Direct Agroinoculation of Maize Seedlings by Injection with Recombinant Foxtail Mosaic Virus and Sugarcane Mosaic Virus Infectious Clones
Summary
An Agrobacterium-based injection (agroinjection) protocol is presented for the inoculation of foxtail mosaic virus and sugarcane mosaic virusclones into maize seedlings. Inoculation in this manner leads to viral infection, virus-induced gene silencing of marker genes, and viral overexpression of GFP.
Abstract
Agrobacterium-based inoculation approaches are widely used for introducing viral vectors into plant tissues. This study details a protocol for the injection of maize seedlings near meristematic tissue with Agrobacterium carrying a viral vector. Recombinant foxtail mosaic virus (FoMV) clones engineered for gene silencing and gene expression were used to optimize this method, and its use was expanded to include a recombinant sugarcane mosaic virus (SCMV) engineered for gene expression. Gene fragments or coding sequences of interest are inserted into a modified, infectious viral genome that has been cloned into the binary T-DNA plasmid vector pCAMBIA1380. The resulting plasmid constructs are transformed into Agrobacterium tumefaciens strain GV3101. Maize seedlings as young as 4 days old can be injected near the coleoptilar node with bacteria resuspended in MgSO4 solution. During infection with Agrobacterium, the T-DNA carrying the viral genome is transferred to maize cells, allowing for the transcription of the viral RNA genome. As the recombinant virus replicates and systemically spreads throughout the plant, viral symptoms and phenotypic changes resulting from the silencing of the target genes lesion mimic 22 (les22) or phytoene desaturase (pds) can be observed on the leaves, or expression of green fluorescent protein (GFP) can be detected upon illumination with UV light or fluorescence microscopy. To detect the virus and assess the integrity of the insert simultaneously, RNA is extracted from the leaves of the injected plant and RT-PCR is conducted using primers flanking the multiple cloning site (MCS) carrying the inserted sequence. This protocol has been used effectively in several maize genotypes and can readily be expanded to other viral vectors, thereby offering an accessible tool for viral vector introduction in maize.
Introduction
Infectious clones of many plant viruses have been engineered for virus-induced gene silencing (VIGS), gene overexpression (VOX), and most recently, virus-enabled gene editing (VEdGE)1,2,3,4,5,6,7,8,9,10,11. As new viral constructs are developed, methods to successfully infect plant tissues with these modified viruses must also be considered. Current methods to launch virus infections in plants include particle bombardment, rub-inoculation of in vitro RNA transcripts or DNA clones, vascular puncture inoculation, or Agrobacterium tumefaciens inoculation (agroinoculation)5,12,13,14,15,16,17. Each of these inoculation methods has inherent advantages and disadvantages, which include cost, need for specialized equipment, and feasibility within a given plant-virus system. Methods that utilize infiltration or injection of Agrobacterium strains containing binary T-DNA constructs designed to deliver recombinant viruses are preferred, because they are simple and inexpensive. However, detailed agroinoculation methods for monocotyledonous species such as Zea mays (maize) are lacking.
One of the first reports of agroinoculation for virus delivery was published in 1986, when the genome of cauliflower mosaic virus (CaMV) was inserted into a T-DNA construct, and the resulting Agrobacterium carrying this construct was rub-inoculated onto turnip plants18. Additional methods for agroinoculation have since been developed. For example, in the case of foxtail mosaic virus (FoMV), Nicotiana benthamiana can be used as an intermediate host to generate virus particles in leaves that provide an inoculum source6. Rub inoculation of maize using infected N. benthamiana leaves is efficient, rapid, and simple, but the use of an intermediate host does not work for all maize-infecting viruses. Sugarcane mosaic virus (SCMV), for example, cannot infect N. benthamiana, requiring the use of alternative inoculum sources for vectors derived from this virus. In 1988, Agrobacterium containing maize streak virus (MSV), a DNA virus, was introduced into maize seedlings by injection (agroinjection), demonstrating Agrobacterium-based inoculation methods are also useful for monocots19. Despite this early success with agroinjection, few studies utilizing this technique in maize have been published, leaving open questions about the applicability of this method for RNA viruses and VIGS, VOX, and VEdGE vectors20,21,22. However, broad use of agroinjection in monocot species is promising, because this general approach has been utilized in orchid, rice, and wheat23,24,25,26,27,28.
This protocol was optimized for FoMV and Agrobacterium strain GV3101 and has also been applied to an SCMV vector. FoMV is a potexvirus with a wide host range that includes 56 monocot and dicot species29. FoMV has a 6.2 kilobase (kb) positive sense, single-stranded RNA genome that encodes five different proteins from five open reading frames (ORFs)30,31,32,33,34,35. FoMV was previously developed into both a VIGS and VOX vector for maize by incorporating an infectious clone onto a T-DNA plasmid backbone6,36,37. The viral genome was modified for VIGS applications by adding a cloning site (MCS1*) immediately downstream of the coat protein (CP) (Figure 1A)36. For VOX and VEdGE applications, the CP promoter was duplicated and a second cloning site (MCS2) was added to enable insertion of sequences of interest between ORF 4 and the CP (Figure 1B)6. The FoMV vector containing both MCS1 and MCS2 with no inserts is FoMV empty vector (FoMV-EV) (Figure 1).
SCMV is an unrelated virus that has been developed for VOX in maize38. It is a member of the Potyviridae family, of which several members have been engineered to express foreign proteins in planta39,40,41,42,43,44. The host range of SCMV includes maize, sorghum, and sugarcane45,46, making it valuable for gene functional studies in these major crop plants36,38. SCMV has a positive sense, single-stranded RNA genome of approximately 10 kb in length47,48. To create the SCMV VOX vector, the well-established P1/HCPro junction was utilized as an insertion site for heterologous sequences38. This cloning site is followed by sequence encoding a NIa-Pro protease cleavage site, leading to the production of proteins independent from the SCMV polyprotein (Figure 1C).
T-DNA plasmids carrying infectious cDNA of these recombinant viruses have been transformed into Agrobacterium strain GV3101. GV3101 is a nopaline type strain, which are well-known to be able to transfer T-DNA to monocotyledonous species, including maize26,28,49. Additionally, previous agroinjection studies have used the strains C58 or its derivative GV3101, as well19,20,22,27.
Three marker genes were used in the development of this protocol: two for gene silencing and one for gene expression. A 329 base-pair (bp) fragment from the maize gene lesion mimic 22 (les22, GRMZM2G044074) was used to construct the silencing vector FoMV-LES22. When les22 is silenced in maize, small, round patches of necrotic cells appear along the vasculature of leaves that expand and coalesce into large areas of necrotic leaf tissue50. FoMV-PDS, containing a 313 bp fragment from the sorghum gene phytoene desaturase (pds, LOC110436156, 96% sequence identity to maize pds, GRMZM2G410515), induces silencing of pds in maize, resulting in small streaks of photobleached cells along the vasculature of the leaves that lengthen over time51. The intact coding sequence for green fluorescent protein (GFP) was used to demonstrate protein expression for both FoMV (FoMV-GFP) and SCMV (SCMV-GFP). GFP expression in the leaves is typically most detectable at 14 days post inoculation (DPI)6. Although there have been previous studies utilizing agroinjection of viral vectors in maize, these experiments have only shown that agroinjection can facilitate viral infection from an infectious clone in maize seedlings and do not expand to recombinant viruses designed for VIGS or VOX applications19,20,21,22. The protocol presented here builds upon previous agroinjection methods, particularly Grismley et al.19. Overall, this agroinjection method is compatible with VIGS and VOX vectors, does not require specialized equipment or alternative hosts as inoculum sources, and decreases the overall time and cost required to set up and perform inoculations relative to other common methods that require biolistics or in vitro transcription. This protocol will facilitate functional genomics studies in maize with applications involving VIGS, VOX, and VEdGE.
Protocol
1. Plasmid construction
NOTE: This protocol can be applied to other viral vectors or Agrobacterium strains, but this may affect the overall success of inoculation by agroinjection. Always perform bacterial inoculation and plating steps in a laminar flow hood.
