Purification of the Membrane Compartment for Endoplasmic Reticulum-associated Degradation of Exogenous Antigens in Cross-presentation
PREPARAÇÃO DO INSTRUTOR
CONCEITOS
PROTOCOLO DO ALUNO
1. Growing Cells and Addition of Exogenous Antigens
- Prepare bOVA using a biotin-protein labeling kit following the manufacturer's protocol.
NOTE: Ordinarily, bOVA contains 2 M biotin per 1 M OVA on average. - Grow DC2.4 cells in RPMI-1640 supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 100 U/mL penicillin-streptomycin, 55 mM 2-mercaptoethanol, 10 mM HEPES (pH 7.5), and 10% fetal calf serum (hereafter RPMI) at 37 °C in 5% CO2 in a humidified incubator (hereafter without mention, cells are incubated at this condition). It is also possible to use DMEM supplemented with 10% fetal calf serum, 3.7 g/L NaHCO3 (hereafter DMEM) in place of RPMI.
- A day before purification, split the cells into RPMI to 1 x 105 cells/mL in a tissue culture plate. Avoid keeping the cells at a confluent state. DC2.4 cells can highly incorporate exogenous antigens until a semi-confluent state, but rapidly lose the ability after reaching an over-confluent state.
- Before the DC2.4 cells are semi-confluent, add 250 µg/mL of bOVA for 1 x 106 cells/mL and incubate for 2 – 4 h.
NOTE: During downstream experiments, all buffers and reagents are kept at 4 °C unless otherwise indicated.
2. Preparation of Microsomes
- Harvest the DC2.4 cells by gentle pipetting from the tissue plate into a new 50 mL conical tube.
- Centrifuge at 1,000 x g for 5 min, 4 °C and carefully remove the medium with bOVA by aspiration.
- Wash the DC2.4 cells twice with 40 mL of PBS buffer (1.37 mM NaCl, 8.1 mM Na2HPO4, 2.68 mM KCl, 1.47 mM KH2PO4) by centrifugation at 1,000 x g for 5 min, 4 °C and discard the supernatant each time by aspiration.
- Resuspend the pellet in step 2.3 in 1 – 2 mL (1/20-1/10 volume of culture medium) of homogenization medium (0.25 M sucrose, 1 mM EDTA, 10 mM HEPES-NaOH [pH 7.4]) with 1/1,000 volume of protease inhibitor cocktails and transfer the resuspended DC2.4 cells into an ice-cold Dounce homogenizer.
- Disrupt the resuspended DC2.4 cells gently by 10 – 20 strokes with the ice-cold Dounce homogenizer.
NOTE: During the disruption step, the Dounce homogenizer is cooled on ice. - Transfer the cell-disrupted suspension to a new 15 mL conical tube.
- Add 8 – 9 mL homogenization medium to 10 mL total and centrifuge the conical tube for 10 min at 2,000 x g and 4 °C.
- Transfer the supernatant to a new 15-mL conical tube to remove unbroken cells and nuclei, and centrifuge the conical tube again for 10 min at 2,000 x g and 4 °C.
- Transfer the supernatant into a new ultracentrifugation tube and centrifuge for 45 min at 100,000 x g and 4 °C.
- Aspirate the supernatant and resuspend the pellet carefully in 1 – 2 mL of homogenization medium with 1/1,000 volume of protease inhibitor cocktails by pipetting the solution up and down several times to make a microsome fraction for downstream experiments.
- Transfer the microsome fraction into a new 5-mL round bottom tube.
NOTE: After this step, it is possible to enrich objective microsomes by iodixanol density gradient centrifugation according to the manufacturer's protocol, to remove non-specific microsomes. The peak fractions for exogenous antigens are collected and then subjected to the next step of purification (step 3).
3. Purification of Microsomes with bOVA Undergoing ERAD
- Add 1/100 volume of fresh SA-magnetic beads to the microsome fraction of step 2.11 in the 5 mL round bottom tube.
NOTE: Before this step, it is possible to pre-clear the microsome fraction by control-magnetic beads to reduce contaminations of non-specific microsomes. - Gently mix well and rotate the round bottom tube slowly for 30 min at 4 °C.
- Add 2-3 mL of homogenization medium to 4 mL total into the round bottom tube and gently mix well.
- Place the round bottom tube on a magnetic stand and incubate for 10 min at 4 °C. Since the beads bound to the vesicles are attracted to the magnet, brown micro-beads will gradually accumulate to the tube wall closest to the magnet.
