Exploring Caspase Mutations and Post-Translational Modification by Molecular Modeling Approaches
Summary
The present protocol uses a biomolecular simulation package and describes the molecular dynamics (MD) approach for modeling the wild-type caspase and its mutant forms. The MD method allows for assessing the dynamic evolution of the caspase structure and the potential effect of mutations or post-translational modifications.
Abstract
Apoptosis is a type of programmed cell death that eliminates damaged cells and controls the development and tissue homeostasis of multicellular organisms. Caspases, a family of cysteine proteases, play a key role in apoptosis initiation and execution. The maturation of caspases and their activity is fine-tuned by post-translational modifications in a highly dynamic fashion. To assess the effect of post-translational changes, potential sites are routinely mutated with residues persistent to any modifications. For example, the serine residue is replaced with alanine or aspartic acid. However, such substitutions could alter the caspase active site’s conformation, leading to disturbances in catalytic activity and cellular functions. Moreover, mutations of other amino acid residues located in critical positions could also break the structure and functions of caspases and lead to apoptosis perturbation. To avoid the difficulties of employing mutated residues, molecular modeling approaches can be readily applied to estimate the potential effect of amino acid substitutions on caspase structure. The present protocol allows the modeling of both the wild-type caspase and its mutant forms with the biomolecular simulation package (Amber) and supercomputer facilities to test the effect of mutations on the protein structure and function.
Introduction
Apoptosis is one of the most widely-studied cellular processes that regulate the morphogenesis and tissue homeostasis of multicellular organisms. Apoptosis can be initiated by a wide range of external or internal stimuli, such as the activation of death receptors, disturbance in the cell cycle signals, DNA damage, endoplasmic reticulum (ER) stress, and various bacterial and viral infections1. The caspases – key apoptotic players – are conventionally classified into two groups: initiators (caspase-2, caspase-8, caspase-9, and caspase-10) and effectors (caspase-3, caspase-6, and caspase-7), depending on their domain structure and the place in the caspase cascade2,3. Upon cell death signals, the initiator caspases interact with adaptor molecules that facilitate proximity-induced dimerization and autoprocessing to form an active enzyme. The effector caspases are activated through cleavage by initiator caspases and perform downstream execution steps by cleaving multiple cellular substrates4.
The maturation and function of the initiator and effector caspases are regulated by a large number of different intracellular mechanisms, among which the post-translational modification plays an indispensable role in cell death modulation5. The addition of modifying groups (phosphorylation, nitrosylation, methylation, or acetylation) or proteins (ubiquitination or SUMOylation) changes the enzymatic activity of caspases or the protein conformation and stability that regulate apoptosis. Site-directed mutagenesis is widely applied to investigate the potential post-translational modification sites and discern their role. A putative modification site is usually replaced by another amino acid, which cannot be further modified. Thus, potentially phosphorylated serine and threonine are mutated to alanine, and lysine ubiquitination sites are replaced with arginine. Another strategy includes substituting an amino acid that particularly mimics post-translational modification (e.g., glutamate and aspartate have been used to mimic phosphorylated serine or threonine)6. However, some of these substitutions located in the high vicinity of an active site or in critical positions could change caspase structure, disturb catalytic activity, and suppress apoptotic cell death7. Similar effects could be observed in cases of tumor-associated missense mutations in caspase genes. For example, the tumor-associated mutation of caspase-6 – R259H – resulted in conformational changes of loops in the substrate-binding pocket, reducing the efficient catalytic turnover of substrates8. The G325A amino acid substitution in caspase-8 identified in head and neck squamous cell carcinoma could hamper caspase-8 activity, which led to the modulation of nuclear factor-kB (NF-kB) signaling and promoted tumorigenesis9.
To assess the potential effect of amino acid substitutions on caspase structure and function, molecular modeling can be applied. The molecular dynamics (MD) approach is described in this work for modeling the wild-type caspase and its mutant forms using the biomolecular simulation package (Amber). The MD method gives a view of the dynamic evolution of the protein structure following the introduction of mutations. Originally developed by Peter Kollman's group, the Amber package became one of the most popular software tools for biomolecular simulations10,11,12,13. This software is divided into two parts: (1) AmberTools, a collection of programs routinely used for system preparation (atom type assignment, adding hydrogens and explicit-water molecules, etc.) and trajectory analysis; and (2) Amber, which is centered around the pmemd simulation program. AmberTools is a free package (and a prerequisite for installing Amber itself), while Amber is distributed with a separate license and fee structure. Parallel simulations on a supercomputer and/or using graphics processing units (GPUs) can substantially improve the performance for the scientific research of protein structure dynamics14. The latest available software versions are AmberTools21 and Amber20, but the described protocols can also be used with the former versions.
