基于荧光的连续核酸内切酶偶联DNA甲基化测定,用于筛选DNA甲基转移酶抑制剂
Summary
DNA甲基转移酶是潜在的癌症药物靶标。在这里,提出了一个协议来评估DNA甲基转移酶抑制的小分子。该测定利用核酸内切酶将DNA甲基化与荧光生成偶联,并允许实时监测酶活性。
Abstract
DNA甲基化是表观遗传基因调控的一种形式,对正常的细胞功能很重要。在细胞中,称为DNA甲基转移酶(DNMT)的蛋白质建立并维持DNA甲基化模式。正常DNA甲基化模式的变化与癌症的发展和进展有关,使DNMT成为潜在的癌症药物靶标。因此,鉴定和表征这些酶的新型小分子抑制剂非常重要。本文提出了一种可用于筛选DNA甲基转移酶抑制剂的方案。连续偶联动力学测定允许在存在和不存在潜在小分子抑制剂的情况下确定DNA甲基化的初始速度。该测定使用甲基敏感的核酸内切酶Gla I将半甲基化DNA底物的甲基化偶联到荧光生成。
这种连续测定允许实时监测酶活性。在微量滴定板中进行小体积测定可降低试剂成本。使用该测定方法,对DNMT1抑制剂进行了小型示例筛选,DNMT1是人类中最丰富的DNMT同工酶。高度取代的蒽醌天然产物乳酸A是一种有效的DNA竞争性DNMT1抑制剂。在这里,我们检查三种潜在的小分子抑制剂 – 蒽醌或具有一到三个取代基的蒽醌样分子 – 在两种浓度下描述测定方案。初始速度用于计算在每个分子存在下观察到的活性百分比。检查的三种化合物中的一种表现出DNMT1活性的浓度依赖性抑制,表明它是DNMT1的潜在抑制剂。
Introduction
DNA甲基化是调节基因表达和染色质结构的重要表观遗传标记。甲基化主要发生在 CpG 二核苷酸中——胞嘧啶,然后是鸟苷;将甲基加入到胞嘧啶的5位。正确的DNA甲基化模式,从而正确的基因表达,对于适当的细胞发育和功能是必需的。许多疾病状态与正常甲基化模式的改变有关1,2,3。例如,癌症的发生和进展以及DNA甲基化模式的改变之间存在联系。通常,癌细胞表现出较低的甲基胞嘧啶总体水平,这会导致基因组不稳定。同时,存在于基因组中的甲基胞嘧啶集中在肿瘤抑制基因的启动子区域,这导致这些重要蛋白质的基因沉默。值得注意的是,表观遗传变化是动态和可逆的,这与与肿瘤发生相关的DNA突变不同。这使得参与表观遗传基因调控的蛋白质成为有趣的药物靶点2,4。
DNA甲基转移酶(DNMT)是负责产生和维持DNA甲基化模式的蛋白质。三种催化活性同工酶DNMT1,DNMT3a和DNMT3b存在于人类中。在发育和分化过程中, 从头 甲基转移酶DNMT3a和DNMT3b建立甲基化模式。两种酶都可以结合催化无活性的DNMT3L蛋白,形成活性增加的复合物1,5。细胞分裂后,子细胞含有半甲基化的DNA——仅在双链的一条链中含有甲基胞嘧啶的DNA——因为新合成的DNA没有甲基化标记。DNMT1的主要功能是甲基化这种半甲基化的DNA,从而重建完整的甲基化模式1,5。
DNMT活动与癌症之间的联系已经确立。DNMT1的过表达,无论是通过转录还是翻译后机制,都是几种常见致癌途径的结果6,7,8,9。使用亚态性等位基因降低DNMT1活性的遗传方法导致Apc(Min)小鼠10的肿瘤形成减少。敲低DNMT1的反义寡核苷酸抑制细胞培养和小鼠肿瘤模型中的肿瘤形成11,12。因此,抑制DNMT1活性似乎是一种有前途的癌症治疗方法。然而,DNMT3同工酶所扮演的角色并不是那么简单。DNMT3a 突变见于急性髓系白血病13 和骨髓增生异常综合征14。已鉴定的突变中至少有一个已被证明会降低酶15的DNA甲基化活性。然而,DNMT3b在乳腺癌16和结直肠癌17中过表达。由于各种DNMT同工酶在致癌作用中发挥不同的作用,因此鉴定同工酶特异性抑制剂至关重要。这些化合物不仅可用于治疗药物的开发,而且同工酶特异性抑制剂也将成为剖析每种DNMT同工酶在癌症病因学中的作用的宝贵工具。