- FoMV silencing construct
NOTE: Luria-Bertani (LB) media (Miller) is used for all media unless otherwise specified. Liquid LB is made by suspending 25 g of granules into 1,000 mL of distilled water and autoclaving for 15 min at 121 °C. Solid LB media is similarly made with the addition of 1.5% agar before autoclaving. Antibiotics are added after LB is cooled to ~60 °C, and the solution is poured into 95 x 15 mm Petri plates. The antibiotic concentrations to use are as follows: rifampicin (rif) at 25 µg/mL, gentamycin (gent) at 50 µg/mL, and kanamycin (kan) at 50 µg/mL.- PCR amplify fragments from the maize gene to be silenced (e.g., les22 or pds) using a forward primer with a PacI restriction site and a reverse primer with an XbaI restriction site. This will enable ligation of the gene fragments into the MCS1* of the FoMV-pCAMBIA1380 binary vector in the antisense orientation.
NOTE: Set up the PCR using a high-fidelity DNA polymerase, forward and reverse primers at 10 µM each, plasmid DNA template, and water, following the DNA polymerase specifications. Amplify for 35 cycles, using an annealing temperature according to the DNA polymerase and primer melting temperature (Tm), and a 30 s extension per kilobase to be amplified. - Perform PCR purification using a PCR purification kit according to the kit specifications.
- Digest the purified PCR product and the FoMV-EV with the restriction enzymes XbaI and PacI. Use 1 µg of plasmid or all of the purified PCR product, 2 µL of 10x buffer, 1 µL of restriction enzyme, and add water to make a 20 µL final reaction volume. Incubate according to enzyme specification.
- Ligate the digested PCR product and FoMV-EV together with T4 DNA ligase according to the manufacturer's protocol.
- Transform the ligated plasmid into DH5α chemically competent E. coli cells using the heat shock method.
- Thaw cells on ice and add 3 µL of plasmid to the tube. Incubate on ice for 30 min, then heat shock for 30 s at 42 °C.
- Place on ice for 5 min, add 200 µL super optimal broth with catabolic repression (SOC) and allow E. coli cells to recover in SOC media for 1 h at 37 °C with shaking at 225 rpm.
- Plate on kanamycin selective LB media and incubate at 37 °C overnight.
- Check colonies for accurate clones by Sanger sequencing using the primers FM-5840F and FM-6138R (Supplemental Table 1). Submit 250 ng of plasmid DNA to a facility that will perform Sanger sequencing. For this experiment samples were sent to Iowa State University DNA Core Facility.
- Inoculate 2 mL of liquid LB with the chosen colony and incubate at 37 °C overnight with shaking at 225 rpm. Extract plasmid DNA from the overnight culture through an alkaline lysis plasmid DNA preparation52.
- Transform plasmid DNA into Agrobacterium strain GV3101 cells using the freeze-thaw method. Allow 100 µL of chemically competent cells to thaw on ice, add 1-5 µL of plasmid and incubate on ice for 30 min. Place in liquid nitrogen for 1 min, then incubate at 37 °C for 3 min. Add 1 mL of SOC, allow to recover for 2-3 h at 28 °C with shaking, plate on rif, gent, and kan selective LB media and incubate at 28 °C for 2 days.
- Screen colonies for the presence of insert with colony PCR. Pick a single bacterial colony and mix it in 30 µL of water. Set up a PCR reaction by adding 12.5 µL of polymerase master mix, 1.25 µL of each 10 µM primer, FM-5840F and FM-6138R, 3 µL of the bacterial colony suspension, and water to a final volume of 25 µL. Cycle 35 times with an annealing temperature of 64 °C and an extension time of 1 min (1 min for every kb amplified).
- Inoculate 2-5 mL of liquid LB (rif, gent, kan) with the correct Agrobacterium colony. Let it grow overnight at 28 °C with shaking at 225 rpm.
- Mix the overnight culture with a 50% glycerol solution 1:1. Store at -80 °C for long-term storage.
- PCR amplify fragments from the maize gene to be silenced (e.g., les22 or pds) using a forward primer with a PacI restriction site and a reverse primer with an XbaI restriction site. This will enable ligation of the gene fragments into the MCS1* of the FoMV-pCAMBIA1380 binary vector in the antisense orientation.
- FoMV expression construct
- PCR amplify the coding sequence of interest including start and stop codons (e.g., GFP) as described in 1.1.1, adding a Bsu36I restriction site on the forward primer and a PspOMI restriction site on the reverse primer to enable directional cloning in the sense orientation into MCS2.
- Perform PCR purification using a PCR purification kit according to the kit's specifications.
- Digest the PCR product and the FoMV-EV with the restriction enzymes Bsu36I and PspOMI, as described in 1.1.3.
- Ligate the digested PCR product and FoMV-EV together with T4 DNA ligase according to the manufacturer's protocol.
- Transform into DH5α chemically competent E. coli cells using the heat shock method as described in 1.1.5. Plate on kanamycin selective LB media and incubate at 37 °C overnight.
- Check colonies for accurate clones by Sanger sequencing as described in 1.1.6 using the primers 5AmuS2 and 5AmuA2 (Supplemental Table 1).
- Inoculate 2 mL of liquid LB with the chosen colony and incubate at 37 °C overnight with shaking at 225 RPM. Extract plasmid DNA from the overnight culture through an alkaline lysis plasmid DNA preparation52.
- Transform plasmid DNA into Agrobacterium strain GV3101 chemically competent cells using the freeze-thaw method as described in 1.1.8. Plate on rif, gent, and kan selective LB media and incubate at 28 °C for 2 days.
- Screen colonies for the presence of insert with colony PCR using the primers 5AmuS2 and 5AmuA2.
- Inoculate 2-5 mL liquid LB (rif, gent, kan) with the correct Agrobacterium colony. Shake overnight at 225 rpm at 28 °C.
- Mix the overnight culture with a 50% glycerol solution 1:1. Store at -80 °C for long-term storage.
- SCMV expression construct
- PCR amplify the gene of interest (e.g., GFP) excluding the stop codon as described in 1.1.1, including a PspOMI digestion site on the forward primer and a SbfI digestion site on the reverse primer to enable directional cloning into the SCMV-pCAMBIA1380 binary vector.
NOTE: Insert must be cloned in frame with the viral polyprotein. - Perform PCR purification using a PCR purification kit according to the kit's specifications.
- Digest the PCR product and the SCMV-EV with the restriction enzymes PspOMI and SbfI, as described in 1.1.3.
- Ligate the digested PCR product and SCMV-EV together with T4 DNA ligase according to the manufacturer's protocol.
- Transform the product into DH5α chemically competent E. coli cells using the heat shock method as described in 1.1.5. Plate on kan selective LB media and incubate at 37 °C overnight.
- Screen colonies for accurate clones by Sanger sequencing as described in 1.1.6 using the primers SC-745F and HCProR1 (Supplemental Table 1).
- Inoculate 2 mL of liquid LB with the chosen colony and incubate at 37 °C overnight with shaking at 225 rpm. Extract the plasmid DNA from the overnight culture through an alkaline lysis plasmid DNA preparation52.
- Transform the plasmid DNA into Agrobacterium strain GV3101 chemically competent cells using the freeze-thaw method as described in 1.1.8. Plate on rif, gent, and kan selective LB media and incubate at 28 °C for 2 days.
- Screen colonies for presence of insert with colony PCR with the primers SC-745F and HCProR1 as described in 1.1.9.
- Inoculate 2-5 mL of liquid LB (rif, gent, kan) with the correct Agrobacterium colony. Shake overnight at 225 rpm at 28 °C.
- Mix the overnight culture with a 50% glycerol solution 1:1. Store at -80 °C for long-term storage.
- PCR amplify the gene of interest (e.g., GFP) excluding the stop codon as described in 1.1.1, including a PspOMI digestion site on the forward primer and a SbfI digestion site on the reverse primer to enable directional cloning into the SCMV-pCAMBIA1380 binary vector.
2. Seedling preparation
- Plant 1-2 maize seeds ('Golden Bantam' sweet corn, FR1064, B73, etc.) in peat-based growing medium in small inserts placed inside trays 4-7 days before injection. Place in a growth chamber under 16 h days at 25 °C and 8 h nights at 22 °C (~185 photosynthetically active radiation (PAR)) or in a greenhouse under 16 h days at 22-25 °C and 8 h nights at 22-25 °C (350-400 PAR).
NOTE: Susceptibility to Agrobacterium varies among maize genotypes, affecting the rates of success. Additionally, some viral vectors may be incompatible with certain maize genotypes. - Water regularly and fertilize once a week with 15-5-15 liquid fertilizer at 330 parts per million (PPM).