Protocol
1. System preparation
NOTE: The molecular models of the native and mutant protein forms are built based on an appropriate crystal structure obtained from the Protein Data Bank15,16.
- To retrieve the selected PDB structure, use the Download Files drop-down list and click on PDB Format. Remove remarks and connectivity data, and insert a TER card between separate protein chains in the PDB file. Optionally, set the protonation state of histidine residues by replacing the residue name HIS with HIE, HID, or HIP (Supplementary File 1).
NOTE: It is reasonable not to remove crystallographically resolved water molecules (if present in the PDB file). - To prepare the starting model, launch the tleap program (AmberTools package) and then enter commands that manipulate tleap objects.
- Launch the program: tleap. Load the ff14SB force field to describe the protein with molecular mechanics: > source leaprc.protein.ff14SB.
- Load force fields for water molecules and atomic ions (Na+, Cl–): > source leaprc.water.tip3p.
- Load the PDB file and build coordinates for hydrogens, creating an object named mol: > mol = loadpdb protein.pdb.
- Check for internal inconsistencies that could cause problems: > check mol.
NOTE: Disulfide bridges, if present, must be specified manually. Replace the names of covalently bound cysteines CYS with CYX and type the following command in tleap to create a bond between SG atoms: bond mol.X.SG mol.Y.SG (X and Y are cysteine residue numbers). - Create a solvent box around the protein (TIP3P water, 12 Å distance): > solvatebox mol TIP3PBOX 12.0.
- Check the total charge: > charge mol, and add counterions (Na+ or Cl–) to neutralize the system:
> addions mol Na+ 0. - Create the topology prmtop file and the coordinate inpcrd file (inputs for running a simulation): > saveamberparm mol protein.prmtop protein.inpcrd. Quit the tleap program: > quit.
NOTE: Amber-format coordinate files can be easily converted to PDB format using the ambpdb tool (refer to the Users' Manual, see Table of Materials).
2. Energy minimization
NOTE: The energy minimization is necessary to remove any bad contacts and overlaps between atoms in the starting system that lead to instability when running MD.
- Carry out the first stage of the energy minimization (2,500 steps of the steepest descent algorithm + 2,500 steps of the conjugate gradient algorithm) to optimize the positions of added hydrogen atoms and water molecules while keeping the protein coordinates fixed by positional restraints on heavy atoms.
- Run the pmemd program as follows:
pmemd -O -i min1.in -p protein.prmtop -c protein.inpcrd -o protein_min1.out
-r protein_min1.rst -ref protein.inpcrd. - Follow the required arguments: -i FILE control data; -p FILE molecular topology, force field parameters, atom names; -c FILE initial coordinates; -o FILE user-readable log output; -r FILE final coordinates; -ref FILE reference coordinates for position restraints.
- Specify options in the input file min1.in:
&cntrl
imin=1, maxcyc=5000, ncyc=2500,
cut=10.0, ntb=1,
ntc=1, ntf=1,
ntpr=10,
ntr=1,
restraintmask=':1-517 & !@H=',
restraint_wt=2.0
/
NOTE: Input options: imin=1 perform minimization; maxcyc=5000 maximum number of minimization cycles; ncyc=2500 minimization method is switched from steepest descent to conjugate gradient after ncyc cycles; cut=10.0 non-bonded cutoff (Å); ntb=1 periodic boundaries are imposed, constant volume; ntc=1 no constraints are applied to bond lengths (for better energy convergence); ntf=1 all interactions are calculated; ntpr=10 energy information is printed to the out file every ntpr steps; ntr=1 flag for restraining specified atoms using a harmonic potential; restraintmask=':1-517 & !@H=' specifies restrained atoms; restraint_wt=2.0 weight (kcal/mol∙Å2) for positional restraints.
- Run the pmemd program as follows:
- Carry out the second stage of the energy minimization (5,000 steepest descent steps + 5,000 conjugate gradient steps) without restraints to optimize the whole system.
