Combining Clearing and Fluorescence Microscopy for Visualising Changes in Gene Expression and Physiological Responses to Plasmodiophora brassicae
Summary
The present protocol describes an optimized method for the histological observation of galls induced by Plasmodiophora brassicae. Vibratome sections of hypocotyls are cleared before fluorescence imaging to study the involvement of transcription factors and phytohormones during disease progression. This protocol overcomes resin embedding limitations, enabling in planta visualization of fluorescent proteins.
Abstract
Infection of Brassica crops by the soilborne protist Plasmodiophora brassicae leads to gall formation on the underground organs. The formation of galls requires cellular reprogramming and changes in the metabolism of the infected plant. This is necessary to establish a pathogen-oriented physiological sink toward which the host nutrients are redirected. For a complete understanding of this particular plant-pathogen interaction and the mechanisms by which host growth and development are subverted and repatterned, it is essential to track and observe the internal changes accompanying gall formation with cellular resolution. Methods combining fluorescent stains and fluorescent proteins are often employed to study anatomical and physiological responses in plants. Unfortunately, the large size of galls and their low transparency act as major hurdles in performing whole-mount observations under the microscope. Moreover, low transparency limits the employment of fluorescence microscopy to study clubroot disease progression and gall formation. This article presents an optimized method for fixing and clearing galls to facilitate epifluorescence and confocal microscopy for inspecting P. brassicae-infected galls. A tissue-clearing protocol for rapid optical clearing was used followed by vibratome sectioning to detect anatomical changes and localize gene expression with promoter fusions and reporter lines tagged with fluorescent proteins. This method will prove useful for studying cellular and physiological responses in other pathogen-triggered structures in plants, such as nematode-induced syncytia and root knots, as well as leaf galls and deformations caused by insects.
Introduction
Plants affected by pathogens or insects may develop abnormal structures (organ deformations or galls), which allow the invader to ingest nutrients and reproduce1. Here, an efficient histopathological approach was undertaken to study the changes taking place in galls that develop on the underground parts of plants infected with the protist Plasmodiophora brassicae (Figure 1). The minor thread associated with this pathogen emerges from the fact that P. brassicae resting spores can retain their ability to invade plants for many years. In the case of large-scale cultivation of oilseed rape (Brassica napus), this is a serious problem since economic factors restrict crop rotation, leading to resting spore accumulation in the soil2. Oilseed rape's resistance to clubroot disease caused by P. brassicae is genetically determined. Sadly, the pathogen often outsmarts resistance because of its biology and the narrow genetic pool from which oilseed rape originated. Therefore, it has become relevant to study the post-infection responses in host plants and their capability to slow disease progression or prevent certain symptoms from developing.
In clubroot disease, severity is generally evaluated based on the development of galls and the degree of root system damage. This is known as the Disease Index-DI3. However, it does not fully capture the true appraisal of this plant-pathogen interaction. In particular, it does not address how the pathogen is distributed within the roots and if the plant can restrain P. brassicae movement within its tissues. Further, it is not easy to anticipate to what extent P. brassicae reprograms underground organ anatomy. Studies on the model plant Arabidopsis thaliana have shown that P. brassicae infection leads to the inhibition of xylogenesis (both the initiation and maturation steps) and the enhancement of phloem differentiation within galls4,5. Moreover, in roots and hypocotyls of infected plants, cambial cell progeny does not quit the mitotic state and proliferates longer than in healthy plants6. This process governs the final size of the gall and determines the number of pathogen resting spores produced within the infected plant. P. brassicae-driven developmental, metabolic, and physiological reprogramming in the host is very complex7; therefore, the application of tools allowing the inspection of internal changes within galls is crucial for properly assessing this interaction. The life cycle progression of P. brassicae is accompanied by reprogramming of the host cell metabolism, which can be observed as starch or lipid deposition7,8. The main obstacle to the successful microscopy of galls comes from their low transparency. Due to this, the majority of histological specimens presenting clubroot-driven changes within galls originate from fixation-embedding (wax or resin) techniques followed by microtome sectioning. Such approaches were successfully used for locating the promoter activity for numerous genes active in clubroot galls4,5 or various staining techniques facilitating the observation of P. brassicae life-cycle progression9. However, it must be noted that the fixing and embedding stages are time-consuming and result in partial or complete washing out of important biomolecules (e.g., lipids), significantly hindering certain observations. Recently, P. brassicae life cycle progression in the host was visualized with the help of the Fluorescent In Situ Hybridization (FISH), in which a SABATH-type methyltransferase (PbBSMT) gene-specific probe was used to mark resting spore formation10. A good alternative is the use of other fluorescence-based methods where the autofluorescence of some cellular components, the activity of 5'- upstream regulatory regions of genes fused to fluorescent protein markers, and the accumulation of particular fluorescently tagged proteins can be seen. However, in addition to the low transparency of the samples, a major drawback associated with such objects is working with unfixed specimens, which significantly decreases the time in which good quality images can be documented. In 2015, Kurihara et al.11 developed a clearing reagent, which allows the preservation of fluorescent proteins and increases the transparency of plant tissue specimens. Additionally, it is compatible with numerous histological stains. Recently, the same technique was successfully applied to visualize different cell wall components in plant tissues12,13. Here, this protocol has been used to analyze various clubroot gall development aspects. The workflow begins with the fixation of galls, vibratome sectioning, tissue clearing, staining, and fluorescence imaging. Depending on one's needs, directly or after particular staining, the resulting sections can be subjected to inspection under an epifluorescence or confocal microscope. This method provides an effective solution to study local changes in gene expression and physiological responses, including phytohormone balance and signaling. Disease progression can be tracked by looking at the distribution pattern of resting spores and maturation dynamics. Furthermore, the protocol can be easily applied for imaging characteristic changes within P. brassicae infected plants, including the inhibition of xylogenesis or the host plant defense responses visible as local lignification in resistant genotypes. Examples in this protocol come from imaging conducted on the Arabidopsis thaliana model; however, the protocol can also be applied to other crop species belonging to the Brassicaceae family. The method described below will facilitate future detailed studies of cellular structures and molecular changes accompanying gall formation in P. brassicae-infected plants.