- With the tube remaining in the magnetic stand, carefully discard the supernatants by aspiration to remove the unbound vesicles.
- Wash the magnetic beads bound to the vesicles twice with 5 mL of homogenization medium by the magnetic stand for 10 min at 4 °C and discard the supernatant each time by carefully aspiration.
NOTE: Without the magnetic stand, purification by centrifugations at 2,000 x g for 10 min, 4 °C is also available. - Resuspend the magnetic beads bound to the vesicles carefully in 100 µL homogenization medium by pipetting the solution up and down several times.
- Transfer the resuspended vesicles into a new microtube as the purified microsome for downstream experiments.
NOTE: Typically, 50 µL microsome fraction containing 5 – 20 µg proteins from 1 x 107 cells can be isolated.
4. Analysis of the Purified Microsomes
- Resuspend the magnetic beads bound to the vesicles from step 3.8 by 100 – 200 µL of TNE buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5 M EDTA, 1% Nonidet P-40) with 1/1,000 volume of protease inhibitor cocktails instead of the homogenization medium.
- Transfer the lysate from step 4.1 to a new 1.5-mL microtube.
- Determine the protein concentration of step 4.2 by using a BCA assay kit per the manufacturer's protocol.
NOTE: Typically, 5 – 10 µL lysate from step 4.2 is enough to determine the protein concentration. - Transfer the lysate from step 4.2 (2 µg of protein for silver staining and 10 µg of protein for Western blotting) into new microtubes.
- Put the microtubes on a heat block at 95 °C and boil proteins in 1x SDS gel-loading buffer (100 mM Tris-HCl pH 6.8, 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol) for 5 min.
- Centrifuge the microtubes for 10 min at 21,500 x g and 4 °C. Collect the supernatants into new microtubes to remove the insoluble fractions.
- Analyze the resolved proteins in step 4.6 (2 µg) by standard SD-PAGE.
- Visualize the protein bands by the silver staining using silver staining kit following the manufacturer's protocol.
- Analyze the resolved proteins in step 4.6 (10 µg) by standard SD-PAGE and Western-blotting. Use the reagent (SA-HRP) per the manufacturer's protocol and visualize by fluorography.
5. In Vitro Reconstitution of ERAD Ubiquitination of bOVA Using Purified Microsomes
- Transfer the purified microsomes (5 – 10 µg of protein) from step 3.8 with a 50% volume of reticulocyte lysate (RL), in 1x reaction buffer (50 mM Tris pH 7.4, 3 mM ATP, 0.5 mM MgCl2), and 0.2 pM Flag-tagged ubiquitin in a new microtube. The final volume of this experiment is 20 – 40 µL.
- Incubate the microtube for 2 h at 37 °C.
NOTE: If the amount of ubiquitinated bOVA in step 5.12 is too small to detect by Western blotting, add 10 µM of MG132 or 2 µM of lactacystin to inhibit the processing of the ubiquitinated bOVA by proteasomes. - Stop the reaction by placing the microtube on ice.
- Resolve the microsomes by adding 500 µL TNE buffer with 1/1,000 volume of protease inhibitor cocktails.
- Centrifuge the microtube for 10 min at 21,500 x g and 4 °C. Collect the supernatants into a new microtube to remove the insoluble fractions, which contain ubiquitinated bOVA in DC2.4 before the in vitro ubiquitination assay.
- Transfer the supernatant to a new microtube and add 1/100 volume of new SA-magnetic beads (usually 5 µL for 500 µL of supernatants).
- Gently mix well and rotate the microtube slowly for 30 min at 4 °C.
- Centrifuge the microtube for 10 min at 21,500 x g and 4 °C. Discard the supernatant by aspiration to recover the bOVA and ubiquitinated bOVA bound with the SA-magnetic beads.
- Wash twice the collected SA-magnetic beads with 1 mL of TNE buffer by centrifugation for 10 min at 21,500 x g and 4 °C. Discard the supernatant each time by aspiration.
- Boil the SA-magnetic beads in 1x SDS gel loading buffer for 5 min at 95 °C on a heat-block to resolve the purified proteins by the SA-magnetic beads.
- Centrifuge the microtube for 10 min at 21,500 x g and 4 °C. Collect the supernatants into a new microtube to remove the insoluble fractions.