3. Preparation of Agrobacterium
- One day before the injection, prepare LB liquid media with the appropriate antibiotic (rif, gent, kan) and inoculate with the Agrobacterium strain carrying the desired viral construct. It is recommended to add 20 µL of glycerol stock into 50 mL of LB, which should yield enough bacterial culture to inoculate >100 plants and can be scaled up or down as needed.
NOTE: Prepare enough inoculum to have a final amount of bacterial suspension of at least 1 mL at an optical density of 600 nm (OD600) of 1.0 for every 4-5 plants. - Shake at 225 rpm at 28 °C for 24 h.
- Pellet bacteria for 10 min at 4,000 x g at room temperature. Discard the supernatant.
- Wash the pellet thoroughly with 1 mL of deionized (DI) water by pipetting or gentle vortexing.
- Repeat step 3.3 to pellet bacteria.
- Resuspend the pellet in 1 mL of 10 mM MgSO4 solution by pipetting or gentle vortexing.
- Optionally, add 200 µM acetosyringone to the solution. Although commonly used, acetosyringone only enhances the transformation ability of some Agrobacterium strains. The authors have not found that the addition of acetosyringone affects efficiency in this protocol (Supplemental Table 2).
NOTE: 10 mM MgSO4 solution can be made from a 1 M stock solution with a pH of 6.3 stored at room temperature. Solution will likely not require pH adjustment.
- Optionally, add 200 µM acetosyringone to the solution. Although commonly used, acetosyringone only enhances the transformation ability of some Agrobacterium strains. The authors have not found that the addition of acetosyringone affects efficiency in this protocol (Supplemental Table 2).
- Measure OD600 of the sample with a spectrophotometer and dilute to 1.0 OD600 with 10 mM MgSO4 solution.
NOTE: This is a safe stopping point. Bacterial suspension can be kept at room temperature for up to 5 h before injection.
4. Injection
NOTE: Maize seedlings from 4-7 days old can be used for injection. Seedling growth rate is greatly affected by growth conditions, amount of PAR (i.e., higher PAR in greenhouse than in growth chamber), and genotype, among other things that can be difficult to control in greenhouse conditions. Plants can be injected as young as 4 days old when they are 2-3 cm tall with no leaves expanded and as old as 7 days when the lowermost rounded-tip leaf is expanded. The success rate of this inoculation methods drops rapidly as plants age beyond 7 days after sowing. The injection site is the same no matter the age of the seedlings.
- Wearing safety goggles, inject the bacterial suspension into the seedlings 2-3 mm above the coleoptilar node using a 25G x 5/8" needle attached to a 1 mL disposable syringe.
NOTE: The coleoptilar node is where crown roots will eventually form. This is the lowest node on the plant. Typically, there will be a color change from green to white at and below the node. The injection location is just above the meristem. Dissecting a few seedlings at this stage may help with visualizing the location of the meristem and consequently the proper injection site. - Apply gentle pressure to the syringe until the suspension fills up the coleoptile or is visible in the whorl, depending on the growth stage of the plants. This is approximately 100-200 µL of suspension.
NOTE: If it is difficult to inject the suspension into the seedling, the injection site may be too low. Moderate pressure is all that should be needed to inject the suspension. - Inject all seedlings, changing syringes and needles for each construct.
5. Continued plant care
- Transplant the injected seedlings to 13 x 13 x 15 cm or larger pots when they are 7-8 days old.
- Maintain growth conditions (16 h photoperiod and fertilizing once per week).
6. Confirmation of infection (Phenotypically and RT-PCR)
- Phenotypically score plants between 14-21 DPI. Lesions from silencing the control genes lesion mimic 22 or phytoene desaturase can easily be seen on the leaves and are distinct from FoMV symptoms. GFP expression can be detected via fluorescent microscope imaging or other UV light imaging.
NOTE: Some constructs/viral vectors may take longer to show symptoms or may not show any symptoms at all. High light conditions greatly increase the phenotypes caused by silencing lesion mimic 22 and phytoene desaturase. Lesions may be less visible or absent if plants are maintained in lower light conditions such as a growth chamber, however the actual infection rate as determined by RT-PCR should not be affected (Table 1). - To confirm infection molecularly, sample leaf 6 between 14-21 DPI and extract total RNA using a phenol-chloroform extraction according to manufacturer's instruction.
- Using the extracted RNA as a template to generate first-strand cDNA.
- Set up the cDNA reaction with up to 5 µg of total RNA, 1 µL of random hexamer primers, 1 µL of oligo (dT)18 primers, 1 µL of dNTPs, 1 µL of of reverse transcriptase and water for a final volume of 14.5 µL.
- Using primers designed for the viral construct and the cDNA as a template, perform PCR on each sample to confirm viral infection and determine the integrity of the gene or gene fragment of interest as described in 1.1.1, except reduce cycles to 25 for FoMV and 30 for SCMV to avoid false positives.
- For FoMV silencing constructs, use primers FM-5840F and FM-6138R to amplify across the MCS1*, which contains the maize gene fragment. For FoMV expression constructs, use primers 5AmuS2 and 5AmuA2 to amplify across the MCS2, which contains the inserted gene.
- For SCMV expression constructs, use primers SC745-F and HCProR1 to amplify across the MCS, which contains the inserted gene (Supplementary Figure 3).
- For an endogenous control gene, use primers ZmActS and ZmActA, which amplify an mRNA fragment of maize actin (GRMZM2G126010) or primers ZmUbiF and ZmUbiR, which amplify an mRNA fragment of maize polyubiquitin (GRMZM2G409726_T01).
- Visualize the PCR product on a 1% agarose gel containing a nucleic acid stain to determine the presence or absence of virus and gene or gene fragment.
Representative Results
The goal of this study was to develop a simple protocol for directly introducing recombinant viruses engineered for gene silencing or gene expression into maize seedlings (Figure 2). The virus vectors carrying inserts are designed and cloned using standard molecular biology techniques. Gene fragments for silencing are inserted into MCS1* in FoMV-EV and coding sequences for expression are inserted into FoMV-EV at MCS2 or SCMV-EV at MCS. The resulting plasmids are transferred to Agrobacterium strain GV3101. Subsequently, maize seedlings are injected within a week or less after planting. Two weeks after injection, plants can be assessed both phenotypically and molecularly for viral infection, gene silencing, and gene expression.
Maize plants are grown in a peat-based medium for 4-7 days. At this stage, the shoot apical meristem is just above the coleoptilar node (Figure 3A). After the coleoptile has extended 2-3 centimeters or up until 7 days after sowing, plants are injected 2-3 mm above the coleoptilar node (Figure 3B-F). At approximately 12 days after injection, plants will begin displaying silencing phenotypes on their leaves, commonly observed near vascular tissue, and these lesions are visually distinct from FoMV viral mosaic symptoms (Figure 4). Both the presence of FoMV and the silencing of target genes is detectable in injected plants (Figure 5). GFP expression can be detected by 2 weeks after injection under a fluorescent microscope and is strongest on leaves 5-7 (Figure 6). When observed under a fluorescence imaging system, GFP expression from FoMV can be visualized as many small, punctate areas of fluorescence distributed across leaves near vascular tissue while GFP expression from SCMV consists of larger patches (Figure 6, Supplemental Figure 1). Although viral mosaic symptoms are often visible on plants infected with FoMV silencing constructs, plants injected with GFP expression constructs that are successfully expressing GFP often do not have these symptoms. As a result, a plant with no visible symptoms may still be positive for virus and GFP expression. Additionally, puncturing the meristem during the agroinjection procedure should be avoided as this can cause morphological defects, but the resulting plants survive and are often symptomatic (Figure 7).
Although this protocol was originally developed using sweet corn, several maize inbred lines can be successfully inoculated with FoMV gene silencing constructs using agroinjection. For example, FR1064 and B73 typically have high rates of viral infection (Table 2). Notably, Mo17, a line with known genetic resistance to FoMV, had a 0% infection efficiency as expected36, 53. Additionally, the construct used influences infection efficiency (Table 3). In the case of FoMV, FoMV-EV and FoMV-LES22 typically have the highest infection efficiencies at 53% and 54%, respectively. FoMV-PDS has a slightly lower efficiency at 38%, and FoMV-GFP is the lowest at 17%. SCMV-GFP has an infection efficiency of 8%. These percentages are averages over several experiments; individual experiments can have higher or lower infection efficiencies.
