Reconstruction of the Blood-Brain Barrier In Vitro to Model and Therapeutically Target Neurological Disease
Summary
The blood-brain barrier (BBB) has a crucial role in sustaining a stable and healthy brain environment. BBB dysfunction is associated with many neurological diseases. We have developed a 3D, stem-cell-derived model of the BBB to investigate cerebrovascular pathology, BBB integrity, and how the BBB is altered by genetics and disease.
Abstract
The blood-brain barrier (BBB) is a key physiological component of the central nervous system (CNS), maintaining nutrients, clearing waste, and protecting the brain from pathogens. The inherent barrier properties of the BBB pose a challenge for therapeutic drug delivery into the CNS to treat neurological diseases. Impaired BBB function has been related to neurological disease. Cerebral amyloid angiopathy (CAA), the deposition of amyloid in the cerebral vasculature leading to a compromised BBB, is a co-morbidity in most cases of Alzheimer's disease (AD), suggesting that BBB dysfunction or breakdown may be involved in neurodegeneration. Due to limited access to human BBB tissue, the mechanisms that contribute to proper BBB function and BBB degeneration remain unknown. To address these limitations, we have developed a human pluripotent stem cell-derived BBB (iBBB) by incorporating endothelial cells, pericytes, and astrocytes in a 3D matrix. The iBBB self-assembles to recapitulate the anatomy and cellular interactions present in the BBB. Seeding iBBBs with amyloid captures key aspects of CAA. Additionally, the iBBB offers a flexible platform to modulate genetic and environmental factors implicated in cerebrovascular disease and neurodegeneration, to investigate how genetics and lifestyle affect disease risk. Finally, the iBBB can be used for drug screening and medicinal chemistry studies to optimize therapeutic delivery to the CNS. In this protocol, we describe the differentiation of the three types of cells (endothelial cells, pericytes, and astrocytes) arising from human pluripotent stem cells, how to assemble the differentiated cells into the iBBB, and how to model CAA in vitro using exogenous amyloid. This model overcomes the challenge of studying live human brain tissue with a system that has both biological fidelity and experimental flexibility, and enables the interrogation of the human BBB and its role in neurodegeneration.
Introduction
The blood-brain barrier (BBB) is a key microvascular network separating the central nervous system (CNS) from the periphery to maintain an ideal environment for proper neuronal function. It has a critical role in regulating the influx and efflux of substances into the CNS by maintaining metabolic homeostasis1,2,3,4, clearing waste4,5,6, and protecting the brain from pathogens and toxins7,8.
The primary cell type of the BBB is the endothelial cell (EC). Endothelial cells, derived from the mesoderm lineage, form the walls of the vasculature1,9. Microvascular ECs form tight junctions with each other to greatly decrease the permeability of their membrane10,11,12,13,14 while expressing transporters to facilitate the movement of nutrients into and out of the CNS1,4,12,14. Microvascular ECs are encircled by pericytes (PCs)-mural cells that regulate microvascular function and homeostasis and are critical for regulating the permeability of the BBB to molecules and immune cells15,16,17. The astrocyte, a major glial cell type, is the final cell type comprising the BBB. Astrocyte end-feet wrap around the EC-PC vascular tubes while the cell bodies extend into the brain parenchyma, forming a connection between neurons and vasculature1. Distinct solute and substrate transporters are localized on astrocyte end-feet (e.g., aquaporin 4 [AQP-4]) that have a critical role in BBB function18,19,20,21.
The BBB is critical in maintaining proper brain health function, and dysfunction of the BBB has been reported in many neurological diseases, including Alzheimer's disease (AD)22,23,24,25, multiple sclerosis7,26,27,28, epilepsy29,30, and stroke31,32. It is increasingly recognized that cerebrovascular abnormalities play a central role in neurodegeneration, contributing to increased susceptibility to ischemic and hemorrhagic events. For example, more than 90% of AD patients have cerebral amyloid angiopathy (CAA), a condition characterized by the deposition of amyloid β (Aβ) along the cerebral vasculature. CAA increases BBB permeability and decreases BBB function, leaving the CNS vulnerable to ischemia, hemorrhagic events, and accelerated cognitive decline33.
We recently developed an in vitro model of the human BBB, derived from patient-induced pluripotent stem cells, which incorporates ECs, PCs, and astrocytes encapsulated in a 3D matrix (Figure 1A). The iBBB recapitulates physiologically relevant interactions, including vascular tube formation and localization of astrocyte end-feet with vasculature24. We applied the iBBB to model the susceptibility of CAA mediated by APOE4 (Figure 1B). This enabled us to identify the causal cellular and molecular mechanisms by which APOE4 promotes CAA, and leverage these insights to develop therapeutic strategies that reduce CAA pathology and improve learning and memory in vivo in APOE4 mice24. Here, we provide a detailed protocol and video tutorial for reconstructing the BBB from human iPSCs and modeling CAA in vitro.
Protocol
1. Differentiating iPSCs into iBBB cells
NOTE: These differentiation protocols have been previously described in Mesentier-Louro et al.34.
- Coating cell culture plates
- Thaw reduced growth factor (GF) membrane matrix overnight at 4 °C. Dilute 500 µL of basement membrane matrix in 49.5 mL of DMEM. Keep this solution cold to prevent premature polymerization of the coating solution.
- Add 1-2 mL per well of a 6-well tissue culture-treated plate. Leave the coating solution on for at least 20 min at 37 °C before replacing it with the appropriate warmed culture medium.
- Differentiation of Human iPSCs into ETV2-inducible endothelial cells
Timing: 8 days
NOTE: This protocol is adapted from Blanchard et al.24 and Qian et al.35. We use the doxycycline-inducible expression of the transcription factor ETV2, as described by Wang et al.36 to improve the yield. ETV2 was introduced via PiggyBac plasmid and the cells were then transfected.- Culture iPSCs on reduced GF basement membrane matrix-coated plates in feeder-free pluripotent stem cell medium until the cells reach ~70% confluency.