文献中报道了几种DNMT抑制剂。已知的DNMT抑制剂可分为两类:核苷和非核苷。核苷抑制剂通常是胞苷类似物。这些化合物被掺入DNA中并共价捕获DNMT。 5-氮杂胞苷和5-氮杂-2′-脱氧胞苷已被批准用于治疗骨髓增生异常综合征和急性髓系白血病4,18。这些化合物的高毒性、低生物利用度和化学不稳定性存在问题。正在进行的工作正在研究下一代核苷抑制剂的功效;SGI-110来源于5-氮杂-2′-脱氧胞苷,就是一个例子19,20。核苷抑制剂不是同工酶特异性的,会使遇到的任何DNMT同工酶失活。因此,用核苷去甲基化剂处理会导致所有DNMT同工酶4,18的耗竭。非核苷抑制剂不需要掺入DNA中即可发挥其抑制作用。相反,这些分子直接与DNMT结合,引入了同工酶特异性抑制的可能性。迄今为止,已经发现了几种非核苷抑制剂,包括SGI-102721,肼屈嗪22,普鲁卡因酰胺23,RG108及其衍生物24,以及天然产物(−)-表没食子儿茶素3-没食子酸酯(EGCG)25和乳酸A26,27。迄今为止发现的大多数非核苷抑制剂不是同工酶选择性的,或者对一种DNMT同工酶表现出弱的偏好。此外,这些分子的效力需要提高,特别是在细胞4,18中。因此,需要发现或开发更有效的同工酶选择性DNMT抑制剂。
发现新的DNMT小分子抑制剂的一个障碍是传统上用于检查DNMT活性的费力测定28。检测通常是不连续的,有多个步骤。DNMT的酶活性仍然常规使用放射性S-腺苷甲硫氨酸(SAM)29,30,31,32,33,34进行测定。DNA甲基化的非放射性测定也已经开发出来。例如,利用甲基敏感限制性核酸内切酶和电泳分离消化产物的测定已经描述35,36。这些类型的不连续、多步骤测定不容易用于药物发现。自 2000 年代中期以来,已经开发了几种具有更高通量的 DNA 甲基化测定28。闪烁邻近测定用于筛选DNMT1抑制剂37。另一种利用甲基敏感限制性核酸内切酶的测定用于筛选DNMT3a抑制剂25,38。虽然这两种检测方法都比传统的DNA甲基化测定具有更高的通量,但这些测定需要多个步骤,并且不允许实时观察甲基化活性。最近,已经描述了一种连续的动力学测定,该测定将甲基化反应的一种产物S-腺苷同型半胱氨酸(SAH)的形成与NADPH氧化39相关的340nm处的光谱变化耦合。该测定利用三种偶联酶产生光谱信号。
我们开发了一种基于荧光的核酸内切酶偶联DNA甲基化测定,该测定利用了单一的市售偶联酶,可以实时生成数据(图1)。含有三个甲基胞嘧啶的发夹寡核苷酸用作底物。底物DNA在5’端包含一个荧光团,在3’端包含一个淬灭剂。半甲基化CpG位点的甲基化产生核酸内切酶Gla I的切割位点 – 完全甲基化的GCGC。产物寡核苷酸的Gla I裂解从淬灭剂中释放荧光团并实时产生荧光。该测定可用于检查DNMT任何亚型的活性;然而,DNMT1观察到更高的活性,因为该同工酶优先甲基化半甲基化的DNA1,5。如果从DNMT1中删除自动抑制性复制病灶靶向序列(RFTS)结构域,则可以观察到更强大的活性。该结构域位于N末端调控区,与催化位点结合并阻止DNA结合。去除前 ~600 个氨基酸会导致截短酶比全长酶更活跃(kcat/Km 增加 ~640 倍)40。这种酶的活化形式,称为缺乏RFTS的DNMT1(氨基酸621-1616),由于其增加的催化能力,可以更容易地鉴定抑制剂。本文提出了一种在测定中利用缺乏RFTS的DNMT1来筛选潜在小分子抑制剂的方案。使用核酸内切酶偶联连续测定,在存在和不存在几个小分子的情况下确定初始速度。在两种浓度下检查每种潜在抑制剂,以寻找浓度依赖性DNMT1抑制。计算在每种情况下在存在小分子的情况下观察到的活性百分比。
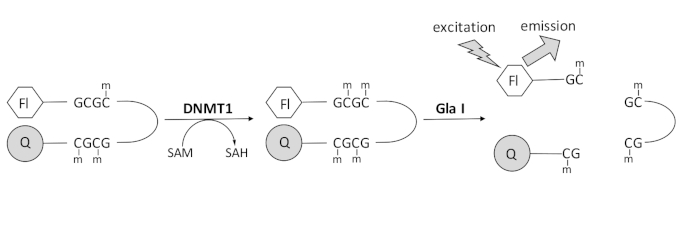
图1:DNA甲基化测定。 使用半甲基化的发夹DNA,5’端有荧光团,3’端有淬灭剂作为底物。DNMT1催化甲基从 S-腺苷甲硫氨酸转移到非甲基化的CpG位点,产生 S-腺苷同型半胱氨酸和完全甲基化的DNA。DNA产物含有核酸内切酶Gla I的切割位点,该位点可切割完全甲基化的GCGC位点。产物DNA的切割从3’淬灭剂中释放5’荧光团,产生荧光。缩写:Fl = 荧光团;Q =淬火;DNMT1 = DNA甲基转移酶1;SAM = S-腺苷蛋氨酸;SAH = S-腺苷同型半胱氨酸。 请点击此处查看此图的大图。
Protocol
Representative Results
Discussion
为了鉴定和表征DNA甲基转移酶的抑制剂,必须测量酶的活性。存在几种检查DNA甲基转移酶活性的方法。通常使用放射性监测活动;SAM标记甲基的转移可以定量为29,30,31,32,33,34。利用甲基敏感核酸内切酶的基于凝胶的测定也已被描述为<sup class="xr…
Declarações
The authors have nothing to disclose.
