用利甘德分析(DRACALA)的微分径向毛细管作用识别小配体的结合蛋白
Summary
利甘德分析(DRaCALA)的微分径向毛细管作用可用于使用ORFeome库识别生物体的小配体结合蛋白。
Abstract
在过去的十年里,细菌生理学中对小信号分子的理解取得了巨大的进步。特别是,在模型生物体中系统地识别和研究了几个核苷酸衍生的二次信使(NSM)的目标蛋白质。这些成就主要归功于一些新技术的发展,包括捕获复合技术和配体检测(DRaCALA)的微分径向毛细管作用,这些技术被用来系统地识别这些小分子的目标蛋白质。本文描述了使用NSM,瓜诺辛五角星和四磷酸盐(p)pppGpp,作为DRACALA技术的例子和视频演示。使用DRaCALA,在模型有机体 Escherichia大肠杆菌 K-12中发现了20种已知蛋白质中的9种,以及12种新靶蛋白(p)pppGpp,显示了这种测定的力量。原则上,DRACALA可用于研究可标有放射性同位素或荧光染料的小配体。这里讨论了 DRACALA 的关键步骤、优点和缺点,以便进一步应用此技术。
Introduction
细菌使用几个小信号分子来适应不断变化的环境1,2。例如,自动诱导剂,N-乙酰异丙胺乳酮及其改良的寡肽,调解细胞间细菌之间的交流,以协调人群行为,这种现象被称为法定人数感应2。另一组小信号分子是NSM,包括广泛研究的环状腺苷单磷酸盐(cAMP)、环二聚氨酸、环状二磷酸盐(环二磷酸盐)和瓜诺辛五磷酸盐和四磷酸盐(p)pppGpp1。细菌产生这些NSM作为对各种不同压力条件的反应。一旦产生,这些分子结合到他们的目标蛋白质,并调节几个不同的生理和代谢途径,以应付遇到的压力,提高细菌的生存能力。因此,识别目标蛋白是破译这些小分子分子功能的必然前提。
在过去的十年里,对这些小信号分子的了解激增,这主要是因为一些技术创新揭示了这些小分子的目标蛋白质。其中包括捕获复合技术3,4,5,和配体检测(DRaCALA)6的微分径向毛细管作用,本文将讨论。
DRaCALA 由文森特·李和同事于 2011年 6 月发明,它部署了硝基纤维素膜的能力,以微分隔离无蛋白质和蛋白质结合的配体。蛋白质等分子不能扩散到亚硝基纤维素膜上,而小配体(如NSM)能够扩散。通过将NSM(例如ppGpp)与要测试的蛋白质混合,并在膜上发现它们,可以预期两种情况(图1):如果(p)ppppp与蛋白质结合,放射性标签(p)pppGpp将由蛋白质保留在点的中心,并且不会向外扩散,从而给出一个强烈的小点(即, 强烈的放射性信号)在磷构体下。然而,如果(p)pppGpp不与蛋白质结合,它将自由向外扩散,产生一个具有均匀背景放射性信号的大点。
此外,如果蛋白质存在足够的量,DRACALA可以检测小分子和整个细胞解剖中未纯化蛋白质之间的相互作用。这种简单性允许使用 DRACALA 使用 ORFeome 表达库快速识别蛋白质靶点。事实上,使用 DRaCALA 系统地识别了 cAMP7、环形二安培8、环形二聚氰化物9、10和 (p) pppGpp 11、12、13 的目标蛋白质。本视频文章以 (p) ppGpp 为例,演示和描述成功进行 DRACALA 筛查的关键步骤和考虑因素。值得注意的是,强烈建议在执行 DRACALA 之前,结合本文阅读对 DRACALA14进行更详尽的描述。
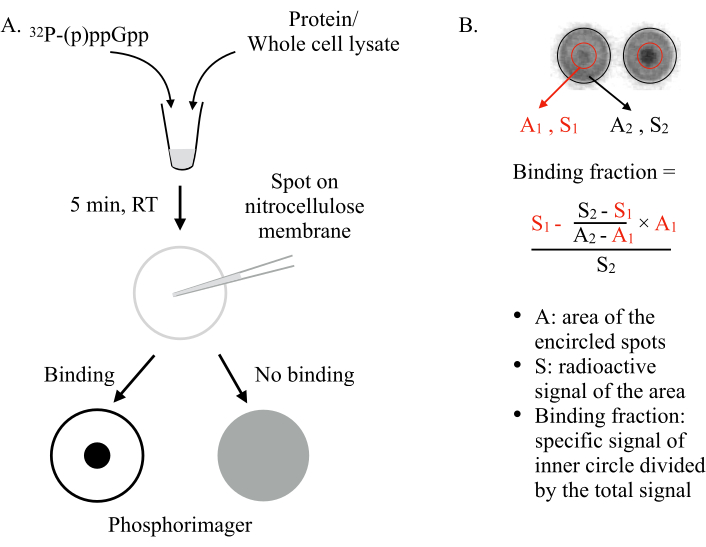
图1:德拉卡拉(A)示意图分析原理。有关详细信息,请参阅文本。(B) 约束分数的量化和计算。有关详细信息,请参阅文本。简言之,DRaCALA 点将通过绘制两个圆圈来分析,这些圆圈将整个点和内部暗点(即由于测试蛋白质的结合而保留的 (p)pPpGpp)。特定的结合信号是减去非特定背景信号(按A1×计算(S 2-S 1)/(A2-A1)后内圈(S1)的放射性信号。结合分数是除以总放射性信号 (S2) 的特定绑定信号。缩写: Dracala = 利甘德分析的差速径向毛细管操作;(p) ppgpp = 瓜诺辛五角星和四磷酸盐;RT = 室温。请单击此处查看此图的较大版本。
Protocol
Representative Results
Discussion
进行 DRACALA 筛查的关键步骤之一是获得良好的全细胞解解。首先,测试的蛋白质应大量和可溶性地生产。其次,细胞的裂解应完成,裂解剂的粘度必须极小。酶的加入和三个冷冻-解冻周期的使用往往足以完全解冻细胞。然而,释放的染色体DNA使溶质粘稠,并产生高背景结合信号,导致误报如图3B,C所示。为了减轻这种情况,可以使用来自S.马塞森的Dnase 1?…
Declarações
The authors have nothing to disclose.
Acknowledgements
这项工作得到向YAZ提供的NNF项目赠款(NNF19OC0058331)以及根据玛丽·斯考多夫斯卡-居里赠款协议(No 801199)向MLS提供的欧洲联盟地平线2020研究和创新方案的支持。

Materials
| 32P-α-GTP | Perkinelmer | BLU006X250UC | |
| 96 x pin tool | V&P Scientific | VP 404 | 96 Bolt Replicator, on 9 mm centers, 4.2 mm Bolt Diameter, 24 mm long |
| 96-well V-bottom microtiter plate | Sterilin | MIC9004 | Sterilin Microplate V Well 611V96 |
| Agar | OXOID – Thermo Fisher | LP0011 | Agar no. 1 |
| ASKA collection strain | NBRP, SHIGEN, JAPAN | Ref: DNA Research, Volume 12, Issue 5, 2005, Pages 291–299. https://doi.org/10.1093/dnares/dsi012 | |
| Benzonase | SIGMA | E1014-25KU | genetically engineered endonuclease from Serratia marcescens |
| Bradford Protein Assay Dye | Bio-Rad | 5000006 | Reagent Concentrate |