The general workflow of the protocol is quite straightforward, and all stages of gall development can be easily imaged and characterized (Figure 2). Since P. brassicae is a soilborne pathogen, all experiments must be carried out in soil-based systems. The pathogen prefers acidic conditions; therefore, non-lime-treated soil substrates must be used. Although P. brassicae does not pose a threat to humans, it is strictly a plant pathogen that can spread through soil and water. Therefore, all parts of the infected plant, as well as the soil, need to be destroyed after the experiment by autoclaving or by treatment with bleach.
Protocol
1. Plant growth conditions
- Grow Arabidopsis thaliana plants in a short-day light regime with 9 h of light at 22 °C and 15 h of dark at 20 °C and an irradiance of 120 µmol·m−2s−1 (measured as the photosynthetically active radiation, PAR at the canopy level, see Table of Materials).
2. Preparation of spore inoculum
NOTE: For details, see Fuchs et al.14.
- Homogenize two to three frozen galls from Chinese cabbage plants (Brassica rapa var. pekinensis cultivar "Granaat") in a blender containing 300 mL of autoclaved distilled water and filter through four layers of sterile gauze.
- Centrifuge the filtrate (at 6,000 x g, for 5 min, and at 4 °C) and mechanically remove the starch layer from the spore pellet using a spatula. Repeat this process until most of the starch is removed.
- Determine the spore concentration using a hemocytometer13 by counting the number of spores in 20 different field areas.
- Ensure that the final concentration of spore suspension used for inoculating is 1 x 106 spores·mL−1 for Arabidopsis thaliana. Inoculate each plant 20 days after germination with 2 mL of calibrated spore suspension.
3. Tissue preparation and fixation
- Prepare the plant tissue.
- Carefully remove soil from the root systems of clubroot-infected and non-infected plants. Clean thoroughly with water and collect hypocotyls and galls (length between 0.5-2 cm) into microcentrifuge tubes.
- Perform PFA fixation (4% paraformaldehyde, PFA in 1x PBS + 0.01% Triton X-100) following the steps below.
- Add 4 g of PFA powder (see Table of Materials) to 100 mL of PBS (phosphate-buffered saline, pH 7.4, Table 1) by stirring at 65 °C (do not boil). Use KOH (10 M and 1M) dropwise to obtain a clear solution with a pH of around 11.
- Adjust the pH with H2SO4 to bring it down to 6.9 and add 0.01% (v/v) Triton X-100 to improve fixation. Aliquot the solution to store at −20 °C (do not refreeze) or store at 4 °C.
- Perform tissue fixation.
- Fix the samples in 200-500 µL of PFA fixative for 1 h at room temperature by applying a constant vacuum (700 mbar) using a vacuum pump. Ensure that the object is completely immersed in the PFA solution.
NOTE: Samples can be stored for a few weeks at 4 °C (fridge) and used later for sectioning.
- Fix the samples in 200-500 µL of PFA fixative for 1 h at room temperature by applying a constant vacuum (700 mbar) using a vacuum pump. Ensure that the object is completely immersed in the PFA solution.
4. Tissue embedding and sectioning
- Use 4% w/v agarose for agarose embedding of non-infected hypocotyls and infected galls. Boil the solution to dissolve agarose and pour it when it cools down while it is still viscous.
- Dry the object slightly on a tissue for a few seconds to remove excess PFA.
- Use toothpicks/forceps to carefully embed and orient the plant object in agarose. To prevent oblique sections, ensure that the object is oriented and molded correctly so that its axis is perpendicular to the plane of the vibratome blade (for radial sections).
- To accelerate agarose solidifying, place the Petri or multi-culture plate at 4 °C for 10 min.
NOTE: If the object is too large (as in the case of larger galls of Arabidopsis at 21 days post-inoculation [DPI]), the object can be sectioned even without embedding in agarose.