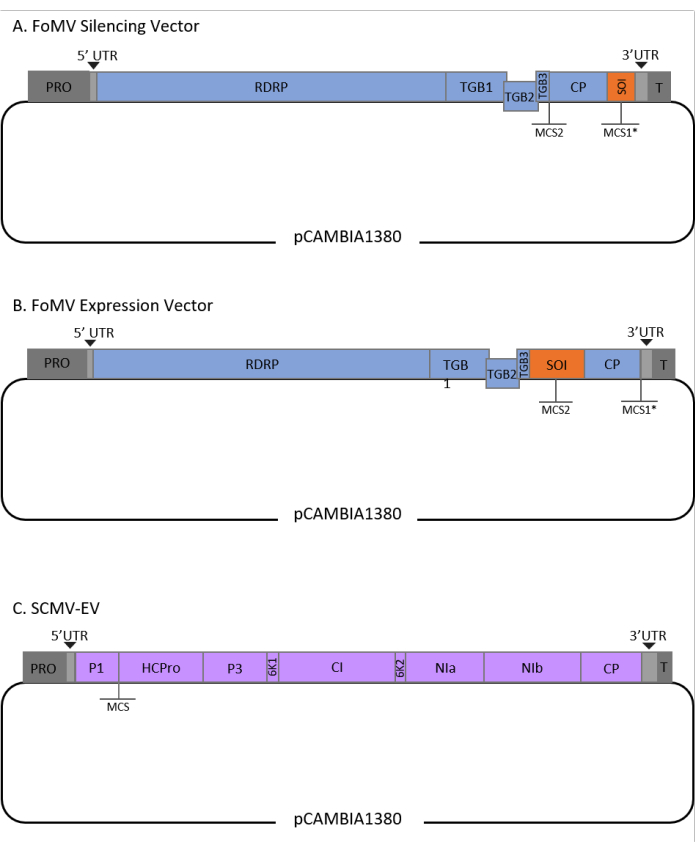
Figure 1: Schematic representations of the FoMV and SCMV T-DNA clones used for agroinjection in maize. The FoMV vector contains two multiple cloning sites (MCS1* and MCS2). The empty vector, FoMV-EV, is 7,269 bp and does not contain any inserts in either MCS. (A) Gene silencing using the FoMV vector can be achieved by inserting gene fragments into MCS1*, designated as sequence of interest (SOI), typically in the anti-sense orientation. (B) Gene expression using the FoMV vector can be accomplished by inserting gene ORFs into the MCS2 in the sense orientation, designated as SOI. (C) The SCMV vector was engineered to have one MCS between P1 and HCPro. The empty vector, SCMV-EV, is 11,015 bp and does not contain any inserts in the MCS. Gene ORFs inserted into the MCS that are in frame with the SCMV polyprotein will be expressed as proteins. Please click here to view a larger version of this figure.
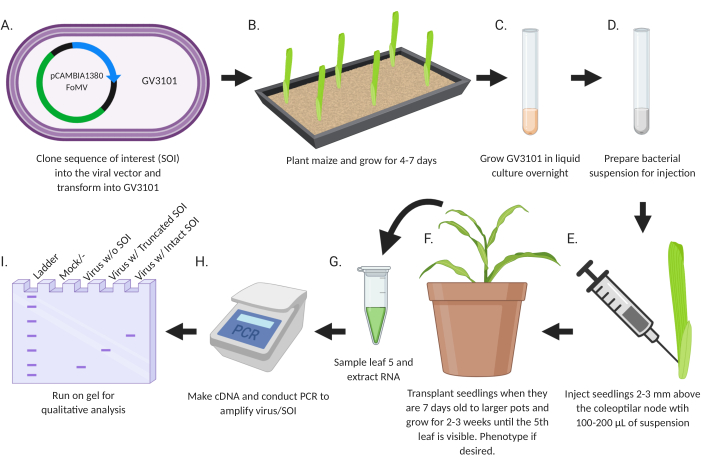
Figure 2: Schematic summary of the agroinjection protocol. (A) Clone SOI, either a CDS or gene fragment, into the viral vector and transform into Agrobacterium strain GV3101. (B) Plant maize and grow for 4-7 days. (C) Grow GV3101 in liquid culture overnight at 28 °C. (D) Prepare bacterial suspension for injection. (E) Inject seedlings 2-3 mm above the coleoptilar node with 100-200 µL of suspension. (F) Transplant seedlings when they are 7 days old to larger pots and grow for 2-3 weeks until the 5th leaf is visible. Phenotype if desired. (G) Sample leaf 5 and extract RNA. (H) Make cDNA and conduct PCR to amplify virus/SOI. (I) Run on gel for qualitative analysis to determine presence/absence of virus and a truncated or intact SOI. This figure was created with BioRender.com. Please click here to view a larger version of this figure.

Figure 3: Agroinjection method of inoculating seedlings just above the coleoptile node. (A) 4-5-day old plants. The coleoptile is fully expanded, and the first true leaf may be partially visible, but is not unfurled. (B) 6-7-day old plants. The first leaf may be expanded but no collars will be visible. The second leaf will also be visible and may be starting to unfurl at this stage. (C) Dissection of 6-7-day old plants showing the location of the shoot apical meristem in relation to the coleoptile node. (D) Injection of 4-5-day old plants. (E) Injection of 6-7-day old plants. (F) Injection of 6-7-day old plants using a dye solution, showing dyed inoculum coming out of the whorl of the seedling. (G) Close-up of injection site of 6-7-day old plants in relation to the coleoptile node. (H) Close-up of a 6-7-day old plant post-injection, showing dyed inoculum in the whorl of the plant. Please click here to view a larger version of this figure.

Figure 4: Symptoms of the silencing control genes used in the agroinjection experiments. (A) A leaf photographed at 17 DPI after the plant was injected with FoMV-LES22. FoMV-LES22 carries a 329 bp insert of the 3' CDS of the lesion mimic 22 maize gene in the antisense orientation. Silencing results in the accumulation of a toxic metabolite which in turn causes the necrotic lesions that first appear as streaks along vasculature and grow into larger patches as shown here. (B) A leaf photographed at 17 DPI after the plant was injected with FoMV-PDS. FoMV-PDS carries a 313 base pair insert of the 3' CDS of the sorghum phytoene desaturase gene in the antisense orientation. Silencing of pds in maize causes a systemic photobleaching phenotype that starts as small, thin streaks along vasculature that grow into longer streaks along the length of the leaf as shown here. Please click here to view a larger version of this figure.