- Use min2.in and protein_min1.rst as inputs: pmemd -O -i min2.in -p protein.prmtop
-c protein_min1.rst -o protein_min2.out -r protein_min2.rst. - Specify options in the input file min2.in:
&cntrl
imin=1, maxcyc=10000, ncyc=5000,
cut=10.0, ntb=1,
ntc=1, ntf=1,
ntpr=10
/
- Use min2.in and protein_min1.rst as inputs: pmemd -O -i min2.in -p protein.prmtop
3. Heating
NOTE: This stage aims to heat the system from 0 K to 300 K. Initial velocities are assigned to atoms since the starting model based on the PDB file does not contain velocity information.
- Carry out the heating process with positional restraints on the protein atoms (50 ps, constant volume).
- Use heat.in and protein_min2.rst as inputs: pmemd -O -i heat.in -p protein.prmtop
-c protein_min2.rst -o protein_heat.out
-r protein_heat.rst -x protein_heat.mdcrd -ref protein_min2.rst - Follow the required arguments: -x FILE coordinate sets saved over MD trajectory.
- Specify options in the input file heat.in:
&cntrl
imin=0, irest=0, ntx=1,
nstlim=25000, dt=0.002,
ntc=2, ntf=2,
cut=10.0, ntb=1,
ntpr=500, ntwx=500,
ntt=3, gamma_ln=2.0,
tempi=0.0, temp0=300.0,
ntr=1, restraintmask=':1-517',
restraint_wt=1.0,
nmropt=1
/
&wt TYPE='TEMP0',
istep1=0, istep2=25000,
value1=0.1, value2=300.0 /
&wt TYPE='END' /
NOTE: Input options: imin=0 run MD; irest=0 run a new simulation; ntx=1 no initial velocities are read from the rst file; nstlim=25000 number of MD steps; dt=0.002 time step (ps); ntc=2 bonds involving hydrogen are constrained using the SHAKE algorithm; ntf=2 forces for the constrained bonds are not calculated; ntwx=500 coordinates are written to the mdcrd file every ntwx steps; ntt=3 Langevin temperature regulation; gamma_ln=2.0 collision frequency for Langevin dynamics (ps−1); tempi=0.0 initial temperature; temp0=300.0 reference temperature; nmropt=1 parameters specified in an &wt namelist are read; TYPE='TEMP0' vary the target temperature.
- Use heat.in and protein_min2.rst as inputs: pmemd -O -i heat.in -p protein.prmtop
4. Equilibration
NOTE: This stage is necessary to adjust the density of water and obtain the equilibrium state of the protein.
- Perform the equilibration at 300 K without any restraints (500 ps, constant pressure).
- Use equil.in and protein_heat.rst as inputs: pmemd -O -i equil.in -p protein.prmtop
-c protein_heat.rst -o protein_equil.out -r protein_equil.rst -x protein_equil.mdcrd. - Specify options in the input file equil.in:
&cntrl
imin=0, irest=1, ntx=5,
nstlim=250000,
dt=0.002,
ntc=2, ntf=2,
cut=10.0, ntb=2, ntp=1, taup=2.0,
ntpr=1000, ntwx=1000, ntwr=50000,
ntt=3, gamma_ln=2.0,
temp0=300.0
/
NOTE: Input options: irest=1 restart the simulation; ntx=5 coordinates and velocities are read from a previously generated rst file; ntb=2 periodic boundaries are imposed, constant pressure; ntp=1 isotropic pressure scaling; taup=2.0 pressure relaxation time (ps); ntwr=50000 every ntwr steps, the rst file is written, ensuring recovery from a crash.
- Use equil.in and protein_heat.rst as inputs: pmemd -O -i equil.in -p protein.prmtop
5. Production dynamics
- After equilibrium has successfully been achieved, carry out a production MD simulation (10 ns or longer) at constant pressure, and generate the trajectory file for subsequent analysis of the protein structure.