5. Vibratome sectioning
NOTE: The vibratome is equipped with a vibrating razor blade to cut through plant organs/tissue. The vibration speed, amplitude, angle of the blade, and section thickness are all parameters that can be adjusted (see Table of Materials).
- Using a blade, carefully cut and mold the object within an agarose block, noting the preferred orientation for sectioning.
- Stick the agarose block/large gall to properly mount it on the specimen holder using cyanoacrylate/instant glue and masking tape (see Table of Materials).
- For Arabidopsis hypocotyls and galls (early stages), keep the thickness of sections between 30-40 µm for obtaining good quality images.
NOTE: For analyzing the later stages of gall progression, the thickness of sections can be between 50-80 µm. - Refrain from cutting thinner sections as it can destroy the gall sample due to the vibrations of the moving blade.
- Adjust the thickness for sections, speed, and vibration amplitude according to the gall's thickness and size (40 µm or 60 µm).
- Add distilled water to the water bath and begin with sectioning.
- Carefully collect sections using forceps or brush and transfer them into a microcentrifuge tube containing 1mL of 1x PBS buffer (pH 7.4).
NOTE: Typically, the higher the vibration amplitude, the better the sectioning quality. For non-infected hypocotyls or galls that do not contain mature resting spores, the vibration amplitude can be kept at 1.2 mm with 0.60 mm·s−1 speed. In case of large galls with partial loss of cellular integrity and visible signs of spore maturation, the vibration amplitude should be decreased to 0.55 mm with 0.45 mm·s−1 speed.
6. Specimen clearing
- Remove 1x PBS from the microcentrifuge tube and add 200-500 µL of clearing solution (Table 1).
- From now on, store the samples at room temperature in the dark. Ensure that the objects are always submerged in the clearing solution at all times.
- Replace the clearing solution with the new one if its color changes after some time during sample processing.
NOTE: The color of the clearing solution can turn yellowish due to sample processing. In such a case, replacing the solution is recommended to improve the clearing of tissues.
7. Staining procedure
- For Calcofluor white staining for the cell walls (for outlaying plant cells), prepare 5% v/v of Calcofluor white stain (see Table of Materials) in the clearing solution. Stain for at least 5 min in the dark.
- For staining lipids with Nile Red (for staining resting spores and oil droplets in infected cells), prepare 1 mg/mL of the stock (see Table of Materials) in clearing solution. Dilute it further to obtain 1:99. Stain for at least 10 min in the dark.
- For Basic Fuchsin lignin staining, prepare 0.2% w/v of the stain (see Table of Materials) in clearing solution. Stain for at least 10 min in the dark.
NOTE: During double staining, samples are first stained with Nile Red for 10 min before Calcofluor white application. The excess staining solutions were removed after each step. If the object seems overstained, remove the excess stain by washing with the clearing solution. Dilute stains using the clearing solution, if required. Always stain the sample with Nile Red/Basic Fuchsin before Calcofluor staining. Staining first with Calcofluor can prevent proper staining of the spores by Nile Red.
8. Microscopy
- Mount cleared sections on the microscopy slide and observe under an epifluorescence or a confocal microscope (see Table of Materials). Use clearing solution as the mounting medium to prevent drying of the sample.
- Use multiple acquisition modes for imaging more than one fluorescence spectrum simultaneously.
NOTE: The excitation/emission spectra used in the presented protocol are as follows: for Nile Red 553/636 nm, for xylem autofluorescence 380/475 nm, for Basic Fuchsin 561/650 nm, for Calcofluor white 405/475 nm, for erRFP 585/608 nm, and for GFP 488/509 nm (Table 2).
Representative Results
With Nile Red that stains lipids and suberin, it is possible to view the pathogen resting spores containing lipids (Figure 3A,B). Hence, using double staining, crisp images can be obtained to look at the pattern of pathogen distribution within the galls. Counterstaining with Calcofluor white creates contrast and helps to simultaneously track xylem development with P. brassicae maturation (Figure 3B).
Xylem formation and development could also be checked by observing autofluorescence in unstained samples (Figure 4A) or by using stains such as Basic Fuchsin that enable fluorescence-based imaging of lignin (Figure 4B).
By using this method, one could track gene expression changes or responses to growth regulators. A perfect example is where Arabidopsis plants harboring pHCA2:erRFP construct were utilized to visualize HIGH CAMBIAL ACTIVITY 2 (HCA2) gene expression in phloem tissue within clubroot galls. HCA2 gene activity has been previously found in meristematicaly active cambium and phloem-lineage cells15. Here, it co-localizes with the phloem in the late stages of P. brassicae-driven gall development, and its activity reflects how P. brassicae increases phloem complexity (Figure 5). The resulting image shows phloem proliferation at the late stage of gall development when the cambium gets fragmented. Figure 5A shows an uncleared hand section of the gall, whereas Figure 5B shows more crisp and localized fluorescent signals obtained by vibratome sectioning followed by tissue clearing. The objects were counterstained with Calcofluor white. Figure 6 compares this image (Figure 6B) with a similar region depicted in resin-embedded and microtome-sectioned galls (Figure 6A) at 21 DPI. Differential cytokinin responses between infected and non-infected plants were assessed by checking the expression of the TCS:GFP (Two Component Signalling) marker16 in developing galls (Figure 7). While imaging weak GFP signals in galls and tissues with secondary thickenings, it is important to note that additional background signal due to autofluorescence of mature xylem cells is also captured during imaging.