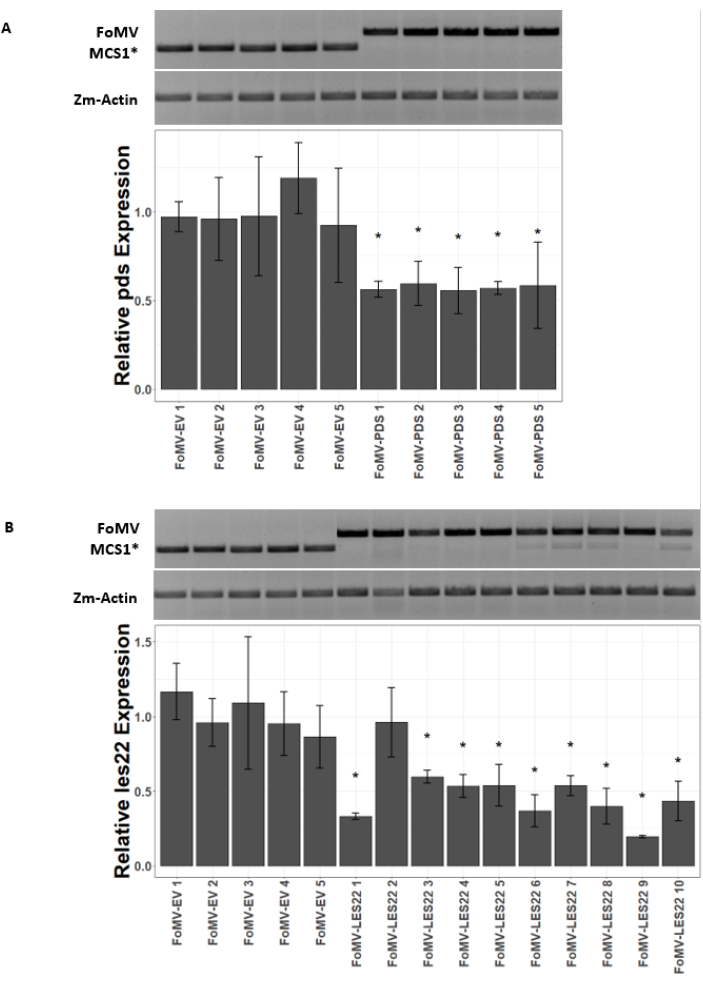
Figure 5: qRT-PCR of plants injected with FoMV gene silencing constructs. Confirmation of systemic FoMV infection and gene silencing induced by the FoMV-LES22 and FoMV-PDS constructs delivered via agroinjection in sweet corn plants (Golden x Bantam). (A) The gel images show RT-PCR analyses confirming the presence of FoMV-MCS1* empty vector (315 bp amplicon) and FoMV-PDS (625 bp amplicon) in leaf 6 of five individual plants. The PCR primers used produce an amplicon that spans MCS1*. The maize gene actin (Zm-Actin) amplicon serves as the reference gene. The bar graph represents the qRT-PCR relative expression values for pds expression in leaf 6 at 37 days post inoculation (DPI) by agroinjection with FoMV-MCS1* or FoMV-PDS. Suppression of pds is detectable in each of the five biological replicates (p=0.003; post hoc Dunnett's test; error bars indicate standard deviation (SD) of three technical replicates). (B) The gel images show RT-PCR analyses confirming the presence of FoMV-MCS1* (315 bp amplicon) in leaf 6 of five individual plants. FoMV-LES22 (625 bp amplicon) was detected in leaf 6 tissue (samples FoMV-LES22 1-5, 38 DPI) and leaf 4 (samples FoMV-LES22 6-10, 20 DPI) for ten individual plants. The Zm-Actin amplicon served as the reference gene. The bar graph represents the qRT-PCR relative expression values for les22 expression in maize tissues by agroinjection of FoMV-MCS1* or FoMV-LES22 viral constructs. Les22 suppression occurs in 9 of 10 biological replicates (p=<0.0001; post hoc Dunnett's test; error bars indicate SD for three technical replicates). Please click here to view a larger version of this figure.
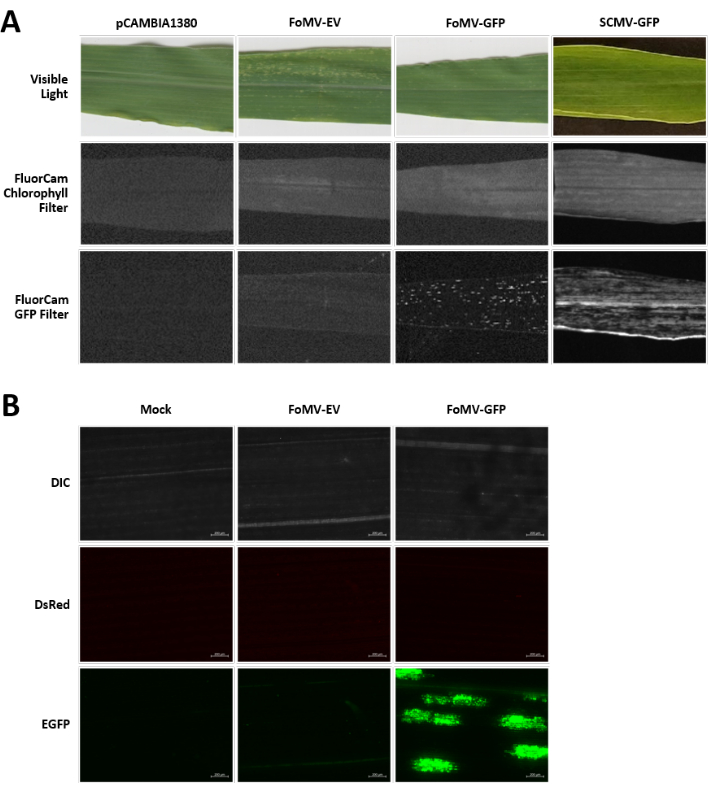
Figure 6. Phenotypes of various constructs used in the agroinjection experiments. All imaged plants were injected when they were 6-7 days old with Agrobacterium strain GV3101 carrying the indicated constructs. Images were taken at 16 DPI. (A) Leaf symptoms of pCAMBIA1380 (empty plasmid backbone), FoMV-EV, FoMV-GFP, and SCMV-GFP in visible light, under the FluorCam chlorophyll filter at 250 µs exposure, and under the FluorCam GFP filter at 10 ms exposure. (B) Fluorescent microscopy images of the leaves of mock-treated (injected with MgSO4 solution only), FoMV-EV, and FoMV-GFP injected plants. The DIC, DsRed, and EGFP channels are shown and were each taken at 1500 ms exposure. Scale bar is 200 µm. Please click here to view a larger version of this figure.

Figure 7. Morphological effects of injection. An example of the more severe morphological effects that can occur from direct injection into meristematic tissue. This injury can result in "shredding" of the leaves and splitting of the stem. Please click here to view a larger version of this figure.
| Virus | Growth Conditions | Genotype | # Infected Plants | Total # of Plants | % Infection | Avg % of Infection |
| FoMV-EV | Growth Chamber | Sweet Corn | 22 | 23 | 96% | 97% |
| B73 | 18 | 18 | 100% | |||
| B104 | 20 | 21 | 95% | |||
| Greenhouse | Sweet Corn | 20 | 23 | 87% | 89% | |
| B73 | 17 | 18 | 94% | |||
| B104 | 16 | 19 | 84% | |||
| SCMV-EV | Growth Chamber | Sweet Corn | 14 | 21 | 67% | 47% |
| B73 | 5 | 18 | 28% | |||
| B104 | 10 | 21 | 48% | |||
| Greenhouse | Sweet Corn | 14 | 23 | 61% | 49% | |
| B73 | 0 | 19 | 0% | |||
| B104 | 19 | 22 | 86% |
Table 1: Effect of greenhouse and growth chamber conditions on agroinjection inoculation efficiency. Seeds were germinated under identical growth conditions. Germinated seedlings were agroinjected and half of them were moved to a growth chamber (25 °C 16 h daylight/ 22C 8 h night; 185 PAR) and the other half were moved to a greenhouse (22-25 °C 16 h daylight/22-25 °C 8 h night; 350-400 PAR). This table reports the rate of infection as a percentage, calculated from the number of plants confirmed by RT-PCR to be infected with the respective virus divided by the total number of agroinjected plants. There is no statistical difference in infection efficiencies between growth chamber and greenhouse conditions (FoMV two tailed t-test p=0.08; SCMV two tailed t-test p=0.96).
| Maize Genotype | FoMV-EV | FoMV-LES22 | Combined Total | ||||
| Infected | Total | % Infected | Infected | Total | % Infected | % Infected | |
| Sweet Corn | 18 | 23 | 78% | 15 | 23 | 65% | 72% |
| MO47 | 7 | 22 | 32% | 1 | 21 | 5% | 19% |
| K55 | 1 | 15 | 7% | 3 | 17 | 18% | 13% |
| W64A | 10 | 22 | 45% | 8 | 20 | 40% | 43% |
| MO17 | 0 | 16 | 0% | 0 | 13 | 0% | 0% |
| B73 | 10 | 18 | 56% | 7 | 17 | 41% | 49% |
| B101 | 12 | 21 | 57% | 8 | 24 | 33% | 44% |
| FR1064 | 4 | 4 | 100% | 4 | 4 | 100% | 100% |
| B104 | 10 | 22 | 45% | 5 | 21 | 24% | 35% |
| WCC22 | 2 | 7 | 29% | 4 | 6 | 67% | 46% |
| A188 | 0 | 3 | 0% | 4 | 6 | 67% | 44% |
Table 2: Infection efficiency of FoMV constructs across maize genotypes. FoMV-EV and FoMV-LES22 were agroinjected into 11 genotypes of maize. After injection, the seedlings were moved to the greenhouse. This table details the rate of infection as a percent, calculated from the number of plants infected with FoMV as confirmed by RT-PCR divided by the total number of agroinjected plants. The combined total rate of infection shows the average rates of infection of each genotype for both FoMV constructs tested.