NOTE: We recommend the inducible iPSCs be maintained with selection for the PiggyBac plasmid (i.e., puromycin) - Day 0: Dissociate the cells with a protease-collagenase mixture for 5 min. Transfer the dissociated cells into a conical tube with a 1:3 ratio of the protease-collagenase mixture to stem cell medium. Spin down cells at 300 × g for 3 min. Resuspend the cell pellet with stem cell medium and plate the cells at 20,800 cells/cm2 on reduced GF basement membrane matrix-coated plates in stem cell medium supplemented with 10 µM Y27632.
- Day 1: Replace the medium with DeSR1 [DMEM/F12 + glutamine supplement, 1x Minimum Essential Medium Non-Essential Amino Acids (MEM-NEAA), 1% penicillin-streptomycin (P/S)], supplemented with 10 ng/mL BMP4, 6 µM CHIR99021, and 5 µg/mL doxycycline.
- Day 3: Replace the medium with DeSR2 [DMEM/F12 + glutamine supplement, 1x MEM-NEAA, 1x N-2, 1x B-27, 1% P/S] supplemented with 5 µg/mL doxycycline.
- Day 5: Replace medium with hECSR [Human Endothelial Serum-Free Medium, 1x MEM-NEAA, 1x B-27, 1% P/S] supplemented with 50 ng/mL VEGF-A, 2 µM forskolin, and 5 µg/mL doxycycline.
- Day 7: Replace medium with hECSR supplemented with 50 ng/mL VEGF-A, 2 µM forskolin, and 5 µg/mL doxycycline.
- Day 8: Split ECs 1:3 with a protease-collagenase mixture and re-seed onto fresh reduced GF basement membrane matrix-coated plates in hECSR supplemented with 50 ng/mL VEGF-A and 5 µg/mL doxycycline.
NOTE: This is the best time point to encapsulate ECs into iBBBs for uniform vasculature formation. - Day 10+: Feed the cells every 2-3 days with hECSR supplemented with 50 ng/mL VEGF-A and 5 µg/mL doxycycline. Continue to split the cells 1:3 with a protease-collagenase mixture when the plates are confluent.
NOTE: iPSC-derived ECs can be frozen at any point starting Day 8. Lift the cells, as if to passage. Resuspend the pellet in EC freezing medium [60% knockout serum replacement (KSR), 30% hECSR, 10% DMSO, 50 ng/mL VEGF-A, and 10 µM Y27632], splitting the cells 1:3, and freeze using a freezing container at -80 °C. Thaw onto reduced GF basement membrane matrix-coated plates in hECSR supplemented with 50 ng/mL VEGF-A, 5 µg/mL doxycycline. - Confirm brain microvascular endothelial cell differentiation by immunocytochemistry using antibodies against CD31/PECAM1 or VE-cadherin (Figure 2A).
- Culture iPSCs on reduced GF basement membrane matrix-coated plates in feeder-free pluripotent stem cell medium until the cells reach ~70% confluency.
- Differentiation of Human iPSCs into pericytes
Timing: 6 days
NOTE: This protocol was adapted from Patsch et al.36.- Culture iPSCs on reduced GF basement membrane matrix-coated plates in feeder-free conditions in stem cell medium until the cells reach ~70% confluency.
- Day 0: Dissociate the cells with a protease-collagenase mixture for 5 min. Transfer the dissociated cells into a conical tube with a 1:3 ratio of the protease-collagenase mixture to stem cell medium. Spin down cells at 300 × g for 3 min. Resuspend the cell pellet with stem cell medium and plate the cells at 37,000-40,000 cells/cm2 (depending on the cell line) on reduced GF basement membrane matrix-coated plates in stem cell medium supplemented with 10 µM Y27632.
- Day 1: Replace the media with N2B27 [50% DMEM/F12 + glutamine supplement, 50% Neurobasal, 1x MEM-NEAA, 0.5x, glutamine supplement, 1x N-2, 1x- B-27, 1% P/S] supplemented with 25 ng/mL BMP4 and 8 uM CHIR99021.
- Days 3-4: Replace the media with N2B27 supplemented with 2 ng/mL Activin A and 10 ng/mL PDGF-BB, and feed daily.
- Day 5: Split PCs with a protease-collagenase mixture and seed onto 0.1% gelatin-coated plates at 35,000 cells/cm2 in N2B27 supplemented with 10 ng/mL PDGF-BB.
NOTE: Only split cells once confluent. Some lines might require a few more days before they are ready to split; continue to change the media daily until cells are confluent. - Expand PCs in N2B27 supplemented with 10 ng/mL PDGF-BB, changing media every 2-3 days. Remove PDGF-BB from the media after the cells once again reach confluency and undergo a second passage.
NOTE: Pericytes are ready to be used in iBBBs after the second passage. Once expanded, PCs can be frozen. Lift the cells as if to passage, resuspend the cell pellet in freezing media [90% KSR and 10% DMSO] and freeze using a freezing container at -80 °C. Thaw onto 0.1% gelatin-coated plates at 35,000 cells/cm2 in N2B27. - Confirm pericyte differentiation by immunocytochemistry using antibodies against PDGFRB and NG2 (Figure 2B).
- Differentiation of Human iPSCs into neural progenitor cells (NPCs) and astrocytes
Timing: 45 days total (15 days for NPCs followed by 30 days for astrocytes)
NOTE: The NPC differentiation protocol was adapted from Chambers et al.38, and the astrocyte differentiation is as described in TCW et al.39.- Culture iPSCs on reduced GF basement membrane matrix-coated plates in feeder-free conditions in stem cell medium until the cells reach ~70% confluency.
- Dissociate the cells with a protease-collagenase mixture for 5 min. Transfer the dissociated cells into a conical tube with a 1:3 ratio of the protease-collagenase mixture to stem cell medium. Spin down cells at 300 × g for 3 min. Resuspend the cell pellet with stem cell medium and plate the cells at 100,000 cells/cm2 on reduced GF basement membrane matrix-coated plates in stem cell medium supplemented with 10 µM Y27632.
- Replace the stem cell medium the following day. Continue to feed the cells every other day until they reach >95% confluency (3-4 days depending on the cell line).
- NPC Day 0: Replace the media with N2B27 [50% DMEM/F12 + glutamine supplement, 50% Neurobasal, 1x MEM-NEAA, 0.5x, glutamine supplement, 1x N-2, 1X- B-27, 1% P/S] supplemented with 10 µM SB43152 and 100 nM LDN193189.