| DMSO | SIGMA | D8418 | ≥99.9% |
| DNase 1 | SIGMA | DN25-1G | |
| gel filtration10x300 column | GE Healthcare | 28990944 | contains 20% ethanol as preservative |
| Glycerol | PanReac AppliChem | 122329.1214 | Glycerol 87% for analysis |
| Hypercassette | Amersham | RPN 11647 | 20 x 40 cm |
| Imidazole | SIGMA | 56750 | puriss. p.a., ≥ 99.5% (GC) |
| IP Storage Phosphor Screen | FUJIFILM | 28956474 | BAS-MS 2040 20x 40 cm |
| Isopropyl β-d-1-thiogalactopyranoside (IPTG) | SIGMA | I6758 | Isopropyl β-D-thiogalactoside |
| Lysogeny Broth (LB) | Invitrogen – Thermo Fisher | 12795027 | Miller's LB Broth Base |
| Lysozyme | SIGMA | L4949 | from chicken egg white; BioUltra, lyophilized powder, ≥98% |
| MgCl2 (Magnesium chloride) | SIGMA | 208337 | |
| MilliQ water | ultrapure water | ||
| multichannel pipette | Thermo Scientific | 4661110 | F1 – Clip Tip; 1-10 ul, 8 x channels |
| NaCl | VWR Chemicals | 27810 | AnalaR NORMAPUR, ACS, Reag. Ph. Eur. |
| Ni-NTA Agarose | Qiagen | 30230 | |
| Nitrocellulose Blotting Membrane | Amersham Protran | 10600003 | Premium 0.45 um 300 mm x 4 m |
| PBS | OXOID – Thermo Fisher | BR0014G | Phosphate buffered saline (Dulbecco A), Tablets |
| PEG3350 (Polyethylene glycol 3350) | SIGMA | 202444 | |
| phenylmethylsulfonyl fluoride (PMSF) | SIGMA | 93482 | Phenylmethanesulfonyl fluoride solution – 0.1 M in ethanol (T) |
| Phosphor-imager | GE Healthcare | 28955809 | Typhoon FLA-7000 Phosphor-imager |
| Pipette Tips, filtered | Thermo Scientific | 94410040 | ClipTip 12.5 μl nonsterile |
| Poly-Prep Chromatography column | Bio-Rad | 7311550 | polypropylene chromatography column |
| Protease inhibitor Mini | Pierce | A32955 | Tablets, EDTA-free |
| screw cap tube | Thermo Scientific | 3488 | Microcentifuge Tubes, 2.0 ml with screw cap, nonsterile |
| SLS 96-deep Well plates | Greiner | 780285 | MASTERBLOCK, 2 ML, PP, V-Bottom, Natural |
| spin column | Millipore | UFC500396 | Amicon Ultra -0.5 ml Centrifugal Filters |
| Thermomixer | Eppendorf | 5382000015 | Thermomixer C |
| TLC plate (PEI-cellulose F TLC plates) | Merck Millipore | 105579 | DC PEI-cellulose F (20 x 20 cm) |
| Tris | SIGMA | BP152 | Tris Base for Molecular Biology |
| Tween 20 | SIGMA | P1379 | viscous non-ionic detergent |
| β-mercaptoethanol | SIGMA | M3148 | 99% (GC/titration) |
Referências
- Kalia, D., et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chemical Society Reviews. 42 (1), 305-341 (2013).
- Camilli, A., Bassler, B. L. Bacterial small-molecule signaling pathways. Science. 311 (5764), 1113-1116 (2006).
- Luo, Y., et al. The cAMP capture compound mass spectrometry as a novel tool for targeting cAMP-binding proteins: from protein kinase A to potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channels. Molecular & Cellular Proteomics. 8 (12), 2843-2856 (2009).