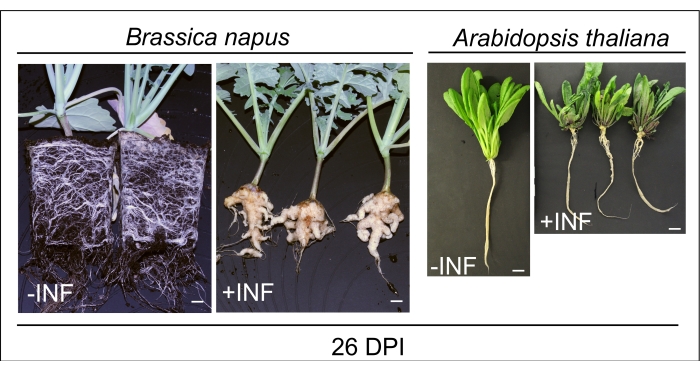
Figure 1: Clubroot disease symptoms on oilseed rape (B. napus) and Arabidopsis thaliana (Columbia-0) at 26 DPI with Plasmodiophora brassicae spores. During the course of disease, large galls develop on the entire root system, making it extremely brittle. It concludes by releasing spores into the surrounding soil to promote future infections. The upper parts of the plant body also show signs of poor growth and development. Finally, the infected plants succumb to the devastating effects on growth metabolism and development once the root system gets completely damaged and the plant can no longer cope with the disease. The scale bar represents 1 cm. The -INF stands for mock-inoculated, whereas +INF stands for P. brassicae-inoculated plants. On this occasion, a picture of oilseed rape plants is provided before the soil removal to present healthy root systems. After washing, only the hypocotyl and upper part of the root are collected. Please click here to view a larger version of this figure.
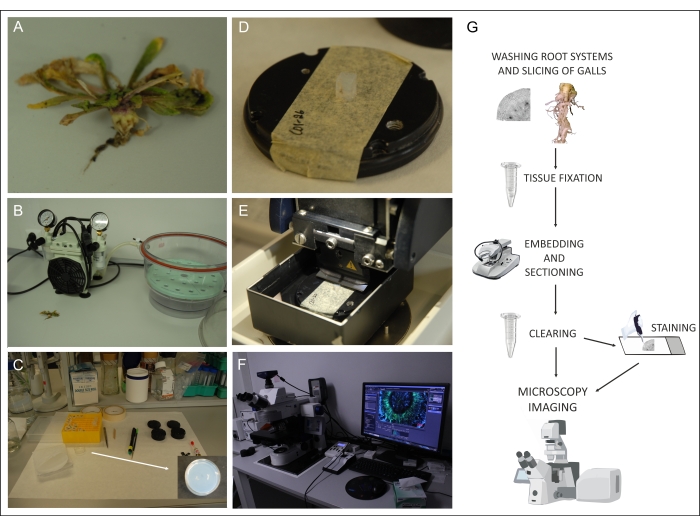
Figure 2: The general workflow. Washed Arabidopsis root systems are (A) dissected, (B) fixed, (C) agarose embedded, (D) mounted, and (E) sectioned on the vibratome. The resulting objects are subjected to tissue clearing (3 days to several weeks at RT in the dark, depending on the tissue type and thickness). (F) Cleared objects can then be stained and inspected under the microscope. (G) Summary of the workflow. Please click here to view a larger version of this figure.