- NPC Days 1-9: Feed the cells daily with N2B27 supplemented with 10 µM SB43152 and 100 nM LDN193189.
- NPC Day 10: Split NPCs 1:3 with the protease-collagenase mixture and seed onto fresh reduced GF basement membrane matrix-coated plates in N2B27 supplemented with 20 ng/mL FGF-basic and 10 µM Y27632.
- NPC Days 11 – 13: Feed NPCs daily with N2B27 supplemented with 20 ng/mL FGF-basic.
- NPC Day 14: Split NPCs 1:3 with the protease-collagenase mixture and reseed onto fresh reduced GF basement membrane matrix-coated plates in N2B27 supplemented with 20 ng/mL FGF-basic and 10 µM Y27632.
- NPC Day 15/Astrocyte Day 0: This is Day 0 of the astrocyte differentiation. Feed the cells with complete Astrocyte medium (AM).
NOTE: NPCs can be maintained in N2B27 supplemented with 20 ng/mL FGF-basic and passaged when confluent. To freeze NPCs, resuspend the cell pellet in freezing media [90% KSR and 10% DMSO], split the cells 1:3, and freeze using a freezing container at -80 °C. Thaw onto reduced GF basement membrane matrix-coated plates in N2B27 supplemented with 20 ng/mL bFGF and 10 µM Y27632. - Astrocyte Days 1-30: Feed the cells every 2-3 days with AM. When cells are >90% confluent, passage using the protease-collagenase mixture and plate at 15,000 cells/cm2 on fresh reduced GF basement membrane matrix-coated plates in AM.
NOTE: Astrocytes should be fully differentiated in 30 days. Astrocytes can be frozen in freezing media [90% KSR and 10% DMSO] in a freezing container at -80 °C. Thaw onto reduced GF basement membrane matrix-coated plates in AM at 25,000-35,000 cells/cm2. - Confirm NPC differentiation by immunocytochemistry using antibodies against Nestin and SOX2. Confirm astrocyte differentiation by immunocytochemistry using antibodies against S100B and CD44 (Figure 2C).
2. Assembling the iBBB
- Thaw reduced growth factor (GF) basement membrane matrix overnight at 4 °C. Do not dilute this matrix in DMEM as the iBBBs are encapsulated in 100% reduced GF basement membrane matrix. Each iBBB requires 50 µL of the matrix.
- Dissociate the differentiated ECs with a protease-collagenase mixture at 37 °C for 5 min. Transfer the dissociated cells into a conical tube with a 1:3 ratio of the protease-collagenase mixture to hECSR (Human Endothelial Serum-Free Medium, 1x MEM-NEAA, 1x B-27, 1% P/S). Spin down cells at 300 × g for 3 min. Resuspend the cell pellet in hECSR.
NOTE: Use a conical tube, either 15 mL or 50 mL, that is large enough to accommodate the volume of dissociated cells plus media. Cells can also be divided among multiple tubes and then recombined upon resuspension. - Dissociate the differentiated PCs with the protease-collagenase mixture at 37 °C for 5 min. Transfer the dissociated cells into a conical tube with a 1:3 ratio of the protease-collagenase mixture to N2B27 (50% DMEM/F12 + glutamine supplement, 50% Neurobasal, 1x MEM-NEAA, 0.5x glutamine supplement, 1x N-2, 1x B-27, 1% penicillin-streptomycin [P/S]). Spin down cells at 300 × g for 3 min. Resuspend the cell pellet in N2B27.
- Dissociate the differentiated astrocytes with the protease-collagenase mixture at 37 °C for 5 min. Transfer the dissociated cells into a conical tube with a 1:3 ratio of the protease-collagenase mixture to complete Astrocyte Medium (AM). Spin down cells at 300 × g for 3 min. Resuspend the cell pellet in AM.
- Using a hemocytometer, count each cell type. Dilute each type of cell suspension to a final concentration of 1 × 106 cells/mL in the appropriate medium.
- Combine the calculated number of cells for each cell type in a conical tube: 50 µL of iBBB requires 2.5 × 105 ECs, 5 × 104 PCs, and 5 × 104 astrocytes. Include 10% more than the calculated number of cells to account for pipetting errors. Spin down the cell mixture at 300 × g for 3 min.
NOTE: It is highly recommended to plate some leftover cells of each cell type in 2D monocultures to fix and stain for quality control of the input cells. Plate each cell type at 35,000 cells/cm2 on previously prepared plates coated with reduced GF basement membrane matrix and fix after 3-4 days. See Table 1 for cell-type specific antibodies. - Aspirate medium from the cell pellet leaving approximately 50 µL of the supernatant above the cell pellet. Using a P200, gently resuspend the cell pellet in residual medium to create a single-cell slurry.
- Mix the cell slurry in enough reduced GF basement membrane matrix supplemented with 10 ng/mL VEGF-A for the desired number of iBBBs at 50 µL of reduced GF basement membrane matrix per iBBB, taking care to mix the cells homogeneously while not introducing any air bubbles.
NOTE: It is critical to keep the cells and reduced GF basement membrane matrix on ice at this stage to prevent premature polymerization. - Pipette 50 µL of reduced GF basement membrane matrix mixture into the well of a 48-well glass-bottom culture dish. Distribute the volume evenly throughout the bottom of the dish (Figure 1C).
- Incubate the iBBBs at 37 °C for 30-40 min to allow the reduced GF basement membrane matrix to polymerize, encapsulating the cells in the matrix.
- Add 500 µL of AM supplemented with 10 ng/mL VEGF-A, 5 µg/mL doxycycline, and 10 µM Y27632.
- Change the medium the following day to AM supplemented with 10 ng/mL VEGF-A. Continue to change the medium every 2-3 days. After 2 weeks, stop supplementing with VEGF-A; iBBBs are ready for downstream assays after 2 weeks.
3. Inducing cerebral amyloid angiopathy (CAA) with Aβ fibrils
- Prepare stock solutions of amyloid-β1-40 and amyloid-β1-42 at 200 µM in PBS.
- Add Aβ to iBBB medium on 2-week iBBBs to a final concentration of 20 nM. Incubate the cells in Aβ-supplemented medium for 96 h (4 days).