| Plant Stage | 4-5 Day Old Plants | 6-7 Day Old Plants | Combined Total | ||||
| Symptomatic | Total Plants | % Infected | Symptomatic | Total Plants | % Infected | % Infected | |
| FoMV-EV | 42 | 72 | 58% | 80 | 170 | 47% | 53% (A) |
| FoMV-PDS | 65 | 157 | 41% | 66 | 184 | 36% | 39% (B C) |
| FoMV-LES22 | 115 | 195 | 59% | 144 | 292 | 49% | 54% (A B) |
| FoMV-GFP | 16 | 103 | 16% | 37 | 217 | 17% | 16% (C) |
| SCMV-GFP | 10 | 95 | 11% | 5 | 82 | 6% | 8% (C) |
Table 3: Summary of injection experiments. This table represents a summary of the injection experiments conducted from August 2017 to August 2018 on Golden Bantam sweet corn seedlings. Plants were assessed for viral symptoms (FoMV-EV), silencing symptoms (pds and les22) or GFP fluorescence (GFP) through visual (FoMV-EV, FoMV-PDS, and FoMV-LES22) or FluorCam (FoMV-GFP and SCMV-GFP) screening. Results are shown individually for 4-5 day old plants and 6-7 day old plants, as well as a summary across all plant ages. There is no significant difference found between 4-5 day old plants and 6-7 day old plants (One-way ANOVA, F=0.6513). There is a difference found between viral construct (Onaway ANOVA, F=<0.0001), with the letters representing the Tukey-Kramer HSD connecting letters report.
Supplemental Table 1: Table listing all primer names and sequences used in this protocol. Please click here to download this Table.
Supplemental Table 2: Acetosyringone test. (A) Initial acetosyringone test, comparing rates of symptoms of mock, FoMV-EV, and FoMV-LES22 injected plants between inoculation suspensions with 200 µM acetosyringone (+) or without acetosyringone (-). (B) Comparing the rates of infection of FoMV-LES22 as determined by RT-PCR between inoculation suspensions without acetosyringone (-), with 200 µM acetosyringone (+), and addition of 20 µM of acetosyringone to the bacterial culture 4 hours prior to resuspension in buffer along with the addition of 200 uM acetosyringone to the final suspension (++). Overall, there was no significant difference found between aceotysyringone treatments (Oneway ANOVA, f=0.5452). Please click here to download this Table.
Supplemental Figure 1: Fluorescence imaging and molecular validation of agroinjected SCMV and expression of heterologous proteins in maize. Maize was agroinjected with a modified SCMV construct containing both CDSs of GFP and nano luciferase (NLuc). (A) Fluorcam imaging was used for screening and detection of GFP. The left is a mock injected plant and the right is SCMV-NLucGFP injected plant. (B) Leaf protein extracts were separated by SDS-PAGE and evaluated for the presence of NLuc, GFP, and SCMV coat protein (CP) by in-gel luciferase assay or immunoblot as indicated. Please click here to download this File.
Discussion
Agrobacterium is an essential tool that facilitates numerous molecular biology techniques in plant-related research. This study provides an agroinjection protocol for inoculating FoMV and SCMV viral vectors directly into maize tissues for VIGS and VOX applications. The main goal is to increase the ease and utility of virus-based technologies for research in monocot crop plants. Although direct agroinoculation of maize has been reported for a few viruses, the authors are not aware of a detailed protocol, and there are no examples of VIGS and VOX applications in those studies19,22.
It has been reported, and was confirmed while developing this protocol, that the injection location is a key factor for successfully launching a systemic viral infection via agroinjection19. Consistently injecting the recommended location on the plant is assumed to be the largest variable, because the exact position of the meristem in maize seedlings is virtually undetectable by eye. To minimize interpersonal variation, dissecting a few maize seedlings down to the meristem is recommended to better visualize its location (Figure 3C). The meristem's position in relation to the coleoptilar node should be roughly the same for plants aged 4-7 days old. Additionally, practicing injection with a dyed liquid provides an easily visible demonstration of how the "inoculum" fills the leaf whorl, and because the injection site is marked with dye, the accuracy of the injection site can be corroborated (Figure 3G,H). Meristematic tissues are the most susceptible to agroinjection, but injecting Agrobacterium suspensions directly into this tissue results in undesirable morphological effects (Figure 6)19. Plants with damaged meristems survive, but the resulting defects are undesirable, and thus, direct injection of this tissue should be avoided.
There are several variables that may impact the successful launch of a systemic viral infection via agroinjection because three complex biological systems (plant, virus, and Agrobacterium strain) must interact in coordination. This complex interplay may be aided by the rapidly-dividing cells of the meristematic region, making it an ideal location for agroinoculation19. The Agrobacterium strain must be able to infect cells of the plant tissues to deliver the T-DNA carrying the viral genome, and the plant must be susceptible to the virus in order to initiate viral replication and systemic infection. Maize genotypes differ in their susceptibility to viruses (e.g., Mo17 is resistant to FoMV) or Agrobacterium strains, but the majority that were tested appear to be susceptible to both FoMV and SCMV (Table 1 and Table 2)53. For example, the inbred line FR1064 and the sweet corn variety Golden Bantam may be particularly susceptible to both GV3101 Agrobacterium and FoMV-based vectors.
The leaf number sampled and the timing of sampling for RT-PCR is critical for accurate assessment of viral infection. In the examples shown here, leaf number was determined by starting at the first rounded leaf (commonly known as the "thumb leaf") and counting upward. Leaves were sampled once they were expanded and the next leaf had begun emerging. However, which leaves are optimal for sampling might vary based on virus species used, growth conditions, and maize genotype. Therefore, an initial time course experiment is recommended when applying this protocol to a new virus system to optimize the sampling strategy with respect to leaves and timing.
The specific construct used significantly affects the efficiency of this protocol. For example, the empty vectors, FoMV-EV and SCMV-EV, and FoMV-PDS and FoMV-LES22,which both contain small inserts (313 bp and 329 bp, respectively), typically produce the highest percentages of plants with viral symptoms in these experiments (Tables 1 and Table 2). However, recombinant viruses carrying larger inserts of the GFP ORF (720 bp) in FoMV-GFP and SCMV-GFP, had much lower infection rates when compared to plants injected with the empty vector or gene silencing constructs. This trend may be due to the negative impacts on viral fitness caused by increasing amounts of exogenous genetic material in the viral genome. Several studies have shown that the insert stability of plant viral vectors is largely dependent on insert size and sequence36,54,55,56,57. Additionally, there was a notable difference in percentage of plants that become infected following inoculation with either the FoMV or SCMV empty vector, suggesting additional work is needed to optimize this protocol for SCMV (Table 1). These results indicate that some troubleshooting may be needed when developing a construct, because the sequence and length of the fragment can both affect efficiency.
Overall, this study has shown that agroinjection of maize seedlings is an effective inoculation method for two different RNA plant viruses, multiple vector configurations, and 11 genotypes of maize. This work with FoMV and SCMV, paired with previous works utilizing injection with maize chlorotic mottle virus (MCMV) or MSV, indicates that agroinjection is suitable for inoculating maize seedlings with infectious clones of both RNA and DNA viruses19,20,21,22. Additionally, this work further shows agroinjection is a viable method for VIGS and VOX vectors and can be applied to plants as young as four days old (Table 3). The protocol presented here is expected to be readily adapted by maize biologists to facilitate research in functional genomics studies involving transient gene silencing (VIGS) and overexpression (VOX). Agroinjection also has the capacity to facilitate virus-based gene editing approaches (VEdGE) that would otherwise be limited by reliance on plant transformation, potentially improving editing efficiency as well as accessibility58,59,60. Given the appropriate Agrobacterium strain, maize genotypes, and viral vectors are thoughtfully combined, inoculation by agroinjection is expected to become a valuable tool for transient gene function analyses in maize.
Declarações
The authors have nothing to disclose.
Acknowledgements
Iowa State University is part of a team supporting DARPA's Insect Allies program HR0011-17-2-0053. This work was also supported by the Iowa State University Plant Sciences Institute, Iowa State University Crop Bioengineering Center, USDA NIFA Hatch project number 3808, and State of Iowa Funds. K.L.H. was also partially supported by the Iowa State University Predictive Plant Phenomics graduate training program funded by the National Science Foundation (DGE #1545453) and by Agricultural and Food Research Initiative grant no. 2019-07318 from the USDA National Institute of Food and Agriculture. The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funders.
