TRUE Gene Silencing: Screening of a Heptamer-type Small Guide RNA Library for Potential Cancer Therapeutic Agents
PREPARAÇÃO DO INSTRUTOR
CONCEITOS
PROTOCOLO DO ALUNO
1. Construction of a Heptamer-type sgRNA Library
- Select a subset of the 16,384 (47) 7-nt sequences unless the full-scale library is to be constructed.
NOTE: The sequences can be random and/or designed to target specific cellular RNAs. For the latter, find hairpin structures resembling the T-arm of tRNA in a target RNA with the aid of an appropriate computer program and/or visually, and select sequences complementary to 7-nt sequences immediately downstream of the hairpin structures4,10,17,18. - Synthesize each of the selected heptamers as a fully 2′-O-methylated, 5′- and 3′-phosphorylated RNA with a DNA/RNA synthesizer. Use the phosphoramidite method on the controlled-pore glass (CPG) support, and leave the 5′-terminal 4,4′-dimethoxytrityl (DMT) protecting group on in the last cycle (the addition of the 5′ phosphate) of the synthesis23.
NOTE: Custom synthetic RNA can be also obtained from oligonucleotide suppliers. - After completion of the last synthesis cycle, transfer the CPG with the synthesized oligonucleotide from the column to a glass vial with a Teflon-liner screw cap, add 1 ml of 29% ammonium hydroxide, and incubate it at 55 °C for 8 hr to cleave the oligonucleotide from the CPG and remove the protecting groups from the bases and phosphates.
- Evaporate ammonium hydroxide with a centrifugal evaporator at 40 °C and re-dissolve the oligonucleotide in 0.2 ml of 0.1 M n-hexylammonium acetate.
- Purify the synthetic oligonucleotide through reverse-phase high-performance liquid chromatography using a polystyrene-divinylbenzene column with a buffer containing 10-50% acetonitrile/0.1 M n-hexylammonium acetate. Run the column at 2.0 ml/min for 25 min at 80 °C.
- Add 0.25-0.5 ml of concentrated acetic acid to the fractionated sample (1-2 ml) to make a 20% acetic acid solution, and incubate this solution for 1 hr at room temperature to remove the DMT group.
- Dry the sample with a centrifugal evaporator at 40 °C, re-dissolve it in 1 ml of 29% ammonium hydroxide, and incubate the solution for 15 min at room temperature to remove the side chain of the 5′ phosphate.
- Reduce the volume (1 ml) of the sample to about 0.3 ml with a centrifugal evaporator at 40 °C.
- Dialyze the entire concentrated sample against distilled water (500 ml) using a dialysis tube of molecular weight cut-off 1,000 at 4 °C overnight.
- Transfer the dialyzed sample from the dialysis tube to a 1.5-ml tube with a pipette, dry it with a centrifugal evaporator at 40 °C, and subsequently re-dissolve it in water-for-injection-grade water, the volume of which should be small enough to make a greater than 100 µM solution.
- Measure an optical density of the RNA sample at 260 nm with a spectrophotometer and adjust its concentration to 100 µM by adding water-for-injection-grade water. Store the RNA solution at below -20 °C before use.
2. Screening of the sgRNA Library for Cell Viability
- Prepare 103 cells/99 μl/well (e.g., HL60 and RPMI-8226) on a 96-well dish in RPMI-1640 media supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. Prepare wells containing the media only for background subtraction. To eliminate edge effects, add sterile water to empty wells surrounding the sample wells.
- Add 1 μl of heptamer-type sgRNA (100 μM) dissolved in water-for-injection-grade water to each well. Assay each sgRNA in triplicate. Incubate the plate at 37 °C in a 5% CO2 humidified incubator for 3 days.
- Add an appropriate volume of a commercially available WST-8 solution to each well. Incubate the plate at 37 °C in the same incubator for 1 to 4 hr.
- Measure absorbance at 450 nm with a microplate reader.
NOTE: In most cases, linearity between absorbance and viable cell number appears to be maintained unless the absorbance value exceeds 1.0.
3. Evaluation of Effective sgRNAs by Flow Cytometry
- Prepare 103 cells/99 µl/well (e.g., HL60) on a 96-well dish in RPMI-1640 media supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. Prepare at least 10 wells for one sgRNA to be tested. To eliminate edge effects, add sterile water to empty wells surrounding the sample wells.
- Add 1 µl of heptamer-type sgRNA (100 µM) dissolved in water-for-injection-grade water to each well. Incubate the plate at 37 °C in a 5% CO2 humidified incubator for 3 days.
- Collect the cells treated with the same sgRNA from at least 10 wells into a 5-ml tube with a pipette. Spin the tube for 5 min in a centrifuge at 300 x g, and discard the media.
- Add 2 ml of 1x phosphate buffered saline (PBS) to the tube, and re-suspend the cells. Spin the tube for 5 min in a centrifuge at 300 x g, and discard PBS. Add 150 µl of 1x annexin V binding buffer to the tube, and re-suspend the cells.