- After 96 h, remove the medium and continue with fixing (section 4).
4. Fixing and staining the iBBB
- To fix the iBBB, remove the medium, wash in 1 mL of PBS, and incubate in 500 µL of 4% paraformaldehyde overnight at 4 °C.
- Remove the 4% paraformaldehyde solution, and rinse for 4 x (at least) 15 min in 1 mL of PBS.
- Permeabilize the samples in 0.3% PBST (PBS with 0.3% Triton X-100) for 1 h at room temperature with gentle shaking.
- Incubate the cultures in blocking buffer (5% Normal Serum in 0.3% PBST) at room temperature for at least 1 h with gentle shaking.
NOTE: The type of serum used is dependent on the host species for the secondary antibodies used. For example, if the host species for the secondary antibodies is goat, use normal goat serum; if the host species is donkey, use normal donkey serum. This can also be done overnight at 4 °C with gentle shaking. - Dilute primary antibodies to the recommended or determined concentration in blocking buffer.
- Replace the blocking solution with the primary antibody dilution. Make sure that the iBBBs are fully submerged in antibody solution for even incubation; 100-150 µL is enough to cover a 50 µL iBBB in a 48-well glass-bottom culture dish. Incubate overnight at 4 °C with gentle shaking.
- Remove the primary antibody. Wash iBBBs for 3 x 10-20 min in 0.3% PBS.
- Dilute the secondary antibody to the recommended or determined concentration in blocking buffer. Additionally, add a nuclear stain, such as Hoechst, to the secondary antibody solution at the recommended or determined concentration.
- Replace the last PBS wash with the secondary antibody dilution. Make sure that the iBBBs are fully submerged in antibody solution for even incubation; 100-150 µL is enough to cover a 50 µL iBBB in a 48-well glass-bottom culture dish. Incubate at room temperature for 2 h with gentle shaking.
NOTE: This can also be done overnight at 4 °C with gentle shaking. - Wash iBBBs 4-5x for at least 1 h per wash in PBS. Store in PBS or anti-fading mounting media at 4 °C until the samples are imaged.
- To image on an inverted microscope, keep iBBBs in glass-bottom plates or dishes. Place a glass coverslip over the glass well to keep the iBBBs in place (Figure 1D).
Representative Results
A properly formed iBBB solidifies into a single translucent disc (Figure 3A). It is normal for the iBBB to detach from the surface onto which it was first pipetted after a few days. This cannot be avoided, but is not a major concern to the proper formation of the iBBB if care is taken when changing media to not accidentally aspirate the iBBB. After 24 h, evenly distributed, single cells can be identified under a brightfield microscope (Figure 3B). After 2 weeks, more distinct structures may be visible, although it is difficult to make out with definition (Figure 3C).
The quality of the iBBB is highly dependent on the quality of the differentiation of the input cells. We highly recommend plating some left-over cells in 2D monocultures to fix and stain for cell-type specific markers. iBBBs formed from endothelial cells that are over 70% positive for PECAM1 or VE-cadherin (Figure 2A), pericytes over 95% positive for PDGFRB (Figure 2B), and astrocytes over 95% positive for S100B and CD44 (Figure 2C) are best for successful iBBBs. The results from this quality check are the earliest indicator for high-quality iBBBs.
Physiologically relevant 3D structures should form after 2 weeks of self-assembly. Upon fixing and staining, we see evidence of tube-like structures that stain positive for the endothelial marker PECAM1, which is critical for tight junction formation (Figure 4A). The greatest variability in iBBB formation is the extent of microvasculature formation. In a "worst-case" scenario, the endothelial cell network appears more fragmented or does not extend throughout the iBBB, while in the "best-case" scenario, the vasculature is uniform and branches throughout the iBBB. Endothelial cell differentiation that is over 70% PECAM1-positive forms more consistent networks. Additionally, aquaporin-4, a protein expressed by astrocytes that localizes to the astrocyte endfeet, aligns with PECAM1 staining, indicating that astrocytes extend their endfeet to contact the endothelial cells (Figure 4B). Finally, we expect to see pericytes around the vasculature (Figure 4C).
The primary readout for cerebral amyloid angiopathy (CAA) in iBBBs is the presence of amyloid-β (Aβ). Aβ can be measured using targeted antibodies or by seeding fluorescently labeled Aβ to induce CAA phenotypes (Figure 5). Treatment of iBBBs with Aβ should increase Aβ staining intensity and area, as the cells of the BBB do not express a lot of endogenous Aβ protein. Alternatively, fixed samples can be stained with Thioflavin T to detect amyloid accumulation. Aβ levels are dependent on the genotype of the stem cells used to generate the iBBB, with some Alzheimer's disease and CAA-associated risk factors increasing the amount of Aβ accumulation and staining (Figure 5B, C)24.
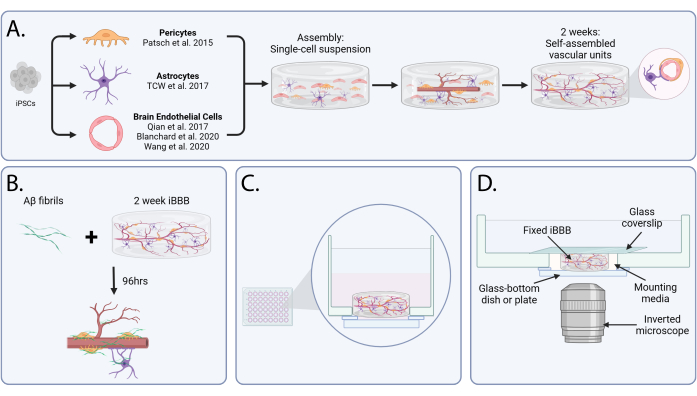
Figure 1: An in vitro blood-brain barrier to model cerebral amyloid angiopathy. (A) Schematic representation of iBBB assembly and maturation. After the differentiation of endothelial cells, astrocytes, and pericytes from induced pluripotent stem cells, cells are encapsulated in a gel matrix in a single-cell suspension. Over the course of 2 weeks, the cells self-assemble into vascular units, similar to the structures identified in vivo. (B) Schematic of the cerebral amyloid angiopathy assay. Amyloid-β is added to matured iBBBs for 96 h to induce Aβ aggregation. (C) Diagram showing the side view of an iBBB after seeding in a glass-bottomed well. (D) A graphic of an iBBB prepared for imaging on an inverted microscope. Abbreviations: BBB = blood-brain barrier; iPSCs = induced pluripotent stem cells; iBBB = an in vitro model of the human BBB derived from a patient iPSC-derived BBB model, which incorporates endothelial cells, pericytes, and astrocytes encapsulated in a 3D matrix; Aβ = Amyloid-β. Please click here to view a larger version of this figure.
