Application of Deep Learning-Based Medical Image Segmentation via Orbital Computed Tomography
Summary
An object segmentation protocol for orbital computed tomography (CT) images is introduced. The methods of labeling the ground truth of orbital structures by using super-resolution, extracting the volume of interest from CT images, and modeling multi-label segmentation using 2D sequential U-Net for orbital CT images are explained for supervised learning.
Abstract
Recently, deep learning-based segmentation models have been widely applied in the ophthalmic field. This study presents the complete process of constructing an orbital computed tomography (CT) segmentation model based on U-Net. For supervised learning, a labor-intensive and time-consuming process is required. The method of labeling with super-resolution to efficiently mask the ground truth on orbital CT images is introduced. Also, the volume of interest is cropped as part of the pre-processing of the dataset. Then, after extracting the volumes of interest of the orbital structures, the model for segmenting the key structures of the orbital CT is constructed using U-Net, with sequential 2D slices that are used as inputs and two bi-directional convolutional long-term short memories for conserving the inter-slice correlations. This study primarily focuses on the segmentation of the eyeball, optic nerve, and extraocular muscles. The evaluation of the segmentation reveals the potential application of segmentation to orbital CT images using deep learning methods.
Introduction
The orbit is a small and complicated space of approximately 30.1 cm3 that contains important structures such as the eyeball, nerves, extraocular muscles, supportive tissues, and vessels for vision and eyeball movements1. Orbital tumors are abnormal tissue growths in the orbit, and some of them threaten patients' vision or eyeball movement, which may lead to fatal dysfunction. To conserve patients' visual function, clinicians must decide on the treatment modalities based on the tumor characteristics, and a surgical biopsy is generally inevitable. This compact and crowded area often makes it challenging for clinicians to perform a biopsy without damaging the normal structure. The deep learning-based image analysis of pathology for determining the condition of the orbit could help in avoiding unnecessary or avoidable injury to the orbital tissues during biopsy2. One method of image analysis for orbital tumors is tumor detection and segmentation. However, the collection of large amounts of data for CT images containing orbital tumors is limited due to their low incidence3. The other efficient method for computational tumor diagnosis4 involves comparing the tumor to the normal structures of the orbit. The number of orbital CT images in normal structures is relatively larger than that in tumors. Therefore, the segmentation of normal orbital structures is the first step to achieve this goal.
This study presents the entire process of deep learning-based orbital structure segmentation, including the data collection, pre-processing, and subsequent modeling. The study is intended to be a resource for clinicians interested in using the current method to efficiently generate a masked dataset and for ophthalmologists requiring information about pre-processing and modeling for orbital CT images.This article presents a new method for orbital structure segmentation and sequential U-Net, a sequential 2D segmentation model based on a representative deep-learning solution in U-Net for medical image segmentation. The protocol describes the detailed procedure of orbit segmentation, including (1) how to use a masking tool for the ground truth of orbit structure segmentation, (2) the steps required for the pre-processing of the orbital images, and (3) how to train the segmentation model and evaluate the segmentation performance.
For supervised learning, four experienced ophthalmologists who had been board certified for over 5 years manually annotated the masks of the eyeball, optic nerve, and extraocular muscles. All the ophthalmologists used the masking software program (MediLabel, see the Table of Materials), which uses super-resolution for efficient masking on CT scans. The masking software has the following semi-automatic features: (1) SmartPencil, which generates super pixel map clusters with similar values of image intensity5; (2) SmartFill, which generates segmentation masks by computing the energy function of the ongoing foreground and background6,7; and (3) AutoCorrection, which makes the borders of the segmentation masks clean and consistent with the original image. Example images of the semi-automatic features are shown in Figure 1. The detailed steps of manual masking are provided in the protocol section (step 1).
The next step is the pre-processing of the orbital CT scans. To obtain the orbital volumes of interest (VOIs), the areas of the orbit where the eyeball, muscle, and nerve are located in normal conditions are identified, and these areas are cropped. The dataset has a high resolution, with <1 mm in-plane voxel resolution and slice thickness, so the interpolation process is skipped. Instead, window clipping is conducted at the 48 HU clipping level and the 400 HU window. After the cropping and window clipping, three serial slices of the orbit VOIs are generated for the segmentation model input8. The protocol section (step 2) provides details on the pre-processing steps.