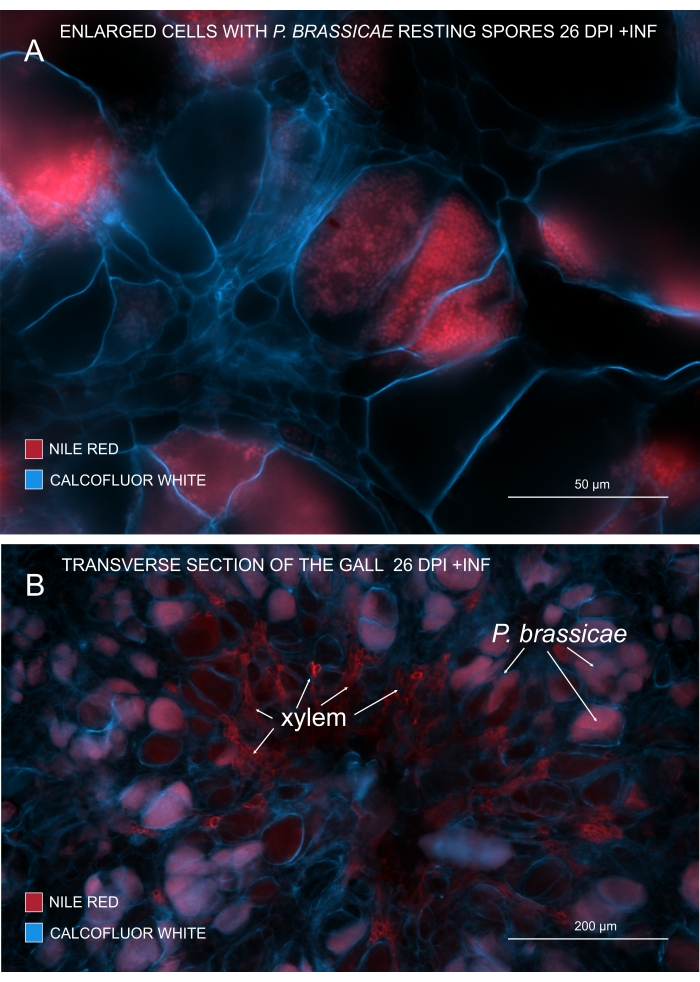
Figure 3: Marking pathogen spores with Nile Red stain. (A,B) Nile Red stains lipids in resting spores, which works perfectly to track P. brassicae maturation. (A) Enlarged cells colonized by P. brassicae and filled with pathogen spores. (B) Mature xylem cells also get stained by Nile Red. The section has been counterstained with Calcofluor white to see the outlay of host cells (A: Objective lens = 20x and section thickness = 60 µm; B: Objective lens = 5x and section thickness = 60 µm). Please click here to view a larger version of this figure.
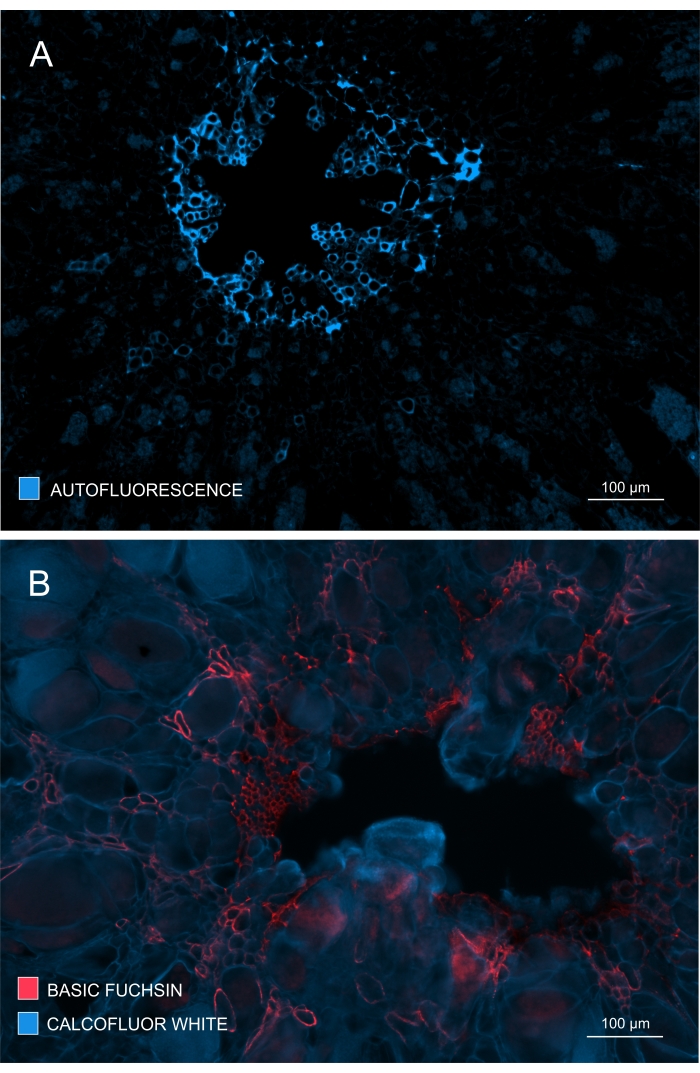
Figure 4: Tracking the extent of xylem development and maturation. (A) Lignin gives strong autofluorescence upon excitation with UV; therefore, mature xylem can be discriminated relatively easily. (B) Double staining with Basic Fuchsin and Calcofluor gives better results since all cells get stained by the latter dye, while mature xylem is distinctly stained with Basic Fuchsin. In this way, double staining provides images with improved contrast depicting noticeable inhibition of xylogenesis (A: Objective lens = 10x and section thickness = 60 µm; B: Objective lens = 10x and section thickness = 60 µm). Please click here to view a larger version of this figure.