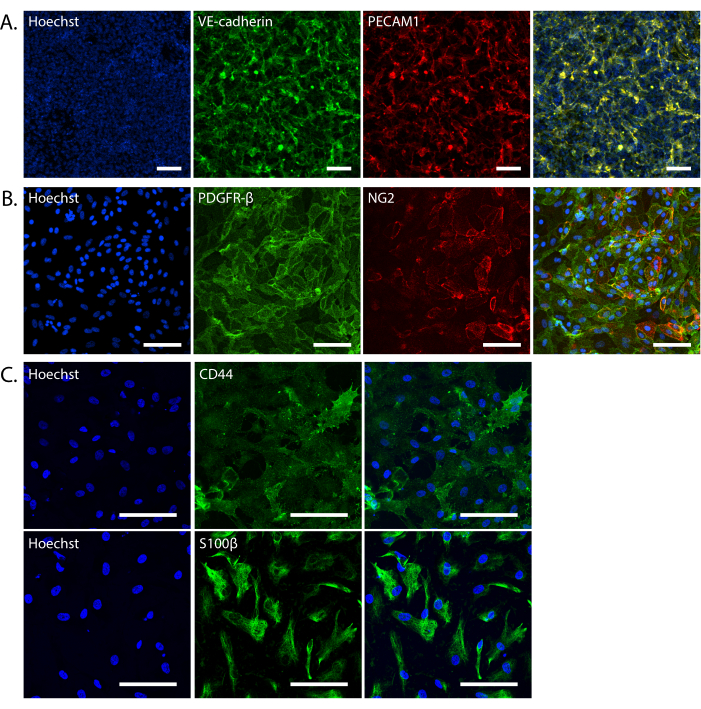
Figure 2: Validation of iPSC-derived iBBB cells. (A) Representative maximum intensity projection of endothelial cells in 2D monocultures stained with VE-cadherin (green) and PECAM1 (red). (B) Representative images of pericytes in 2D monocultures stained with PDGFRB (green) and NG2 (red). (C) Representative images of astrocytes in 2D monocultures stained with CD44 (top; green) and S100B (bottom; green). All nuclei are stained with Hoechst 33342. Images were taken on a Nikon Eclipse Ti2-E at 20x magnification (A,B) or a Leica Stellaris 8 at 40x magnification (C). All scale bars are 100 µm. Abbreviations: BBB = blood-brain barrier; iPSCs = induced pluripotent stem cells; iBBB = an in vitro model of the human BBB derived from a patient iPSC-derived BBB model, which incorporates endothelial cells, pericytes, and astrocytes encapsulated in a 3D matrix; VE-cadherin = vascular endothelial cadherin; PECAM1 = platelet and endothelial cell adhesion molecule 1; PDGFRB = platelet-derived growth factor receptor beta; NG2 = neuron glial antigen 2; S100B = S100 calcium-binding protein beta. Please click here to view a larger version of this figure.
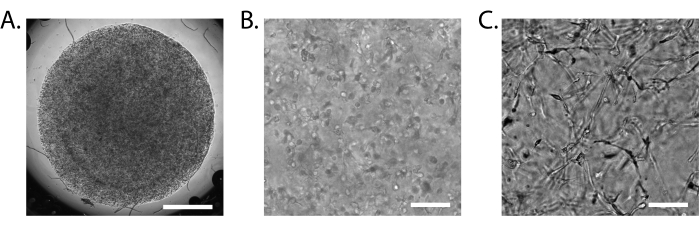
Figure 3: Assembly of the iBBB. (A) Brightfield image of a 15 µL iBBB 24 h after plating at 2x magnification. (B) Brightfield image of an iBBB 24 h after plating at 10x magnification. (C) Brightfield image of an iBBB 2 weeks after plating at 10x magnification. Scale bar = 1 mm (A), 100 µm (B,C). Images taken on an inverted Nikon Eclipse Ts2R-FL. Abbreviations: BBB = blood-brain barrier; iPSCs = induced pluripotent stem cells; iBBB = an in vitro model of the human BBB derived from a patient iPSC-derived BBB model, which incorporates endothelial cells, pericytes, and astrocytes encapsulated in a 3D matrix. Please click here to view a larger version of this figure.
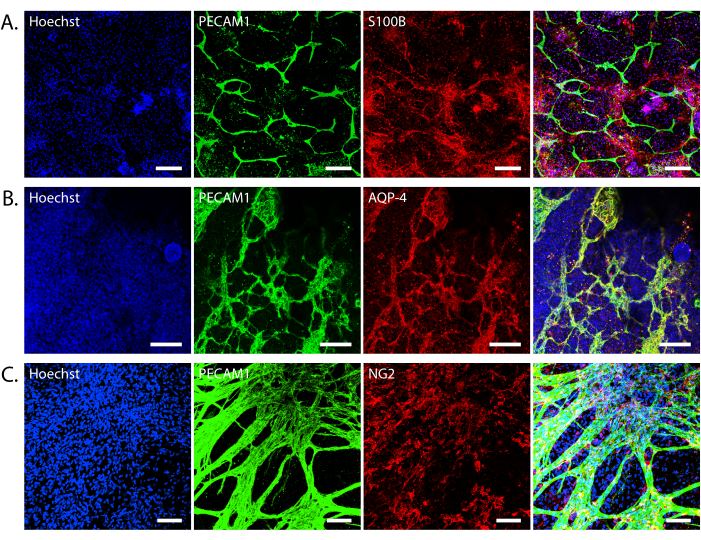
Figure 4: Cell interactions in the iBBB. Representative images of endothelial cells, pericytes, and astrocytes in the iBBB. (A) Endothelial cells (PECAM1) and astrocytes (S100β). (B) Endothelial cells (PECAM1) and astrocyte endfeet (AQP-4) co-localization. (C) endothelial cells (PECAM1) and pericytes (NG2). All nuclei are stained with Hoechst 33342. Confocal Z-stack images were taken on a Leica Stellaris 8 at 20x magnification (A,B) or a Nikon Eclipse Ti2-E at 20x magnification (C). Scale bars = 200 µm (A), 100 µm (B,C). Abbreviations: BBB = blood-brain barrier; iPSCs = induced pluripotent stem cells; iBBB = an in vitro model of the human BBB derived from a patient iPSC-derived BBB model, which incorporates endothelial cells, pericytes, and astrocytes encapsulated in a 3D matrix; PECAM1 = platelet and endothelial cell adhesion molecule 1; AQP-4 = aquaporin-4; NG2 = neuron glial antigen 2. Please click here to view a larger version of this figure.