U-Net9 is a widely used segmentation model for medical images. The U-Net architecture comprises an encoder, which extracts the features of the medical images, and a decoder, which presents the discriminative features semantically. When employing U-Net for CT scans, the convolutional layers consist of 3D filters10,11. This is a challenge because the computation of 3D filters requires a large memory capacity. To reduce the memory requirements for 3D U-Net, SEQ-UNET8, wherein a set of sequential 2D slices are used in the U-Net, was proposed. To prevent the loss of spatiotemporal correlations between the 2D image slices of the 3D CT scan, two bi-directional convolutional long-term short memories (C-LSTMs)12 are employed in basic U-Net. The first bi-directional C-LSTM extracts the inter-slice correlations at the end of the encoder. The second bi-directional C-LSTM, after the output of the decoder, transforms the semantic segmentation information in the dimensions of the slice sequence into a single image segmentation. The architecture of SEQ-UNET is shown in Figure 2. The implementation codes are available at github.com/SleepyChild1005/OrbitSeg, and the usage of the codes is detailed in the protocol section (step 3).
Protocol
The present work was performed with the approval of the Institutional Review Board (IRB) of the Catholic Medical Center, and the privacy, confidentiality, and security of the health information were protected. The orbital CT data were collected (from de-identified human subjects) from hospitals affiliated with the College of Medicine, the Catholic University of Korea (CMC; Seoul St. Mary's Hospital, Yeouido St. Mary's Hospital, Daejeon St. Mary's Hospital, and St. Vincent Hospital). The orbital CT scans were obtained from January 2016 to December 2020. The dataset contained 46 orbital CT scans from Korean men and women ranging in age from 20 years to 60 years. The runtime environment (RTE) is summarized in Supplementary Table 1.
1. Masking the eyeball, optic nerve, and extraocular muscles on the orbital CT scans
- Run the masking software program.
NOTE: The masking software program (MediLabel, see the Table of Materials) is a medical image labeling software program for segmentation, which requires few clicks and has high speed. - Load the orbital CT by clicking on the open file icon and selecting the target CT file. Then, the CT scans are shown on the screen.
- Mask the eyeball, optic nerve, and extraocular muscles using super pixels.
- Run the SmartPencil by clicking on the SmartPencil wizard in MediLabel (Video 1).
- Control the resolution of the super pixel map if necessary (e.g., 100, 500, 1,000, and 2,000 super pixels).
- Click on the cluster of super pixels of the eyeball, optic nerve, and extraocular muscles on the super pixel map, where pixels of similar image intensity values are clustered.
- Refine the masks with the autocorrection functions in MediLabel.
- Click on the SmartFill wizard after masking some of the super pixels on the slices (Video 2).
- Click on the AutoCorrection icon, and ensure that the corrected mask labels are computed (Video 3).
- Repeat step 1.3 and step 1.4 until the refinement of the masking is complete.
- Save the masked images.
2. Pre-processing: Window clipping and cropping the VOIs
- Extract the VOIs with preprocessing_multilabel.py (the file is downloadable from GitHub).
- Run preprocessing_multilabel.py.
- Check the scans and masks, which are cropped and saved in the VOIs folder.
- Transform the VOIs to the set of three sequential CT slices for the input to SEQ-UNET with builder_multilabel.py (the file is downloadable from GitHub).
- Run sequence_builder_multilabel.py.
- Ensure that the slices and masks are resized to 64 pixels by 64 pixels during the transformation.
- During the transformation, perform clipping with the 48 HU clipping level and the 400 HU window.
- Check the saved transformed CT scans (nii file) and masks (nii file) in the scan folder and the mask folder under pre-processed folders, respectively.
3. Four cross-validations of the orbital segmentation model
- Build the model following the steps below.
- Run main.py.
- When running main.py, give the fold number of the four cross-validations by "-fold num x", where x is 0, 1, 2, or 3.
- When running main.py, use the epoch, which is the number of training iterations, as an option, such as "-epoch x", where x is the epoch number. The default number is 500.
- When running main.py, set the batch size, which is the number of training samples in a single training session. The default number is 32.