Acknowledgements
作者感谢巴克内尔大学和化学系对这项工作的支持。
Materials
| 96-well Half Area Black Flat Bottom Polystyrene Not Treated Microplate | Corning | 3694 | |
| 96-Well Polystyrene Conical Bottom Plates | ThermoFisher | 249570 | |
| Bovine Serum Albumin | NEB | B9000S | |
| compound 1 | ChemBridge | 5812086 | screening compound; resuspended in DMSO to 10 mM |
| compound 2 | ChemBridge | 6722175 | screening compound; resuspended in DMSO to 10 mM |
| compound 3 | ChemBridge | 5249376 | screening compound; resuspended in DMSO to 10 mM |
| Dithiothreitol | Sigma | D0632 | |
| Gla I | SibEnzyme | E494 | methyl-sensitive endonuclease |
| Glycerol | RPI | G22025 | |
| Magnesium Chloride | Sigma | M0250 | |
| Oligonucleotide (5'-FAM-CCTATGCGmCATCAGTTTTCTGATGmCGmCATAGG-3'-Iowa Black Quencher) | IDT | custom synthesized | internally quenched hairpin DNA (substrate) |
| Potassium Glutamate | Sigma | G1501 | |
| S-adenosylmethionine | Sigma | A4377 | methyl-donating co-factor (substrate) |
| Tris Base | RPI | T60040 |
Referências
- Jurkowska, R. Z., Jurkowski, T. P., Jeltsch, A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 12 (2), 206-222 (2011).
- Hamidi, T., Singh, A. K., Chen, T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 7 (2), 247-265 (2015).
- Norvil, A. B., Saha, D., Dar, M. S., Gowher, H. Effect of disease-associated germline mutations on structure function relationship of DNA methyltransferases. Genes. 10 (5), 369 (2019).
- Foulks, J. M., et al. Epigenetic drug discovery: targeting DNA methyltransferases. Journal of Biomolecular Screening. 17 (1), 2-17 (2012).
- Goll, M. G., Bestor, T. H. Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry. 74, 481-514 (2005).
- Bigey, P., Ramchandani, S., Theberge, J., Araujo, F. D., Szyf, M. Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene. 242 (1-2), 407-418 (2000).
- Detich, N., Ramchandani, S., Szyf, M. A conserved 3′-untranslated element mediates growth regulation of DNA methyltransferase 1 and inhibits its transforming activity. Journal of Biological Chemistry. 276 (27), 24881-24890 (2001).
- MacLeod, A. R., Rouleau, J., Szyf, M. Regulation of DNA methylation by the Ras signaling pathway. Journal of Biological Chemistry. 270 (19), 11327-11337 (1995).
- Slack, A., Cervoni, N., Pinard, M., Szyf, M. DNA methyltransferase is a downstream effector of cellular transformation triggered by simian virus 40 large T antigen. Journal of Biological Chemistry. 274 (15), 10105-10112 (1999).
- Eads, C. A., Nickel, A. E., Laird, P. W. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic mice. Pesquisa do Câncer. 62, 1296-1299 (2002).
- MacLeod, A. R., Szyf, M. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. Journal of Biological Chemistry. 270 (14), 8037-8043 (1995).
- Ramchandani, S., MacLeod, A. R., Pinard, M., von Hofe, E., Szyf, M. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proceedings of the National Academy Sciences of the United States of America. 94 (2), 684-689 (1997).
- Ley, T. J., et al. DNMT3A mutations in acute myeloid leukemia. New England Journal of Medicine. 363, 2424-2433 (2010).
- Walter, M. J., et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 25 (7), 1153-1158 (2011).
- Russler-Germain, D. A., et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 25 (4), 442-454 (2014).
- Roll, J. D., Rivenbark, A. G., Jones, W. D., Coleman, W. B. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Molecular Cancer. 7, 15 (2008).