We thank Nick Lauter (USDA-ARS, Ames, IA) for seed of maize inbred lines, Christian F. Montes-Serey (Iowa State University) for making the FoMV-GFP clone, and Tyler Austin (Iowa State University) for technical assistance.
Materials
| 1 mL syringes | Fisher Scientific | 14955450 | alternatively, BD 309659 |
| 15 mL Falcon Tubes | Corning Science | 352059 | |
| 1kb+ Ladder | ThermoFisher Scientific | 10787018 | For assessing sizes of PCR products |
| 25G x 5/8" PrecisionGlide Needles | Becton, Dickinson and Company (BD) | 305122 | |
| 28°C Incubator | For Agrobacterium | ||
| 37°C Incubator | For E. coli | ||
| Acetosyringone | MilliporeSigma | D134406 | Optional |
| Agar | MilliporeSigma | A4800 | |
| Agarose | GeneMate | E-3120 | For making gels to check for virus/insert stability |
| Agrobacterium tumefaciens Strain GV3101 | Carries vir plasmid encoding T-DNA transfer machinery, RifR, GmR, from lab stock | ||
| Bsu36I | New England Biolabs | R0524 | |
| cDNA Kit | ThermoFisher Scientific | K1672 | Maxima First Strand cDNA Synthesis Kit with Dnase |
| Chloroform | Fisher Scientific | C298 | For RNA extraction |
| Cuvettes | Fisher Scientific | 14955127 | 1.5 mL |
| D-(+)-Glucose | MilliporeSigma | G7528 | Alkaline Lysis |
| DH5alpha Competent E. coli Cells | New England Biolabs | C2987 | |
| DNA Ligase | ThermoFisher Scientific | K1422 | Rapid DNA Ligation Kit |
| EDTA (Ethylenediamine Tetraacetic Acid, Disodium Salt Dihydrate) | Fisher Scientific | S311 | Alkaline Lysis |
| Ethanol | For RNA extraction | ||
| Fertilizer | Peters Fertilizer 15-15-15 Concentrate | ||
| Flat Inserts | T.O. Plastics | 715357C | For germinating seeds in trays |
| Flats | T.O. Plastics | 710245C | For germinating seeds in trays |
| FluorCam | Photon Systems Instruments | To assess maize plants for GFP expression before microscope | |
| Fluorescence Microscope | |||
| Gel Electrophoresis Box | |||
| Gentamycin Sulfate | Fisher Scientific | BP918 | |
| Glacial Acetate | Fisher Scientific | A38 | Alkaline Lysis |
| Glycerol | Fisher Scientific | G33-500 | For saving frozen stocks of bacteria |
| Go-Taq, 2X | Promega | M7123 | |
| Hydrochloric Acid | Fisher Scientific | A144 | for pHing solutions |
| Isopropanol | Sigma-Aldrich | 109827 | For RNA extraction |
| Kanamycin, Monosulfate | Fisher Scientific | BP906 | |
| Large Pots | Kordlok | SQL0550 | 5x5x4" or bigger. For transplanting seedlings. |
| Luria Bertani (LB) Broth, Miller | Himedia | M1245 | |
| Magnesium Sulfate Heptahydrate | Amresco | 662 | |
| Maize Golden Bantam Sweet Corn Seed | American Meadows, West Coast Seeds | ||
| Maize Inbred Seed | Our seed comes from our institution, but we are not able to provide this for other researchers. | ||
| Maxima H Minus Reverse Transcriptase | ThermoFisher Scientific | EP0753 | |
| MilliQ | Elga | Purelab Ultra | |
| Monarch PCR & DNA Cleanup Kit | New England Biolabs | T1030 | |
| PacI | New England Biolabs | R0547 | |
| Peters Excel 15-5-15 Fertilizer | ICL Specialty Fertilizers | G99140 | |
| Petri Dish, 95 mm x 15 mm | Fisher Scientific | FB0875714G | |
| pH Meter | |||
| Potassium Acetate | Fisher Scientific | P171 | Alkaline Lysis |
| Primers | Our primers were synthesized through our institutional DNA facility or through IDT | ||
| PspOMI | New England Biolabs | R0653 | |
| Q5 High-Fidelity DNA Polymerase | New England Biolabs | M0491 | |
| Rifampicin | EMD Millipore Corp | 557303 | |
| Rnase A | ThermoFisher Scientific | 12091021 | Alkaline Lysis |
| SbfI | New England Biolabs | R0642 | |
| Scale | For weighing chemicals for media or buffers | ||
| SDS (Sodium Dodecyl Sulfate) | Fisher Scientific | BP166 | Alkaline Lysis |
| Sodium Hydroxide | Fisher Scientific | S318 | Alkaline Lysis |
| Soil Substrate | SunGro Horticulture | SS#1-F1P | Sunshine Mix #1/Fafard-1P, any soil mix that maize grows well in is sufficient |
| Spectrophotometer | For measuring OD600 | ||
| Sybr Safe, 10,000X | Invitrogen | S33102 | For making gels to check for virus/insert stability |
| Thermocycler | For PCR | ||
| Tris Base | Fisher Scientific | BP154 | Alkaline Lysis |
| Trizol | Ambion | 15596018 | For RNA extraction |
| Weigh Paper | For weighing chemicals for media or buffers | ||
| XbaI | New England Biolabs | R0145 |
Referências
- Zaidi, S. E. A., Mansoor, S. Viral vectors for plant genome engineering. Frontiers in Plant Science. 8, 539 (2017).
- Kant, R., Dasgupta, I. Gene silencing approaches through virus-based vectors: speeding up functional genomics in monocots. Plant Molecular Biology. 100, 3-18 (2019).
- Hu, J., et al. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Molecular Plant Pathology. 20 (10), 1463-1474 (2019).
- Pasin, F., Menzel, W., Daròs, J. A. Harnessed viruses in the age of metagenomics and synthetic biology: an update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnology Journal. 17 (6), 1010-1026 (2019).
- Cody, W. B., Scholthof, H. B. Plant virus vectors 3.0: Transitioning into synthetic genomics. Annual Review of Phytopathology. 57 (1), 211-230 (2019).
- Mei, Y., et al. Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct. 3 (11), 00181 (2019).
- Ruiz, M. T., Voinnet, O., Baulcombe, D. C. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 10 (6), 937-946 (1998).
- Bekele, D., Tesfaye, K., Fikre, A. Applications of virus induced gene silencing (VIGS) in plant functional genomics studies. Journal of Plant Biochemistry & Physiology. 07 (01), 1000229 (2019).
- Scholthof, H. B., Scholthof, K. B. G., Jackson, A. O. Plant virus gene vectors for transient expression of foreign proteins in plants. Annual Review of Phytopathology. 34 (1), 299-323 (1996).
- Holzberg, S., Brosio, P., Gross, C., Pogue, G. P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant Journal. 30 (3), 315-327 (2002).
- Wang, R., et al. An efficient virus-induced gene silencing vector for maize functional genomics research. Plant Journal. 86 (1), 102-115 (2016).
- Redinbaugh, M. G., et al. Transmission of viral RNA and DNA to maize kernels by vascular puncture inoculation. Journal of Virological Methods. 98 (2), 135-143 (2001).
- Scholthof, H. B. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Molecular Plant-Microbe Interactions. 6 (3), 309 (1993).
- Scholthof, H. B., Scholthof, K. B. G., Kikkert, M., Jackson, A. O. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 213 (2), 425-438 (1995).
- Scholthof, H. B. Rapid delivery of foreign genes into plants by direct rub-inoculation with intact plasmid dna of a tomato bushy stunt virus gene vector. Journal of Virology. 73 (9), 7823-7829 (1999).
- Zhang, J., et al. Vacuum and co-cultivation agroinfiltration of (germinated) seeds results in tobacco rattle virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize. Frontiers in Plant Science. 8, 393 (2017).
- Vaghchhipawala, Z., Rojas, C. M., Senthil-Kumar, M., Mysore, K. S. Agroinoculation and agroinfiltration: simple tools for complex gene function analyses. Methods in Molecular Biology. 678, 65-76 (2011).