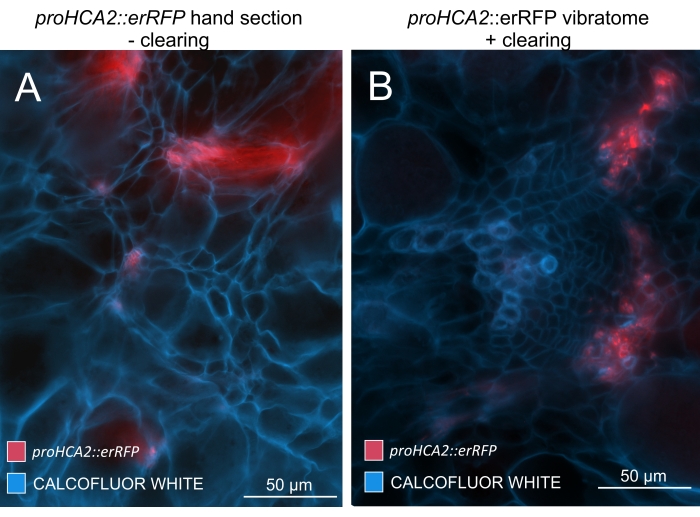
Figure 5: Phloem-specific signal for the HCA2 gene in hypocotyls of P. brassicae-infected plants at 21 DPI. Promoter activity for proHCA2::erRFP harboring transgenic Arabidopsis thaliana can be seen in (A) and (B). Differences can be observed between a non-cleared hand section in panel (A) where the erRFP signal appears diffused due to superimposition and overlapping cell layers, especially in uneven hand sections. On the other hand, (B) shows a vibratome section post-tissue-clearing where the erRFP signal precisely marks the phloem cells in a mature vascular bundle (Objective lens = 20x and section thickness = 60 µm). Please click here to view a larger version of this figure.
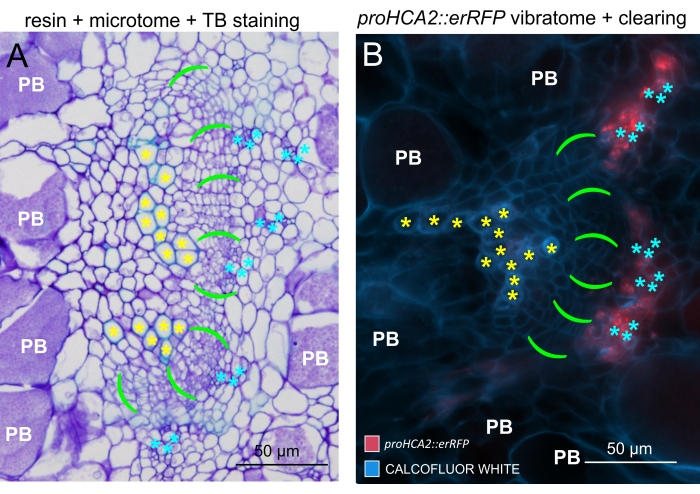
Figure 6: Comparison between TB, resin-embedded, and microtome-sectioned galls with the help of fluorescence. (A) A comparison between images of Toluidine Blue (TB), resin-embedded, and microtome-sectioned galls, and (B) a representative object (also presented in Figure 5B) acquired with the help of fluorescence. Xylem cells are labeled with yellow asterisks, the cambial area with vivid green brackets, phloem with cyan asterisks, and Plasmodiophora brassicae-colonized cells with a white PB symbol. Resin-embedded sections (A) provide a good resolution for studying the distribution of resting spores in hypertrophied organs, degree of disease progression, and other processes such as local lignification in resistant plants. However, the protocol described here (B) enables the sensitive observation of gene expression or protein accumulation and the visualization of other physiological changes and important molecules such as lipids (in spores). Please click here to view a larger version of this figure.
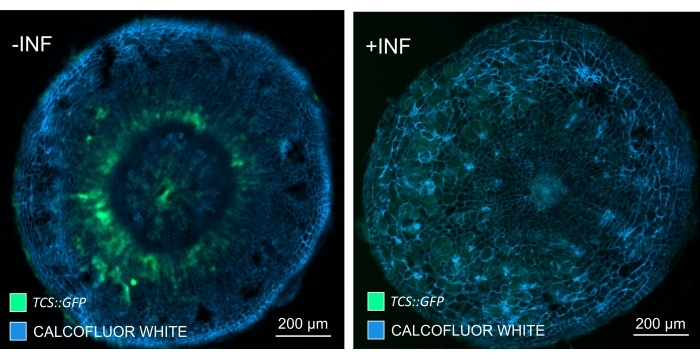
Figure 7: Tracking cytokinin signaling responses in hypocotyls of non-infected (-INF) and P. brassicae-infected (+INF) Arabidopsis plants at 16 DPI. TCS::GFP marker was used for characterizing in planta cytokinin responses. Vibratome sections were subjected to clearing treatment followed by staining with Calcofluor white. Based on the image, at 16 DPI, cytokinin responses appear to be largely diminished in infected galls (right panel), while they remain strong, especially in the phloem pool (cells that will eventually differentiate to form phloem tissue), in non-infected plants (left panel) (Objective lens = 5x and thickness = 30 µm). Certain levels of xylem autofluorescence can also be visible. Please click here to view a larger version of this figure.