- In main.py, load the CT scans and masks, and initialize the SEQ-UNET with the pre-trained parameters using the LIDC-IDRI dataset (downloadable from the cancer imaging archive).
- In main.py, perform the testing of the model after training. Calculate the evaluation metrics, dice score, and volume similarity, and save them in the metrics folder.
- Check the results in the segmented folder.
Representative Results
For the quantitative evaluation, two evaluation metrics were adopted, which were used in the CT image segmentation task. These were two similarity metrics, including dice score (DICE) and volume similarity (VS)13:
DICE (%) = 2 × TP/(2 × TP + FP + FN)
VS (%) = 1 − |FN − FP|/(2 × TP + FP + FN)
where TP, FP, and FN denote the true positive, false positive, and false negative values, respectively, when the segmentation result and the segmentation mask are given.
The performance of SEQ-UNET for orbital structure segmentation was evaluated by four cross-validations. The results are shown in Table 1. The eyeball segmentation using SEQ-UNET achieved a dice score of 0.86 and a VS of 0.83. The segmentation of the extraocular muscles and optic nerve achieved low dice scores (0.54 and 0.34, respectively). The dice score of the eyeball segmentation was over 80% because it had a large portion of the VOIs and little heterogeneity between CT scans. The dice scores of the extraocular muscles and optic nerve were relatively low because they infrequently appeared in the CT volume and were found in a relatively small number of the CT slices. However, the visual similarity scores of the extraocular muscles and optic nerve (0.65 and 0.80, respectively) were higher than their dice scores. This result indicates that the specificity of segmentation was low. Overall, the dice score and visual similarity of SEQ-UNET for the segmentation of all the orbital substructures were 0.79 and 0.82, respectively. Examples of the visual results of orbital structure segmentation are shown in Figure 3. In Figure 3A–C, blue is the predicted segmentation result, and red is the ground truth mask. In Figure 3D, red, green, and orange are the eyeball, optic muscle, and nerve segmentation, respectively.
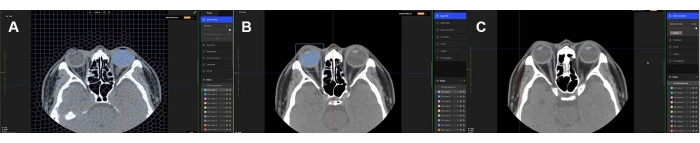
Figure 1: Semi-automatic masking features. Masking the eyeball, extraocular muscles, and optic nerve on orbital CT scans using (A) SmartPencil, (B) SmartFill, and (C) AutoCorrection. The mask of the eyeball is labeled by SmartPencil, which computes the super pixels of the slices, and the mask is made by clicking on the super pixels. After clicking some of the eyeball super pixels, the entire eyeball mask can be computed by SmartFill. In the case of masking the optic nerve, the masking refinement is made by AutoCorrection. Blue color labeled eyeballs are shown in (A) and (B). Please click here to view a larger version of this figure.
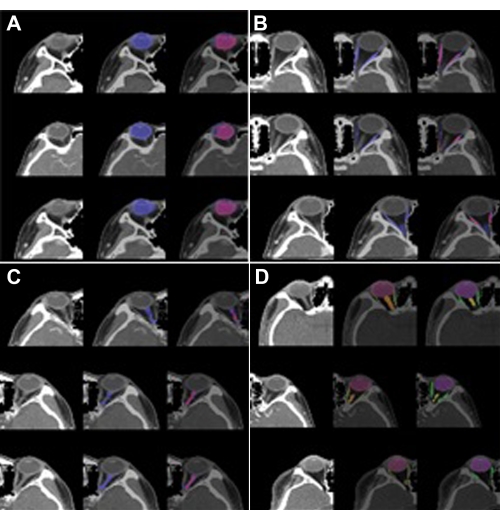
Figure 2: SEQ U-Net architecture. Sequential 2D slices as input and output; two bi-directional C-LSTMs are applied to the end of the encoding and decoding blocks based on the U-Net architecture. Please click here to view a larger version of this figure.