- Nosho, K., et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clinical Cancer Research. 15 (11), 3663-3671 (2009).
- Erdmann, A., Halby, L., Fahy, J., Arimondo, P. B. Targeting DNA methylation with small molecules: what’s next. Journal of Medicinal Chemistry. 58 (6), 2569-2583 (2015).
- Chuang, J. C., et al. S110, a 5-Aza-2′-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Molecular Cancer Therapeutics. 9 (5), 1443-1450 (2010).
- Issa, J. J., et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncology. 16 (9), 1099-1110 (2015).
- Datta, J., et al. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Pesquisa do Câncer. 69 (10), 4277-4285 (2009).
- Zambrano, P., et al. A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer. 5, 44 (2005).
- Lee, B. H., Yegnasubramanian, S., Lin, X., Nelson, W. G. Procainamide is a specific inhibitor of DNA methyltransferase 1. Journal of Biological Chemistry. 280 (49), 40749-40756 (2005).
- Asgatay, S., et al. Synthesis and evaluation of analogues of N-phthaloyl-l-tryptophan (RG108) as inhibitors of DNA methyltransferase 1. Journal of Medicinal Chemstry. 57 (2), 421-434 (2014).
- Ceccaldi, A., et al. C5-DNA methyltransferase inhibitors: from screening to effects on zebrafish embryo development. Chembiochem. 12 (9), 1337-1345 (2011).
- Fagan, R. L., Wu, M., Chédin, F., Brenner, C. An ultrasensitive high throughput screen for DNA methyltransferase 1-targeted molecular probes. PLoS One. 8 (11), 78752 (2013).
- Fagan, R. L., Cryderman, D. E., Kopelovich, L., Wallrath, L. L., Brenner, C. Laccaic acid A is a direct, DNA-competitive inhibitor of DNA methyltransferase 1. Journal of Biological Chemistry. 288 (33), 23858-23867 (2013).
- Eglen, R. M., Reisine, T. Screening for compounds that modulate epigenetic regulation of the transcriptome: an overview. Journal of Biomolecular Screening. 16 (10), 1137-1152 (2011).
- Holz-Schietinger, C., Matje, D. M., Reich, N. O. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. Journal of Biological Chemistry. 287 (37), 30941-30951 (2012).
- Norvil, A. B., et al. Dnmt3b methylates DNA by a noncooperative mechanism, and its activity is unaffected by manipulations at the predicted dimer interface. Bioquímica. 57 (29), 4312-4324 (2018).
- Bashtrykov, P., Ragozin, S., Jeltsch, A. Mechanistic details of the DNA recognition by the Dnmt1 DNA methyltransferase. FEBS Letters. 586 (13), 1821-1823 (2012).
- Bashtrykov, P., et al. Targeted mutagenesis results in an activation of DNA methyltransferase 1 and confirms an autoinhibitory role of its RFTS domain. Chembiochem. 15 (5), 743-748 (2014).
- Berkyurek, A. C., et al. The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. Journal of Biological Chemistry. 289 (1), 379-386 (2014).
- Kanada, K., Takeshita, K., Suetake, I., Tajima, S., Nakagawa, A. Conserved threonine 1505 in the catalytic domain stabilizes mouse DNA methyltransferase 1. Journal of Biochemistry. 162 (4), 271-278 (2017).
- Bashtrykov, P., et al. Specificity of Dnmt1 for methylation of hemimethylated CpG sites resides in its catalytic domain. Chemistry & Biology. 19 (5), 572-578 (2012).
- Dolen, E. K., McGinnis, J. H., Tavory, R. N., Weiss, J. A., Switzer, R. L. Disease-associated mutations G589A and V590F relieve replication focus targeting sequence-mediated autoinhibition of DNA methyltransferase 1. Bioquímica. 58 (51), 5151-5159 (2019).
- Kilgore, J. A., et al. Identification of DNMT1 selective antagonists using a novel scintillation proximity assay. Journal of Biological Chemistry. 288 (27), 19673-19684 (2013).
- Ceccaldi, A., et al. Identification of novel inhibitors of DNA methylation by screening of a chemical library. ACS Chemical Biology. 8 (3), 543-548 (2013).
- Duchin, S., Vershinin, Z., Levy, D., Aharoni, A. A continuous kinetic assay for protein and DNA methyltransferase enzymatic activities. Epigenetics & Chromatin. 8, 56 (2015).
- Syeda, F., et al. The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. Journal of Biological Chemistry. 286 (17), 15344-15351 (2011).
- Switzer, R. L., Medrano, J., Reedel, D. A., Weiss, J. Substituted anthraquinones represent a potential scaffold for DNA methyltransferase 1-specific inhibitors. PLoS One. 14 (7), 0219830 (2019).