- Use prod.in and protein_equil.rst as inputs: pmemd -O -i prod.in -p protein.prmtop
-c protein_equil.rst -o protein_prod.out -r protein_prod.rst -x protein_prod.mdcrd. - Specify options in the input file prod.in:
&cntrl
imin=0, irest=1, ntx=5,
nstlim=5000000,
dt=0.002,
ntc=2, ntf=2,
cut=10.0, ntb=2, ntp=1, taup=2.0,
ntpr=1000, ntwx=1000, ntwr=50000,
ntt=3, gamma_ln=2.0,
temp0=300.0, ig=-1
/
NOTE: The parallel version of pmemd (pmemd.MPI) or GPU-accelerated version (pmemd.cuda) can be used on computer clusters and supercomputers. A long MD simulation may be broken into several segments and performed sequentially. It is strongly recommended to set ig=-1 (random seed option) for every simulation restart. The ptraj or cppptraj programs can be used for the analysis and processing of coordinate trajectories. For more information, see the software Users' Manual.
- Use prod.in and protein_equil.rst as inputs: pmemd -O -i prod.in -p protein.prmtop
Representative Results
The present protocol can be readily applied in studies of post-translational modification of caspases or pathogenic mutations. In this section, the MD modeling workflow is illustrated (Figure 1), which has been successfully used in the study of caspase-27. Using in vitro site-directed mutagenesis of potential phosphorylation sites (Ser/Thr to Ala) and biochemical approaches, it was demonstrated that the Ser384Ala mutation prevented caspase-2 processing and blocked enzymatic activity and the induction of apoptotic cell death (Supplementary Figure 1). However, neither mass-spectrometry analysis nor Phos-Tag technology confirmed that Ser384 undergoes phosphorylation7. To understand how Ser384 regulates caspase-2 activity, MD modeling of the three-dimensional protein structure was performed (Figure 1).
The molecular models of wild-type and mutant forms of caspase-2 were constructed using the 1pyo crystal structure (available caspase-2 structures are listed in Supplementary Table 1). The Ser384Ala mutant was created by removing the Oγ atom in the Ser384 residue (in PDB, Ser and Ala residues are distinguished by having the Oγ atom). Hydrogen coordinates are usually absent in crystal structures. Therefore, hydrogen atoms were added to the protein structure, and then it was solvated by 12 Å thick layer of TIP3P water to enable further explicit-solvent simulations. Sodium ions were added to neutralize the system. The obtained starting models of caspase-2 were subjected to energy minimization, equilibration, and subsequent 10 ns MD simulation according to the MD modeling protocol, using the min1.in, min2.in, heat.in, equil.in, and prod.in control data files.
In the crystal structure of caspase-2, Ser384, together with Arg219 and Arg378, are involved in forming the cavity's surface in the active site. Arg219 and Arg378 form hydrogen bonds with the substrate's carboxyl group, while Ser384 does not intervene in direct interactions with the substrate. MD simulations confirmed that the Ser384Ala substitution did not affect the catalytic residues Cys320 (nucleophile) and His277 (general base) but induced a major conformational change in Arg378. Its guanidinium group turned toward the bulk solvent so that the Nε atom could not form an essential hydrogen bond with the substrate's carboxyl group (Figure 1). In the native enzyme, an electrostatic interaction occurs between the Oγ atom of Ser384 (partial negative charge) and the Arg378 guanidinium group (positive charge). However, the serine substitution apparently disrupts this interaction. Thus, it was shown that the Ser384Ala substitution affected the substrate recognition by the arginine residues in the caspase-2 active site, impairing enzymatic activity and the ability to trigger apoptotic cell death. The discovered mechanism seems evolutionarily conserved and common for other caspase family members. Thus, the implementation of MD simulations allowed for confirming the biochemical results and obtaining new insights into the molecular structure of the active center of caspases.