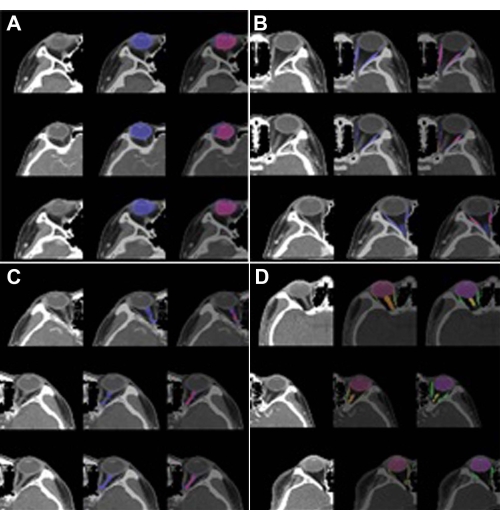
Figure 3: Segmentation results of the orbital structures. (A) Eyeball (label 1), (B) optic muscle (label 2), (C) optic nerve (label 3), and (D) multi-label (labels 1, 2, and 3). The left image is the VOI of the orbit, the center image is the predicted segmentation, and the right image is the ground truth. In (A), (B), and (C), blue is the predicted segmentation result, and red is the ground truth mask. In (D), red, green, and orange are the eyeball, extraocular muscle, and optic nerve segmentation, respectively. The predicted segmentation showed high performance (DICE: 0.86 vs. 0.82) in the case of the eyeball but low performance in the case of theextraocular muscle (DICE: 0.54 vs. 0.65) and optic nerve(DICE: 0.34 vs. 0.8). Please click here to view a larger version of this figure.
| Multi-Label | Label 1 (Eyeball) | Label 2 (Extraocular muscle) | Label 3 (Optic nerve) | |||||
| DICE | VS | DICE | VS | DICE | VS | DICE | VS | |
| SEQ-UNET | 0.79 | 0.82 | 0.86 | 0.83 | 0.54 | 0.65 | 0.34 | 0.8 |
Table 1: Segmentation results for the dice score and visual similarity. The eyeball, which has a relatively large number of slices, was segmented well with a DICE of 0.8, but the extraocular muscle and optic nerve, which have small numbers of slices and line shape, were partially segmented with DICE values of 0.54 and 0.34, respectively.
Video 1: SmartPencil wizard in the masking software program. A demonstration of annotating multiple pixels for eyeball masking. The masking tasks are enabled with one click on clustered super pixels. Please click here to download this Video.
Video 2: SmartFill wizard in the masking software program. A demonstration of annotating multiple pixels for eyeball masking. After selecting some pixels in the annotating area, this function generates full segmentation masks with similar intensities to the selected pixels. Please click here to download this Video.
Video 3: AutoCorrection in the masking software program. A demonstration of the automatic correction of a masked pixel using a pre-trained convolutional neural network algorithm. Please click here to download this Video.
Supplementary Table 1: Runtime environment (RTE) of masking, pre-processing, and segmentation modeling. Please click here to download this Table.
Discussion
Deep learning-based medical image analysis is widely used for disease detection. In the ophthalmology domain, detection and segmentation models are used in diabetic retinopathy, glaucoma, age-related macular degeneration, and retinopathy of prematurity. However, other rare diseases apart from those in ophthalmology have not been studied due to the limited access to large open public datasets for deep learning analysis. When applying this method in situations when no public dataset is available, the masking step, which is a labor-intensive and time-consuming task, is unavoidable. However, the proposed masking step (protocol section, step 1) helps to generate masking with high accuracy within a short time. Using super pixels and neural network-based filling, which cluster pixels that are similar in low-level image properties, clinicians can label the masks by clicking the groups of pixels instead of pointing out the specific pixels. Also, the automatic correction functions help refine the mask processes. This method’s efficiency and effectiveness will help generate more masked images in medical research.
Among the many possibilities in pre-processing, extracting VOIs and window clipping are effective methods. Here, extracting VOIs and window clipping are introduced in step 2 of the protocol. When the clinicians prepare the dataset, extracting the VOI from the given dataset is the most important step in the process because most segmentation cases focus on small and specific regions in the whole medical image. Regarding the VOIs, the regions of the eyeball, optic nerve, and extraocular muscles are cropped based on the location, but more effective methods for extracting VOIs have the potential to improve segmentation performance14.