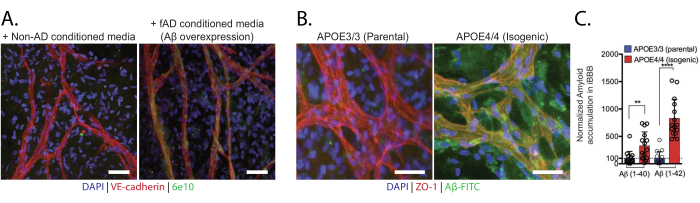
Figure 5: Cerebral amyloid angiopathy in vitro. (A) Non-AD iBBBs exposed to conditioned media from control or Aβ-overexpression neurons. 6e10 antibody recognizes Aβ. (B) Representative images of iBBBs from isogenic APOE3 and APOE4 cell lines treated with 20 nM Aβ-FITC1-42 for 96 h. (C) Quantification of amyloid in iBBBs from isogenic APOE3 and APOE4 cell lines treated with 20 nM Aβ-FITC1-40 or Aβ-FITC1-42.Scale bars = 50 µm (A), 10 µm (B). This figure was adapted from Blanchard et al24. Abbreviations: BBB = blood-brain barrier; iPSCs = induced pluripotent stem cells; iBBB = an in vitro model of the human BBB derived from a patient iPSC-derived BBB model, which incorporates endothelial cells, pericytes, and astrocytes encapsulated in a 3D matrix; Aβ = Amyloid-β. APOE = ApolipoproteinE. Please click here to view a larger version of this figure.
| Markers | Company | Catalog Number | Dilution | |
| Endothelial Cells | PECAM1 (CD31) | R&D Systems | AF806 | 1:500 |
| VE-cadherin (CD144) | R&D systems | AF938 | 1:500 | |
| ZO-1 | Invitrogen | MA3-39100-A488 | 1:500 | |
| Pericytes | PDGFRβ | R&D Systems | AF385 | 1:500 |
| NG2 | Abcam | ab255811 | 1:500 | |
| Astrocytes | S100β | Sigma-Aldrich | S2532-100uL | 1:500 |
| CD44 | Cell Signaling Technology | 3570S | 1:400 | |
| AQP-4 | Invitrogen | PA5-53234 | 1:300 | |
| GFAP | ||||
| ALDH1L1 | ||||
| EAAT1 | ||||
| EAAT2 | ||||
| Amyloid-β | 6e10 | Biolegend | SIG-39320 | 1:1,000 |
| Thioflavin T | Chem Impex | 22870 | 25 µM |
Table 1: Recommended cell markers for differentiation quality checks. Cell markers for the different cell types of the BBB that can be used to check the quality of the differentiations and to identify the cells in the formed iBBB. The markers used in this paper are bolded.
Discussion
BBB dysfunction is a co-morbidity, and potentially, a cause or exacerbating factor in numerous neurological diseases7,40,41. However, it is nearly impossible to study the molecular and cell biology underlying the dysfunction and breakdown of the BBB in humans with neurovascular disease. The inducible-BBB (iBBB) presented in this protocol provides an in vitro system that recapitulates important cell interactions of the BBB, including vascular tube formation and localization of astrocyte end-feet with vasculature. The iBBB can be used to study the molecular pathways involved at any stage of BBB dysfunction and can model neurovascular disease phenotypes, such as amyloid-β aggregation, as seen in cerebral amyloid angiopathy.
The assembly of the iBBB is straightforward, although the quality of the iBBBs is highly dependent on the quality of the iPSC-derived cells that are used. While the iBBB provides a multi-cellular niche that can promote the maturation of the component cells, each cell type needs to be properly patterned before being encapsulated. It is critical to perform quality checks on each differentiation by performing immunofluorescence staining for cell-type specific markers on the individual monocultures (Table 1). Every iPSC line behaves differently and some conditions, such as seeding density or the number of days in each patterning medium, might need to be determined empirically and adjusted to optimize differentiation efficiency.
The described protocol suggests a 50 µL size, but the iBBB can be scaled down to as small as 5 µL, depending on cell availability, the number of desired iBBBs, and the downstream application. Larger iBBBs contain more cells, making them ideal for protein, lipid, or nucleic acid collection. Smaller iBBBs can facilitate drug screenings or other scalable assays.
The iBBB is a very versatile tool for studying the BBB in vitro. Because each cell type is differentiated independently, iBBBs can be assembled from different genetic backgrounds, allowing us to study how genetic risk factors affect specific cell types to contribute to BBB dysfunction. This strategy was applied to show that APOE4, the most common risk factor for Alzheimer's disease and cerebral amyloid angiopathy, acts in part through a pathogenic mechanism specifically in pericytes24. Further use of this tool would allow us to dissect the individual contributions of ECs, PCs, and astrocytes in maintaining BBB integrity and how each cell type falters during disease development.
Currently, the most common method of modeling the BBB in vitro is by using a transwell system, seeded with ECs, and sometimes co-cultured with pericytes and/or astrocytes, to form an impermeable monolayer42,43,44. The 3D structure of the iBBB enables the self-assembly of the vascular unit and allows the formation of tubular structures that more resemble physiological vasculature. A drawback of seeding iBBBs in a well-plate, as described in this protocol, is they do not experience the sheer stresses caused by constant flow vasculature in an in vivo system. To overcome this, the iBBB can be seeded into a microfluidic chip that can generate a dynamic flow45,46,47. This system can also be used to test vasculature permeability and perfusion of small molecules.