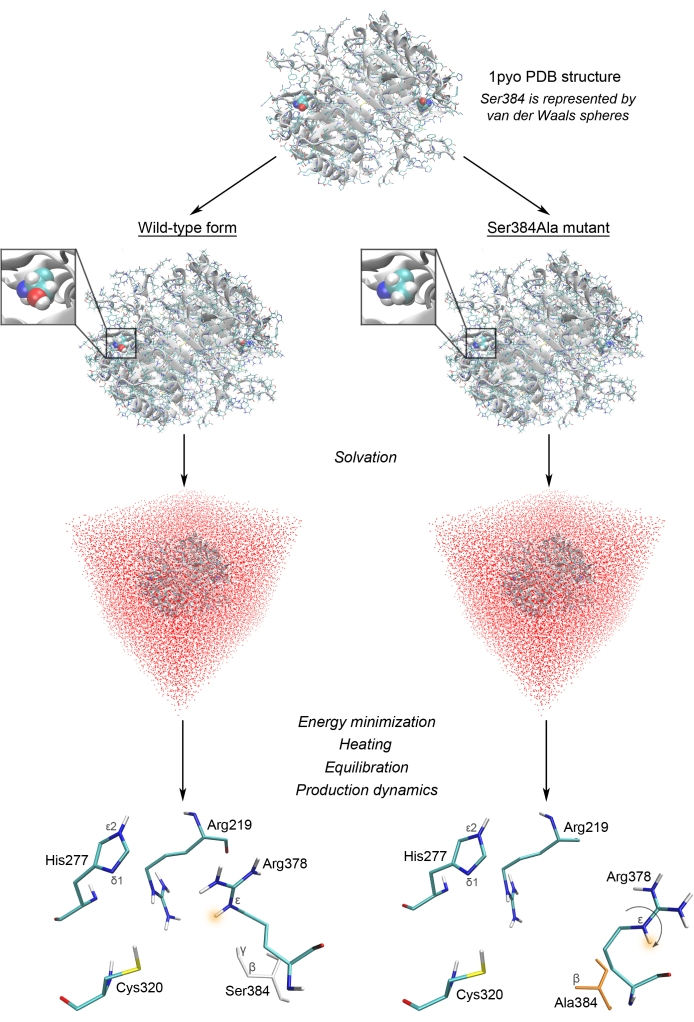
Figure 1: MD modeling workflow used to study the caspase structure. The wild-type caspase-2 and its Ser384Ala mutant were investigated following the instructions in the protocol section. It was revealed that the Ser384Ala substitution induces an important conformational change in the active site residue Arg378. Please click here to view a larger version of this figure.
Supplementary Figure 1: Function of caspase-2 in cell death. In response to extrinsic and intrinsic stimuli, wild-type caspase-2 can be activated by an autoproteolytic mechanism. Active caspase-2 cleaves Bid, which, in turn, promotes mitochondrial outer membrane permeabilization and apoptotic cell death. The Ser384Ala mutation prevents caspase-2 activation and the induction of cell death. Please click here to download this File.
Supplementary Table 1: Caspase-2 structures available in the Protein Data Bank. The structures are sorted by release date. Please click here to download this Table.
Supplementary File 1: Different steps in editing a PDB file. Please click here to download this File.
Discussion
The described MD approach allows for modeling both the wild-type and mutant forms of caspase using the biomolecular simulation packages. Several important issues of the methodology are discussed here. First, a representative crystal structure of caspase needs to be selected from the Protein Data Bank. Importantly, both monomeric and dimeric forms of caspase are acceptable. Choosing high-resolution structures with a minimum number of missing residues is a good idea. The protonation state of some residues can be set manually in the input PDB file. For example, in Figure 1, a hydrogen atom was attached to the Nε2 atom (HIE form) of the catalytic residue His277. Second, a solvent box must be created around the protein for further explicit-solvent simulations. Energy minimization is required to optimize the coordinates of added hydrogen atoms and water molecules. Third, the heating and equilibration must be carried out prior to the production dynamics simulation. At the heating stage, one should ensure that the cavities and binding pockets on the protein surface fill up with water molecules (if necessary, one can increase the number of MD steps). At the equilibration stage, the acquisition of the equilibrium conformation of the protein needs to be confirmed by analyzing the root-mean-square deviation of its backbone atoms from the initial positions12. Fourth, monitoring the conformational changes of the protein structure during the production dynamics simulation is required. For this purpose, one should superimpose trajectory frames onto the starting structure by fitting the backbone atoms and then analyze the conformational changes using a molecular visualization tool (VMD, PyMOL, etc.)17. If necessary, one can increase the number of MD steps and/or break up the simulation into several segments performed sequentially. Fifth, it is recommended to use the GPU-accelerated version of the MD simulation program (pmemd.cuda), whose performance significantly exceeds that achievable by the traditional CPU implementation14,18.
A possible limitation of the described method is the availability of certain caspase structures in the Protein Data Bank. However, more than 900 PDB entries are found under the "caspase" request, indicating that caspase structures have been investigated in detail. Some difficulties may arise when a crystal structure is obtained at low resolution (i.e., at a coarse level of detail) or lacks some fragments important for the caspase function.