For the segmentation, SEQ-UNET is employed in the study. The 3D medical images have large volumes, so deep neural network models require large memory capacities. In SEQ-UNET, the segmentation model is implemented with a small number of slices to reduce the required memory size without losing the features of the 3D information.
The model was trained with 46 VOIs, which is not a large number for model training. Due to the small number of training datasets, the performance of optic nerve and extraocular muscle segmentation is limited. Transfer learning15 and domain adaptation8 could provide a solution for improving the segmentation performance.
The whole segmentation process introduced here is not limited to orbital CT segmentation. The efficient labeling method helps create a new medical image dataset for when the application domain is unique to the research area. The python codes of GitHub concerning the pre-processing and segmentation modeling can be applied to other domains with the modification of the cropping region, the window clipping level, and the model hyper-parameters, such as the number of sequential slices, the U-Net architectures, and so on.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF), grant funded by the Ministry of Science and ICT of Korea (MSIT) (number: 2020R1C1C1010079). For the CMC-ORBIT dataset, the central Institutional Review Board (IRB) of the Catholic Medical Center provided approval (XC19REGI0076). This work was supported by 2022 Hongik University Research Fund.
Materials
| GitHub link | github.com/SleepyChild1005/OrbitSeg | ||
| MediLabel | INGRADIENT (Seoul, Korea) | a medical image labeling software promgram for segmentation with fewer click and higher speed | |
| SEQ-UNET | downloadable from GitHub | ||
| SmartFil | wizard in MediLabel | ||
| SmartPencil | wizard in MediLabel |
Referências
- Li, Z., et al. Deep learning-based CT radiomics for feature representation and analysis of aging characteristics of Asian bony orbit. Journal of Craniofacial Surgery. 33 (1), 312-318 (2022).
- Hamwood, J., et al. A deep learning method for automatic segmentation of the bony orbit in MRI and CT images. Scientific Reports. 11, 1-12 (2021).
- Kim, K. S., et al. Schwannoma of the orbit. Archives of Craniofacial Surgery. 16 (2), 67-72 (2015).
- Baur, C., et al. Autoencoders for unsupervised anomaly segmentation in brain MR images: A comparative study. Medical Image Analysis. 69, 101952 (2021).
- Trémeau, A., Colantoni, P. Regions adjacency graph applied to color image segmentation. IEEE Transactions on Image Processing. 9 (4), 735-744 (2000).
- Boykov, Y. Y., Jolly, M. -. P. Interactive graph cuts for optimal boundary & region segmentation of objects in ND images. Proceedings of Eighth IEEE International Conference on Computer Vision. International Conference on Computer Vision. 1, 105-122 (2001).
- Rother, C., Kolmogorov, V., Blake, A. "GrabCut" interactive foreground extraction using iterated graph cuts. ACM Transactions on Graphics. 23 (3), 309-314 (2004).
- Suh, S., et al. Supervised segmentation with domain adaptation for small sampled orbital CT images. Journal of Computational Design and Engineering. 9 (2), 783-792 (2022).
- Ronneberger, O., Fischer, P., Brox, T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention.Medical Image Computing and Computer-Assisted Intervention – MICCAI. , 234-241 (2015).
- Qamar, S., et al. A variant form of 3D-UNet for infant brain segmentation. Future Generation Computer Systems. 108, 613-623 (2020).
- Nguyen, H., et al. Ocular structures segmentation from multi-sequences MRI using 3D UNet with fully connected CRFS. Computational Pathology and Ophthalmic Medical Image Analysis. , 167-175 (2018).
- Liu, Q., et al. Bidirectional-convolutional LSTM based spectral-spatial feature learning for hyperspectral image classification. Remote Sensing. 9 (12), 1330 (2017).
- Yeghiazaryan, V., Voiculescu, I. D. Family of boundary overlap metrics for the evaluation of medical image segmentation. Journal of Medical Imaging. 5 (1), 015006 (2018).
- Zhang, G., et al. Comparable performance of deep learning-based to manual-based tumor segmentation in KRAS/NRAS/BRAF mutation prediction with MR-based radiomics in rectal cancer. Frontiers in Oncology. 11, 696706 (2021).
- Christopher, M., et al. Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs. Scientific Reports. 8, 16685 (2018).