- Add 2 µl of phycoerythrin (PE)-conjugated annexin V solution and 1 µl of 7-aminoactinomycin D (7AAD) solution to the tube, mix them well, and incubate the tube for 15 min at room temperature in the dark. Analyze the sample using a flow cytometer24.
TRUE Gene Silencing: Screening of a Heptamer-type Small Guide RNA Library for Potential Cancer Therapeutic Agents
Learning Objectives
The six heptamer-type sgRNAs 5′-AUCUUCA-3′ (H1885), 5′-ACACACA-3′ (H3277), 5′-GGGGGCG-3′ (H10927), 5′-GGGGCCC-3′ (H10944), 5′-GCCCCCG-3′ (H12287), and 5′-CACCAGC-3′ (H13260) were chemically synthesized as 2′-O-methyl RNA containing 5′- and 3′-phosphates.
These sgRNAs were examined for the effects on viability of a human leukemia cell line, HL60, and a human myeloma cell line, RPMI-8226. Representative results of the cell viability assays are shown in Figure 1. The four sgRNAs H1885, H3277, H10927, and H13260 were very effective and reduced the viability of HL60 and RPMI-8226 cells by >80% and >70%, respectively. The heptamers H3277 and H10927 were also tested for the effects on non-cancerous HEK293 cell viability, and have been shown to hardly affect the cell viability22.
The flow cytometry with annexin V and 7AAD double staining was performed for HL60 cells treated with the heptamer H1885, H3277, H10927, or H13260. In each sgRNA, a total cell number (56-95%) in early and late apoptotic stages was much higher than that (4-6%) in mock (Figure 2). These observations suggest that the reduction of HL60 cell viability is due to apoptosis.
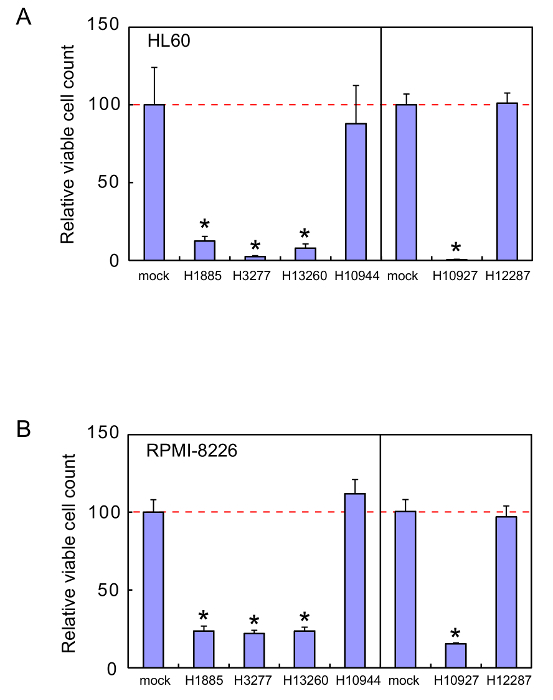
Figure 1. Cell viability assays. Cell viability was measured 72 hr after HL60 cells (A) or RPMI-8226 cells (B) were cultured in the absence or presence of 1 µM of the naked heptamer-type sgRNA H1885, H3277, H10927, H10944, H12287, or H13260. The relative living cell numbers in the absence of sgRNAs are adjusted to 100. Error bars indicate SD (n = 3). *, P < 0.002. Reprinted from Leukemia Research, Vol. 38, Takahashi M. et al., Screening of a heptamer-type sgRNA library for potential therapeutic agents against hematological malignancies, pp 808-815, Fig. 1, Copyright (2014), with permission from Elsevier. Please click here to view a larger version of this figure.
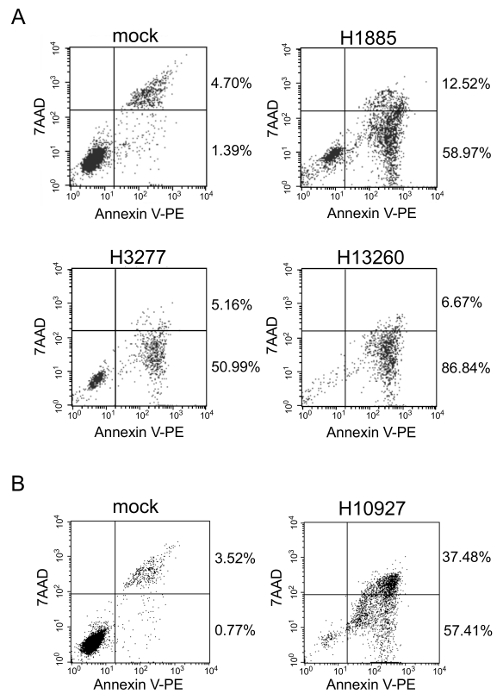
Figure 2. Flow cytometry. With respect to HL60 cells, flow cytometry was performed with annexin V and 7AAD double staining. The cells were analyzed 72 hr after cultured in the absence or presence of 1 µM of the naked heptamer-type sgRNA H1885 (A), H3277 (A), H13260 (A), or H10927 (B). Reprinted from Leukemia Research, Vol. 38, Takahashi M. et al., Screening of a heptamer-type sgRNA library for potential therapeutic agents against hematological malignancies, pp 808-815, Fig. 2, Copyright (2014), with permission from Elsevier. Please click here to view a larger version of this figure.