- Grimsley, N., Hohn, B., Hohn, T., Walden, R. “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proceedings of the National Academy of Sciences. 83 (10), 3282-3286 (1986).
- Grimsley, N. H., Ramos, C., Hein, T., Hohn, B. Merisfematic tissues of maize plants are most suscepnsle to agroinfection with maize streak virus. Bio/Technology. 6 (2), 185-189 (1988).
- Martin, D. P., Rybicki, E. P. Improved efficiency of Zea mays agroinoculation with Maize streak virus. Plant Disease. 84 (10), 1096 (2000).
- Martin, D. P., Willment, J. A., Rybicki, E. P. Evaluation of maize streak virus pathogenicity in differentially resistant Zea mays genotypes. Phytopathology. 89 (8), 695-700 (1999).
- Wang, Q., et al. Further characterization of Maize chlorotic mottle virus and its synergistic interaction with Sugarcane mosaic virus in maize. Scientific Reports. 7, 39960 (2017).
- Hsieh, M. H., et al. Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Science. 201-202 (1), 25-41 (2013).
- Hsieh, M. H., et al. Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. Journal of Experimental Botany. 64 (12), 3869-3884 (2013).
- Zenna, N. S., et al. Genetic analysis of tolerance to rice tungro bacilliform virus in rice (Oryza sativa L.) through agroinoculation. Journal of Phytopathology. 154 (4), 197-203 (2006).
- Marks, M. S., Kemp, J. M., Woolston, C. J., Dale, P. J. Agroinfection of wheat: A comparison of Agrobacterium strains. Plant Science. 63 (2), 247-256 (1989).
- Dasgupta, I., et al. Rice tungro bacilliform virus DNA independently infects rice after Agrobacterium-mediated transfer. Journal of General Virology. 72 (6), 1215-1221 (1991).
- Boulton, M. I., Buchholz, W. G., Marks, M. S., Markham, P. G., Davies, J. W. Specificity of Agrobacterium-mediated delivery of maize streak virus DNA to members of the Gramineae. Plant Molecular Biology. 12 (1), 31-40 (1989).
- Paulsen, A. Q. Purification and properties of foxtail mosaic virus. Phytopathology. 77 (11), 1346 (1977).
- Bancroft, J. B., Rouleau, M., Johnston, R., Prins, L., Mackie, G. A. The entire nucleotide sequence of foxtail mosaic virus RNA. Journal of General Virology. 72 (9), 2173-2181 (1991).
- Bruun-Rasmussen, M., Madsen, C. T., Johansen, E., Albrechtsen, M. Revised sequence of foxtail mosaic virus reveals a triple gene block structure similar to potato virus X. Archives of Virology. 153 (1), 223-226 (2008).
- Rouleau, M., Bancroft, J. B., Mackie, G. A. Partial purification and characterization of foxtail mosaic potexvirus RNA-dependent RNA polymerase. Virology. 197 (2), 695-703 (1993).
- Rouleau, M., Smith, R. J., Bancroft, J. B., Mackie, G. A. Purification, properties, and subcellular localization of foxtail mosaic potexvirus 26-kDa protein. Virology. 204 (1), 254-265 (1994).
- Samuels, T. D., et al. Subcellular targeting and interactions among the potato virus X TGB proteins. Virology. 367 (2), 375-389 (2007).
- Cho, S. Y., Kim, K. H. Identification of the capsid protein-binding region of the SL1(+) RNA located at the 5′ region of the potato virus X genome. Plant Pathology Journal. 28 (1), 75-80 (2012).
- Mei, Y., Zhang, C., Kernodle, B. M., Hill, J. H., Whitham, S. A. A foxtail mosaic virus vector for virus-induced gene silencing in maize. Plant Physiology. 171 (2), 760-772 (2016).
- Bouton, C., et al. Foxtail mosaic virus: A viral vector for protein expression in cereals. Plant Physiology. 177 (4), 1352-1367 (2018).
- Mei, Y., Liu, G., Zhang, C., Hill, J. H., Whitham, S. A. A sugarcane mosaic virus vector for gene expression in maize. Plant Direct. 3 (8), 00158 (2019).
- Gal-On, A., Meiri, E., Huet, H., Hua, W. J., Raccah, B., Gaba, V. Particle bombardment drastically increases the infectivity of cloned DNA of zucchini yellow mosaic potyvirus. Journal of General Virology. 76 (12), (1995).
- Gao, R., et al. Construction of an infectious cDNA clone and gene expression vector of Tobacco vein banding mosaic virus (genus Potyvirus). Virus Research. 169 (1), 276-281 (2012).
- López-Moya, J. J., García, J. A. Construction of a stable and highly infectious intron-containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Research. 68 (2), (2000).
- Choi, I. R., French, R., Hein, G. L., Stenger, D. C. Fully biologically active in vitro transcripts of the eriophyid mite-transmitted wheat streak mosaic tritimovirus. Phytopathology. 89 (12), (1999).
- Kim, K. S., et al. Infectivity of in vitro transcripts of Johnsongrass mosaic potyvirus full-length cDNA clones in maize and sorghum. Archives of Virology. 148 (3), 563-574 (2003).
- Stewart, L. R., Bouchard, R., Redinbaugh, M. G., Meulia, T. Complete sequence and development of a full-length infectious clone of an Ohio isolate of Maize dwarf mosaic virus (MDMV). Virus Research. 165 (2), 219-224 (2012).
- Wylie, S. J., et al. ICTV virus taxonomy profile: Potyviridae. Journal of General Virology. 98 (3), 352-354 (2017).
- Shukla, D. D. taxonomy of potyviruses infecting maize, sorghum, and sugarcane in Australia and the United States as determined by reactivities of polyclonal antibodies directed towards virus-specific N-termini of coat proteins. Phytopathology. 79 (2), 223 (1989).
- Shukla, D. D., Ward, C. W. Amino Acid sequence homology of coat proteins as a basis for identification and classification of the potyvirus group. Journal of General Virology. 69 (11), 2703-2710 (1988).
- Chung, B. Y. W., Miller, W. A., Atkins, J. F., Firth, A. E. An overlapping essential gene in the Potyviridae. Proceedings of the National Academy of Sciences of the United States of America. 105 (15), 5897-5902 (2008).
- Jarchow, E., Grimsley, N. H., Hohn, B. virF, the host-range-determining virulence gene of Agrobacterium tumefaciens, affects T-DNA transfer to Zea mays. Proceedings of the National Academy of Sciences of the United States of America. 88 (23), 10426-10430 (1991).
- Hu, G., Yalpani, N., Briggs, S. P., Johal, G. S. A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. The Plant Cell. 10 (7), 1095 (2007).
- Qin, G., et al. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Research. 17 (5), 471-482 (2007).
- Jones, P. Isolation of plasmid DNA from E. coli. Encyclopedia of Life Sciences. , (2003).
- Ji, Q., Yang, B., Lee, M., Chen, Y., Lübberstedt, T. Mapping of quantitative trait loci/locus conferring resistance to foxtail mosaic virus in maize using the intermated B73-×-Mo17 population. Plant Breeding. 129 (6), 721-723 (2010).
- Pacak, A., et al. The brome mosaic virus-based recombination vector triggers a limited gene silencing response depending on the orientation of the inserted sequence. Archives of Virology. 155 (2), 169-179 (2010).
- Miché, L., Battistoni, F., Gemmer, S., Belghazi, M., Reinhold-Hurek, B. Host-dependent expression of Rhizobium leguminosarum bv. viciae hydrogenase is controlled at transcriptional and post-transcriptional levels in legume nodules. Molecular Plant-Microbe Interactions. 19 (5), 1323-1331 (2018).
- Yamagishi, M., Masuta, C., Suzuki, M., Netsu, O. Peanut stunt virus-induced gene silencing in white lupin (lupinus albus). Plant Biotechnology. 32 (3), 181-191 (2015).
- Avesani, L., et al. Stability of Potato virus X expression vectors is related to insert size: Implications for replication models and risk assessment. Transgenic Research. 16 (5), 587-597 (2007).
- Ali, Z., et al. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Molecular Plant. 8 (8), 1288-1291 (2015).
- Cody, W. B., Scholthof, H. B., Mirkov, T. E. Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant Physiology. 175 (1), 23-35 (2017).
- Ali, Z., Eid, A., Ali, S., Mahfouz, M. M. Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Research. 244, 333-337 (2018).