In conclusion, this method provides a flexible, 3D, iPSC-derived model of the BBB that can be used as a platform to study countless aspects of the BBB on a cellular level, including the ability to recapitulate neurovascular disease phenotypes and the potential to screen drug BBB permeability for a wide range of applications.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work is supported by NIH 3-UG3-NS115064-01, R01NS14239, Cure Alzheimer's Fund, NASA 80ARCO22CA004, Chan-Zuckerberg Initiative, MJFF/ASAP Foundation, and Brain Injury Association of America. C.G. is supported by NIH F31NS130909. Figure 1A was created with BioRender.com.
Materials
| 6e10 amyloid-β antibody | Biolegend | SIG-39320 | Used at 1:1000 |
| Accutase | Innovative Cell Technologies | AT104 | |
| Activin A | Peprotech | 20-14E | |
| Alexa Fluor 488, 555, 647 secondary antibodies | Invitrogen | Various | Used at 1:1000 |
| Amyloid-beta 40 fibril | AnaSpec | AS-24235 | |
| Amyloid-beta 42 fibril | AnaSpec | AS-20276 | |
| Aquaporin-4 antibody | Invitrogen | PA5-53234 | Used at 1:300 |
| Astrocyte basal media and supplements | ScienCell | 1801 | |
| B-27 serum-free supplement | Gibco | 17504044 | |
| BMP4 | Peprotech | 120-05ET | |
| CHIR99021 | Cyamn Chemical | 13112 | |
| DMEM/F12 with GlutaMAX medium | Gibco | 10565018 | |
| Doxycycline | Millipore-Sigma | D3072-1ML | |
| FGF-basic | Peprotech | 100-18B | |
| Fluoromount-G slide mounting medium | VWR | 100502-406 | |
| Forskolin | R&D Systems | 1099/10 | |
| GeltrexTM LDEV-Free hESC-qualified Reduced Growth Factor Basement | Gibco | A1413302 | |
| Glass Bottom 48-well Culture Dishes | Mattek Corporation | P48G-1.5-6-F | |
| GlutaMAX supplement | Gibco | 35050061 | |
| Hoechst 33342 | Invitrogen | H3570 | |
| Human Endothelial Serum-free medium | Gibco | 11111044 | |
| LDN193189 | Tocris | 6053 | |
| Minimum Essential Medium Non-essential Amino Acid Solution (MEM-NEAA) | Gibco | 11140050 | |
| N-2 supplement | Gibco | 17502048 | |
| Neurobasal medium | Gibco | 21103049 | |
| Normal Donkey Serum | Millipore-Sigma | S30-100mL | Use serum to match secondary antibody host |
| Paraformaldehyde (PFA) | ThermoFisher | 28908 | |
| PDGF-BB | Peprotech | 100-14B | |
| PDGFRB (Platelet-derived growth factor receptor beta) antibody | R&D Systems | AF385 | Used at 1:500 |
| Phosphate Buffered Saline (PBS), pH 7.4 | Gibco | 10010031 | |
| Pecam1 (Platelet endothelial cell adhesion molecule 1) antibody | R&D Systems | AF806 | Used at 1:500 |
| Penicillin-Streptomycin | Gibco | 15140122 | |
| PiggyBac plasmid (PB_iETV2_P2A_GFP_Puro) | AddGene | Catalog #168805 | |
| S100B antibody | Sigma-Aldrich | S2532-100uL | Used at 1:500 |
| SB43152 | Reprocell | 04-0010 | |
| Thioflavin T | Chem Impex | 22870 | Used at 25uM |
| Triton X-100 | Sigma-Aldrich | T8787-250mL | |
| VE-cadherin (CD144) antibody | R&D systems | AF938 | Used at 1:500 |
| VEGF-A | Peprotech | 100-20 | |
| Y27632 | R&D Systems | 1254/10 | |
| ZO-1 | Invitrogen | MA3-39100-A488 | Dilution = 1:500 |
Referências
- Daneman, R., Prat, A. The blood-brain barrier. Cold Spring Harbor Perspectives in Biology. 7 (1), a020412 (2015).
- Segarra, M., Aburto, M. R., Acker-Palmer, A. Blood-brain barrier dynamics to maintain brain homeostasis. Trends in Neurosciences. 44 (5), 393-405 (2021).
- Campos-Bedolla, P., Walter, F. R., Veszelka, S., Deli, M. A. Role of the blood-brain barrier in the nutrition of the central nervous system. Archives of Medical Research. 45 (8), 610-638 (2014).
- Hladky, S. B., Barrand, M. A. Fluid and ion transfer across the blood-brain and blood-cerebrospinal fluid barriers; a comparative account of mechanisms and roles. Fluids and Barriers of the CNS. 13 (1), 19 (2016).
- Kaur, J., et al. Waste clearance in the brain. Frontiers in Neuroanatomy. 15, 665803 (2021).
- Verheggen, I. C. M., Van Boxtel, M. P. J., Verhey, F. R. J., Jansen, J. F. A., Backes, W. H. Interaction between blood-brain barrier and glymphatic system in solute clearance. Neuroscience and Biobehavioral Reviews. 90, 26-33 (2018).
- Weiss, N., Miller, F., Cazaubon, S., Couraud, P. O. The blood-brain barrier in brain homeostasis and neurological diseases. Biochimica et Biophysica Acta. 1788 (4), 842-857 (2009).
- Prinz, M., Priller, J. The role of peripheral immune cells in the cns in steady state and disease. Nature Neuroscience. 20 (2), 136-144 (2017).
- Weksler, B. B., et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB Journal. 19 (13), 1872-1874 (2005).
- Liu, W. Y., Wang, Z. B., Zhang, L. C., Wei, X., Li, L. Tight junction in blood-brain barrier: An overview of structure, regulation, and regulator substances. CNS Neuroscience & Therapeutics. 18 (8), 609-615 (2012).
- Siegenthaler, J. A., Sohet, F., Daneman, R. Sealing off the cns’: Cellular and molecular regulation of blood-brain barriergenesis. Current Opinion in Neurobiology. 23 (6), 1057-1064 (2013).
