https://research.fhcrc.org/content/dam/stripe/hahn/methods/mol_biol/SPRIselect%20User%20Guide.pdf (2012)
- . Qubit 4 Fluorometer User Guide Available from: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0017209_Qubit_4_Fluorometer_UG.pdf (2018)
- . Agilent High Sensitivity DNA Kit Guide Available from: https://www.agilent.com/cs/library/usermanuals/public/High%20Sensitivity_DNA_KG.pdf (2016)
- Butler, A., Hoffman, P., Smibert, P., Papalexi, E., Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 36 (5), 411 (2018).
- Weber, R. J., Liang, S. I., Selden, N. S., Desai, T. A., Gartner, Z. J. Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules. 15 (12), 4621-4626 (2014).
- . Single Cell Protocols Cell Preparation Guide Available from: https://support.10xgenomics.com/single-cell-gene-expression/sample-prep (2017)