| Clearing Solution (toxic) | ||
| Components | Percentage (%) | in 100 mL distilled water |
| Xylitol | 10% | 10g |
| Sodium Deoxycholate | 15% | 15g |
| Urea | 25% | 25g |
| 10x PBS (Phosphate buffered saline) | 1x PBS (100 mL) | |
| NaCl | 8 g | 10 mL 10x PBS + 90 mL distilled water |
| KCl | 0.2 g | |
| KH2PO4 | 0.24 g | |
| Na2HPO4 · 2H2O | 1.81 g | |
| distilled water | 100 mL | |
| pH | pH adjusted to 7.4 using HCl | |
| Autoclave and store at 4 °C. | ||
Table 1: Composition of the Clearing solution and Phosphate-buffered saline (PBS).
| Fluorescent stain/ Tag | Excitation/Emission Wavelengths | Microscope filter set used |
| Nile Red | 553/636 nm | filter set 43 |
| Xylem Autofluorescence | 380/475 nm | filter set 49 |
| Calcofluor White | 405/475 nm | filter set 49 |
| Basic Fuchsin | 561/650 nm | filter set 43 |
| erRFP | 585/608 nm | filter set 43 |
| GFP | 488/509 nm | filter set 38 |
Table 2: Excitation/emission spectra selected for the present study.
Discussion
Applying the clearing solution on vibratome-cut sections of galls surely enhances the ability to study the biotrophic interaction between P. brassicae and the host plant. Although the clearing protocol applies even to hand sections, it works better with vibratome sections. Fixing samples in PFA fixative acts as a critical step in the protocol as samples can then be stored at 4 °C for a few days before proceeding with sectioning. This provides flexibility to store samples for a limited period without compromising the expression and preservation of fluorescent proteins during fixation.
Nile Red (in DMSO or methanol) is incompatible with resin sections due to its hydrophobicity, which dissolves resin and destroys resin-embedded sections17. Thus, vibratome sections prove instrumental for studying the pathogen distribution and its life-cycle within developing galls, wherein Nile Red staining can be easily used.
The clearing solution used in this protocol is highly versatile12, allowing a variety of fluorescence stains to be used in different combinations to stain various biomolecules/components of the cell walls (suberin, lignin, cellulose, and chitin in fungal interactions). It is also possible to counterstain sections of fluorescent GFP marker lines and thereby correlate the promoter activity or protein accumulation pattern with the presence of the pathogen in particular cells or regions of the gall. However, background autofluorescence from xylem and pathogen-filled giant cells could not be eliminated even after the clearing protocol. This presents a limitation for looking at fluorescent markers during the later stages of gall formation, especially when using an epifluorescence microscope and imaging weak signals.
Due to the low levels of expression/accumulation of fluorescent signals, transcription factors are difficult to detect, but, with this technique, it is possible to obtain satisfactory images for them. Overall, combining vibratome sectioning with the tissue-clearing approach expands the toolkit for histological observations of complex gall tissues. The flexibility of this protocol eases the process of tissue fixation and reduces the time required for cutting and imaging fresh tissue samples to observe fluorescent proteins and promoter activities. With further improvements and by utilizing other fluorescent dyes specific for various biomolecules, this method will mark greater advances in histological studies and image analysis of dense, opaque tissues with complex tissue organization. In recent times, the presented tissue clearing method has emerged as a popular and widely used protocol to combine and enable the simultaneous acquisition of different fluorescent signals11,12,13. Future development and modifications in such techniques will greatly improve the image resolution for observing plant-pathogen interactions at the cellular level.
Declarações
The authors have nothing to disclose.
Acknowledgements
The work was supported by the National Science Centre Poland OPUS17 grant No. 2019/33/B/NZ9/00751 "Long distance Vascular Coordination in Plants Infected by Plasmodiophora brassicae". We thank Prof. Yrjö Helariutta (Sainsbury Laboratory, University of Cambridge) for sharing the proHCA2::erRFP line.