In summary, MD modeling approaches have proven effective in predicting structural changes of proteins after mutations of their genes, modifications, or intermolecular interactions. The application of MD simulation in the field of cell death opens great opportunities to study the molecular mechanisms of caspase regulation by post-translational modifications. In this regard, long-term MD simulation could assess the dynamic behavior of functional regions and the potential effect of amino acid substitutions to elucidate the molecular causes associated with mutated or modified caspase-2 as well as other caspases. For example, missense mutations of initiator caspase genes (caspase-2/caspase-8/caspase-9/caspase-10) were detected in gastrointestinal cancers and in tumors of the central and peripheral nervous systems19. Together, MD modeling of caspases is a powerful in silico approach for understanding the mechanisms regulating programmed cell death and associated disorders and for developing new effective therapies.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant from the Russian Science Foundation (17-75-20102, the protocol development). Experiments described in the representative results section (analysis of phosphorylation) were supported by the Stockholm (181301) and Swedish (190345) Cancer Societies.
Materials
| Amber20 | University of California, San Francisco | Software for molecular dynamics simulation http://ambermd.org |
|
| AmberTools21 | University of California, San Francisco | Software for molecular modeling and analysis http://ambermd.org |
Referências
- Olsson, M., Zhivotovsky, B. Caspases and cancer. Cell Death and Differentiation. 18 (9), 1441-1449 (2011).
- Lavrik, I. N., Golks, A., Krammer, P. H. Caspase: Pharmacological manipulation of cell death. Journal of Clinical Investigation. 115 (10), 2665-2672 (2005).
- Degterev, A., Boyce, M., Yuan, J. A decade of caspases. Oncogene. 22 (53), 8543-8567 (2003).
- Pop, C., Salvesen, G. S. Human caspases: Activation, specificity, and regulation. Journal of Biological Chemistry. 284 (33), 21777-21781 (2009).
- Zamaraev, A. V., Kopeina, G. S., Prokhorova, E. A., Zhivotovsky, B., Lavrik, I. N. Post-translational modification of caspases: The other side of apoptosis regulation. Trends in Cell Biology. 27 (5), 322-339 (2017).
- Pearlman, S. M., Serber, Z., Ferrell, J. E. A mechanism for the evolution of phosphorylation sites. Cell. 147 (4), 934-946 (2011).
- Zamaraev, A. V., et al. Requirement for Serine-384 in Caspase-2 processing and activity. Cell Death and Disease. 11 (10), 825 (2020).
- Dagbay, K. B., Hill, M. E., Barrett, E., Hardy, J. A. Tumor-associated mutations in caspase-6 negatively impact catalytic efficiency. Bioquímica. 56 (34), 4568-4577 (2017).
- Ando, M., et al. Cancer-associated missense mutations of caspase-8 activate nuclear factor-κB signaling. Cancer Science. 104 (8), 1002-1008 (2013).
- Case, D. A., et al. . AMBER 2020. , (2020).
- Salomon-Ferrer, R., Case, D. A., Walker, R. C. An overview of the Amber biomolecular simulation package. WIREs Computational Molecular Science. 3 (2), 198-210 (2013).
- Roe, D. R., Cheatham, T. E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation. 9 (7), 3084-3095 (2013).
- Maier, J. A., et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation. 11 (8), 3696-3713 (2015).
- Salomon-Ferrer, R., Götz, A. W., Poole, D., Le Grand, S., Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. Journal of Chemical Theory and Computation. 9 (9), 3878-3888 (2013).
- Berman, H. M., et al. The Protein Data Bank. Nucleic Acids Research. 28 (1), 235-242 (2000).
- Burley, S. K., et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Research. 49, 437-451 (2021).
- Martinez, X., et al. Molecular graphics: Bridging structural biologists and computer scientists. Structure. 27 (11), 1617-1623 (2019).
- Lee, T. S., et al. GPU-accelerated molecular dynamics and free energy methods in Amber18: Performance enhancements and new features. Journal of Chemical Information and Modeling. 58 (10), 2043-2050 (2018).
- Jäger, R., Zwacka, R. M. The enigmatic roles of caspases in tumor development. Cancers. 2 (4), 1952-1979 (2010).