List of Materials
| Custom heptamer RNA | Nippon Bioservice | ||
| OligoSep Prep HC Cartridge | Transgenomic | 99-3860 | |
| Water-for-injection-grade Water | Otsuka Pharmaceutical | 7131400A2129 | |
| RPMI-1640 medium | Wako | 189-02025 | |
| Fetal Bovine Serum | SIGMA-ALDRICH | 172012-500ML | |
| Penicillin-Streptomycin Mixed Solution | Nacalai Tesque | 26253-84 | |
| 96 Well Plate | Greiner Bio-one | 655 180 | |
| Cell Counting Kit-8 | DOJINDO | 343-07623 | WST-8 solution |
| Microplate Reader, Sunrise Thermo RC-R | TEKAN | 510-82851 | |
| FACS Round-Bottom Tube | BD Falcon | 60819-820 | |
| Phosphate Buffered Saline | SIGMA-ALDRICH | P7059-1L | |
| PE Annexin V Binding Buffer | BD Biosciences | 51-66121E | |
| PE Annexin V | BD Biosciences | 51-65875X | |
| 7AAD | SIGMA-ALDRICH | A9400-1MG | |
| Flow Cytometer, FACSCalibur | BD Biosciences | 342973 |
Preparação do Laboratório
TRUE gene silencing (termed after tRNase ZL–utilizing efficacious gene silencing) is one of the RNA-directed gene silencing technologies, which utilizes an artificial small guide RNA (sgRNA) to guide tRNA 3′ processing endoribonuclease, tRNase ZL, to recognize a target RNA. sgRNAs can be taken up by cells without any transfection reagents and can downregulate their target RNA levels and/or induce apoptosis in human cancer cells. We have screened an sgRNA library containing 156 heptamer-type sgRNAs for the effect on viability of human myeloma and leukemia cells, and found that 20 of them can efficiently induce apoptosis in at least one of the cancer cell lines. Here we present a protocol for screening of a heptamer-type sgRNA library for potential therapeutic drugs against blood cancers. The protocol includes how to construct the sgRNA library, how to assess the effect of each sgRNA on cell viability, and how to further evaluate the effective sgRNAs by flow cytometry. Around 2,000 hits would be expected to be obtained by screening the full-scale sgRNA library composed of 16,384 heptamers.
TRUE gene silencing (termed after tRNase ZL–utilizing efficacious gene silencing) is one of the RNA-directed gene silencing technologies, which utilizes an artificial small guide RNA (sgRNA) to guide tRNA 3′ processing endoribonuclease, tRNase ZL, to recognize a target RNA. sgRNAs can be taken up by cells without any transfection reagents and can downregulate their target RNA levels and/or induce apoptosis in human cancer cells. We have screened an sgRNA library containing 156 heptamer-type sgRNAs for the effect on viability of human myeloma and leukemia cells, and found that 20 of them can efficiently induce apoptosis in at least one of the cancer cell lines. Here we present a protocol for screening of a heptamer-type sgRNA library for potential therapeutic drugs against blood cancers. The protocol includes how to construct the sgRNA library, how to assess the effect of each sgRNA on cell viability, and how to further evaluate the effective sgRNAs by flow cytometry. Around 2,000 hits would be expected to be obtained by screening the full-scale sgRNA library composed of 16,384 heptamers.
Procedimento
TRUE gene silencing (termed after tRNase ZL–utilizing efficacious gene silencing) is one of the RNA-directed gene silencing technologies, which utilizes an artificial small guide RNA (sgRNA) to guide tRNA 3′ processing endoribonuclease, tRNase ZL, to recognize a target RNA. sgRNAs can be taken up by cells without any transfection reagents and can downregulate their target RNA levels and/or induce apoptosis in human cancer cells. We have screened an sgRNA library containing 156 heptamer-type sgRNAs for the effect on viability of human myeloma and leukemia cells, and found that 20 of them can efficiently induce apoptosis in at least one of the cancer cell lines. Here we present a protocol for screening of a heptamer-type sgRNA library for potential therapeutic drugs against blood cancers. The protocol includes how to construct the sgRNA library, how to assess the effect of each sgRNA on cell viability, and how to further evaluate the effective sgRNAs by flow cytometry. Around 2,000 hits would be expected to be obtained by screening the full-scale sgRNA library composed of 16,384 heptamers.