Materials
| 2N Sulfuric acid (H2SO4) | Roth | UN2796 | pH adjustment |
| Agarose | PRONA | BGQT100 | Embedding |
| Basic Fuchsin | BIOSHOP | BSF410.5 | Fluorescent dye |
| Calcofluor White | Sigma Aldrich | 18909-100ML-F | Fluorescent dye |
| Commercial Bleach | Domestos | ||
| Cyanoacrylate/ Instant glue | Kropelka | Adhesive | |
| Dimethyl Sulfoxide (DMSO) | BIOSHOP | DMS555.500 | Solvent |
| Epifluorescence microscope | Carl Zeiss M2 automated epifluorescence microscope with Colibri LED system | Carl Zeiss M2 | Carl Zeiss Filter Set filter set 38, 43, 49 used |
| Fully automated Vibratome | Leica | VT1200 S | |
| Lightmeter /Photometer | LI-COR Biosciences | LI-250A + LI-190R quantum sensor | For measuring light intensity within the 400-700nm (PAR) waveband |
| Masking tape | For sticking agarose block on mould | ||
| Murashige & Skoog Medium (MS Medium) | Duchefa Biochemie | MO222.0050 | Plant Growth Medium |
| Nile Red | Sigma Aldrich | N3013-100MG | Fluorescent dye |
| Paraformaldehyde PFA | Sigma Aldrich | 158127-100G | Fixative |
| Potassium Chloride (KCl) | POCH | 739740114 | PBS component |
| Potassium Hydroxide (KOH) | Sigma Aldrich | P1767-250G | pH adjustment |
| Potassium Phosphate Monobasic (KH2PO4) | BIOSHOP | PPM302.500 | PBS component |
| Sodium chloride (NaCl) | BIOSHOP | SOD001.1 | PBS component |
| Sodium Deoxycholate | Sigma Aldrich | D6750-25G | Clearing Solution |
| Sodium Phosphate Dibasic (Na2HPO4 · 2H2O) | POCH | 799490116 | PBS component |
| Triton X-100 | BIOSHOP | TRX506.100 | Fixative |
| Urea | Sigma Aldrich | U5378-100G | Clearing Solution |
| Vacuum/Pressure pump and Dessicator | Welch by Gardner Denver | 2522C-02 | For Vacuum Infilteration |
| Xylitol | Sigma Aldrich | X3375-25G | Clearing Solution (componenet) |
Referências
- Harris, M. O., Pitzschke, A. Plants make galls to accommodate foreigners: Some are friends, most are foes. New Phytologist. 225 (5), 1852-1872 (2020).
- Peng, G., et al. Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Canadian Journal of Plant Pathology. 36 (1), 99-112 (2014).
- Siemens, J., Nagel, M., Ludwig-Müller, J., Sacristán, M. D. The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: Parameters for disease quantification and screening of mutant lines. Journal of Phytopathology. 150 (11-12), 592-605 (2002).
- Malinowski, R., Smith, J. A., Fleming, A. J., Scholes, J. D., Rolfe, S. A. Gall formation in clubroot-infected Arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. The Plant Journal. 71 (2), 226-238 (2012).
- Walerowski, P., et al. Clubroot disease stimulates early steps of phloem differentiation and recruits SWEET sucrose transporters within developing galls. The Plant Cell. 30 (12), 3058-3073 (2018).
- Olszak, M., et al. Transcriptional profiling identifies critical steps of cell cycle reprogramming necessary for Plasmodiophora brassicae-driven gall formation in Arabidopsis. Plant Journal. 97 (4), 715-729 (2019).
- Malinowski, R., Truman, W., Blicharz, S. Genius architect or clever thief-How Plasmodiophora brassicae reprograms host development to establish a pathogen-oriented physiological sink. Molecular Plant-Microbe Interactions. 32 (10), 1259-1266 (2019).
- Bi, K., et al. Integrated omics study of lipid droplets from Plasmodiophora brassicae. Scientific Reports. 6, 36965 (2016).
- Schuller, A., Ludwig-Müller, J. Histological methods to detect the clubroot pathogen Plasmodiophora brassicae during its complex life cycle. Plant Pathology. 65 (8), 1223-1237 (2016).
- Badstöber, J., Gachon, C. M. M., Ludwig-Müller, J., Sandbichler, A. M., Neuhauser, S. Demystifying biotrophs: FISHing for mRNAs to decipher plant and algal pathogen-host interaction at the single cell level. Scientific Reports. 10, 14269 (2020).
- Kurihara, D., Mizuta, Y., Sato, Y., Higashiyama, T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 142 (23), 4168-4179 (2015).
- Ursache, R., Andersen, T. G., Marhavý, P., Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. The Plant Journal. 93 (2), 399-412 (2018).
- Sexauer, M., Shen, D., Schön, M., Andersen, T. G., Markmann, K. Visualizing polymeric components that define distinct root barriers across plant lineages. Development. 148 (23), (2021).
- Fuchs, H., Sacristan, M. Identification of a gene in Arabidopsis thaliana controlling resistance to clubroot (Plasmodiophora brassicae) and characterization of the resistance response. Molecular Plant-Microbe Interactions. 9 (2), 91-97 (1996).
- Guo, Y., Qin, G., Gu, H., Qu, L. -JDof56HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. The Plant Cell. 21 (11), 3518-3534 (2009).
- Liu, J., Müller, B., Kleine-Vehn, J., Sauer, M. Imaging TCSn::GFP, a Synthetic Cytokinin Reporter, in Arabidopsis thaliana. Plant Hormones: Methods and Protocols. , 81-90 (2017).
- Suzuki, M., Shinohara, Y., Fujimoto, T., Taatjes, D. J., Roth, J. Histochemical Detection of Lipid Droplets in Cultured Cells. Cell Imaging Techniques: Methods and Protocols. , 483-491 (2013).