- Brightman, M. W., Reese, T. S. Junctions between intimately apposed cell membranes in the vertebrate brain. Journal of Cell Biology. 40 (3), 648-677 (1969).
- Reese, T. S., Karnovsky, M. J. Fine structural localization of a blood-brain barrier to exogenous peroxidase. Journal of Cell Biology. 34 (1), 207-217 (1967).
- Mahringer, A., Fricker, G. Abc transporters at the blood-brain barrier. Expert Opinion on Drug Metabolism & Toxicology. 12 (5), 499-508 (2016).
- Armulik, A., Genove, G., Betsholtz, C. Pericytes: Developmental, physiological, and pathological perspectives, problems, and promises. Developmental Cell. 21 (2), 193-215 (2011).
- Armulik, A., et al. Pericytes regulate the blood-brain barrier. Nature. 468 (7323), 557-561 (2010).
- Daneman, R., Zhou, L., Kebede, A. A., Barres, B. A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 468 (7323), 562-566 (2010).
- Abbott, N. J., Ronnback, L., Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nature Reviews. Neuroscience. 7 (1), 41-53 (2006).
- Heithoff, B. P., et al. Astrocytes are necessary for blood-brain barrier maintenance in the adult mouse brain. Glia. 69 (2), 436-472 (2021).
- Verkman, A. S. Aquaporin water channels and endothelial cell function. Journal of Anatomy. 200 (6), 617-627 (2002).
- Wolburg, H., Lippoldt, A. Tight junctions of the blood-brain barrier: Development, composition and regulation. Vascular Pharmacology. 38 (6), 323-337 (2002).
- Sagare, A. P., Bell, R. D., Zlokovic, B. V. Neurovascular dysfunction and faulty amyloid beta-peptide clearance in alzheimer disease. Cold Spring Harbor Perspectives in Medicine. 2 (10), a011452 (2012).
- Kapasi, A., Schneider, J. A. Vascular contributions to cognitive impairment, clinical alzheimer’s disease, and dementia in older persons. Biochimica et Biophysica Acta. 1862 (5), 878-886 (2016).
- Blanchard, J. W., et al. Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of apoe4 in pericytes. Nature Medicine. 26 (6), 952-963 (2020).
- Huang, Z., et al. Blood-brain barrier integrity in the pathogenesis of alzheimer’s disease. Frontiers in Neuroendocrinology. 59, 100857 (2020).
- Morgan, L., et al. Inflammation and dephosphorylation of the tight junction protein occludin in an experimental model of multiple sclerosis. Neurociência. 147 (3), 664-673 (2007).
- Kirk, J., Plumb, J., Mirakhur, M., Mcquaid, S. Tight junctional abnormality in multiple sclerosis white matter affects all calibres of vessel and is associated with blood-brain barrier leakage and active demyelination. Journal of Pathology. 201 (2), 319-327 (2003).
- Balasa, R., Barcutean, L., Mosora, O., Manu, D. Reviewing the significance of blood-brain barrier disruption in multiple sclerosis pathology and treatment. International Journal of Molecular Sciences. 22 (16), 8370 (2021).
- Marchi, N., Granata, T., Ghosh, C., Janigro, D. Blood-brain barrier dysfunction and epilepsy: Pathophysiologic role and therapeutic approaches. Epilepsia. 53 (11), 1877-1886 (2012).
- Kiani, L. Blood-brain barrier disruption following seizures. Nature Reviews. Neurology. 19 (4), 2023 (2023).
- Knowland, D., et al. Stepwise recruitment of transcellular and paracellular pathways underlies blood-brain barrier breakdown in stroke. Neuron. 82 (3), 603-617 (2014).
- Okada, T., Suzuki, H., Travis, Z. D., Zhang, J. H. The stroke-induced blood-brain barrier disruption: Current progress of inspection technique, mechanism, and therapeutic target. Current Neuropharmacology. 18 (12), 1187-1212 (2020).
- Gireud-Goss, M., Mack, A. F., Mccullough, L. D., Urayama, A. Cerebral amyloid angiopathy and blood-brain barrier dysfunction. Neuroscientist. 27 (6), 668-684 (2021).
- Mesentier-Louro, L. A., Suhy, N., Broekaart, D., Bula, M., Pereira, A. C., Blanchard, J. W. Modeling the blood-brain barrier using human-induced pluripotent stem cells. Methods in Molecular Biology. 2683, 135-151 (2023).
- Qian, T., et al. Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells. Science Advances. 3 (11), e1701679 (2017).
- Wang, K., et al. Robust differentiation of human pluripotent stem cells into endothelial cells via temporal modulation of etv2 with modified mrna. Science Advances. 6 (30), eaba7606 (2020).
- Patsch, C., et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nature Cell Biology. 17 (8), 994-1003 (2015).
- Chambers, S. M., et al. Highly efficient neural conversion of human es and ips cells by dual inhibition of smad signaling. Nature Biotechnology. 27 (3), 275-280 (2009).
- Tcw, J., et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports. 9 (2), 600-614 (2017).
- Zlokovic, B. V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 57 (2), 178-201 (2008).
- Daneman, R. The blood-brain barrier in health and disease. Annals of Neurology. 72 (5), 648-672 (2012).
- Cecchelli, R., et al. Modelling of the blood-brain barrier in drug discovery and development. Nature Reviews. Drug Discovery. 6 (8), 650-661 (2007).
- Helms, H. C., et al. In vitro models of the blood-brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. Journal of Cerebral Blood Flow & Metabolism. 36 (5), 862-890 (2016).
- Erickson, M. A., Wilson, M. L., Banks, W. A. In vitro modeling of blood-brain barrier and interface functions in neuroimmune communication. Fluids Barriers CNS. 17 (1), 26 (2020).
- Musafargani, S., et al. Blood brain barrier: A tissue engineered microfluidic chip. Journal of Neuroscience Methods. 331, 108525 (2020).
- Hajal, C., et al. Engineered human blood-brain barrier microfluidic model for vascular permeability analyses. Nature Protocols. 17 (1), 95-128 (2022).
- Oddo, A., et al. Advances in microfluidic blood-brain barrier (bbb) models. Trends in Biotechnology. 37 (12), 1295-1314 (2019).