- Nesper, J., Reinders, A., Glatter, T., Schmidt, A., Jenal, U. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. Journal of Proteomics. 75 (15), 4874-4878 (2012).
- Laventie, B. J., et al. Capture compound mass spectrometry–a powerful tool to identify novel c-di-GMP effector proteins. Journal of Visual Experiments. (97), e51404 (2015).
- Roelofs, K. G., Wang, J., Sintim, H. O., Lee, V. T. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proceedings of the National Academy of Sciences of the United States of America. 108 (37), 15528-15533 (2011).
- Zhang, Y., et al. Evolutionary adaptation of the essential tRNA methyltransferase TrmD to the signaling molecule 3 ‘,5 ‘-cAMP in bacteria. Journal of Biological Chemistry. 292 (1), 313-327 (2017).
- Corrigan, R. M., et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proceedings of the National Academy of Sciences of the United States of America. 110 (22), 9084-9089 (2013).
- Roelofs, K. G., et al. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathogen. 11 (10), 1005232 (2015).
- Fang, X., et al. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Molecular Microbiology. 93 (3), 439-452 (2014).
- Corrigan, R. M., Bellows, L. E., Wood, A., Grundling, A. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proceedings of the National Academy of Sciences of the United States of America. 113 (12), 1710-1719 (2016).
- Zhang, Y., Zbornikova, E., Rejman, D., Gerdes, K. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. Mbio. 9 (2), 02188 (2018).
- Yang, J., et al. The nucleotide pGpp acts as a third alarmone in Bacillus, with functions distinct from those of (p) ppGpp. Nature Communications. 11 (1), 5388 (2020).
- Orr, M. W., Lee, V. T. Differential radial capillary action of ligand assay (DRaCALA) for high-throughput detection of protein-metabolite interactions in bacteria. Methods in Molecular Biology. 1535, 25-41 (2017).
- Kitagawa, M., et al. Complete set of ORF clones of Escherichia coli ASKA library (A complete Set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Research. 12 (5), 291-299 (2005).
- Hochstadt-Ozer, J., Cashel, M. The regulation of purine utilization in bacteria. V. Inhibition of purine phosphoribosyltransferase activities and purine uptake in isolated membrane vesicles by guanosine tetraphosphate. Journal of Biological Chemistry. 247 (21), 7067-7072 (1972).
- Zhang, Y. E., et al. p)ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Molecular Cell. 74 (6), 1239-1249 (2019).
- Wang, B., et al. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nature Chemical Biology. 15 (2), 141-150 (2019).