- Analyze the SA-magnetic beads bound to the proteins by standard SD-PAGE and Western-blotting. The use of antibodies (anti-Flag, anti-multi-Ub, and anti-mouse IgG-HRP) and the reagent (SA-HRP) is per the manufacturer's protocol and visualized by fluorography.
Purification of the Membrane Compartment for Endoplasmic Reticulum-associated Degradation of Exogenous Antigens in Cross-presentation
Learning Objectives
To elucidate the molecular mechanism of CP, it is necessary to identify the cellular compartments, where exogenous antigens undergo ERAD-like transport and processing. While observations by immunofluorescent microscopy or by electron microscopy identified the cellular compartment where exogenous antigens accumulated16,17,18,19,30,31,32,33, the cellular compartments for ERAD-like processing of exogenous antigens are not clearly defined. Recently it was shown that non-classical endosomes with ER resident molecules were responsible for CP34, but these cellular compartments were unpurified. The difficulty in isolating and purifying the cellular compartments can be attributed to the fact that exogenous antigens are localized both in the endosome and ER-like compartments and that there is no identifying molecule for these endocytic compartments other than exogenous antigens. However, the condition of exogenous antigens undergoing ERAD-like transport is different from the steady state; in the transport across lipid bimolecular membrane, exogenous antigens penetrate the membrane via translocons, such as Sec61 (Figure 2A). Thus, when using bOVA as an exogenous antigen, bOVA should be associated with Sec61. Since membrane-associated bOVA specifically bound with SA, the microsome prepared from DC2.4, which was pretreated with bOVA, could be isolated distinctively by SA-magnetic beads (Figure 2A). Then equivalent amounts of proteins from purified microsomes with or without pre-incubation of bOVA were resolved by SDS-PAGE followed by silver staining and Western blotting with SA-HRP (Figure 2B, 2C). As shown in Figure 2B and Figure 2C, the isolated microsomes contained several unique proteins that were purified dependent upon exogenously added bOVA and SA-magnetic beads. In addition to these unique proteins, isolated microsomes also contained nonspecific proteins, which bound to SA-magnetic beads with or without bOVA. Treatment of the microsome with trypsin before purification by the magnet prevented the purification of microsomes (Figure 2D), indicating that the purification methods depended on the presence of membrane-penetrating bOVA.
The purified microsomes showed the ability to ubiquitinate the incorporated bOVA in vitro, under the presence of RLs (Figure 3A). The amounts of bOVA and poly-ubiquitinated bOVA were augmented in the presence of MG132 (Figure 3B), indicating that the incorporated bOVA was processed by the ERAD system and that our purified microsomes contained ERAD machinery proteins.
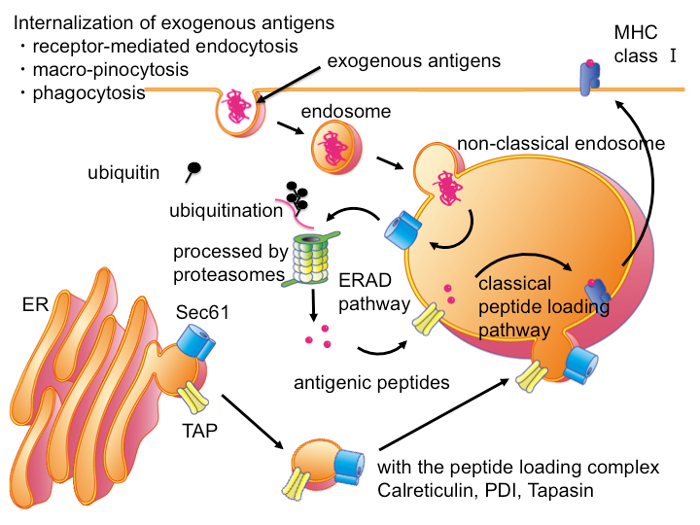
Figure 1: Intracellular Pathways for CP in DCs. In DCs, exogenous antigens are transported into non-classical endocytic compartments, which also contain ER-resident molecules in addition to molecules of the classical late endosome. In this compartment, exogenous antigens are exported into cytosol through translocons such as Sec61. In the cytosol, exogenous antigens are processed by the ubiquitin-proteasome system into antigenic peptides as ERAD substrates. Antigenic peptides are transported into same or adjacent non-classical endocytic compartments, or adjacent ER through TAP transporter and then loaded on the MHC I molecules by PLC. Please click here to view a larger version of this figure.
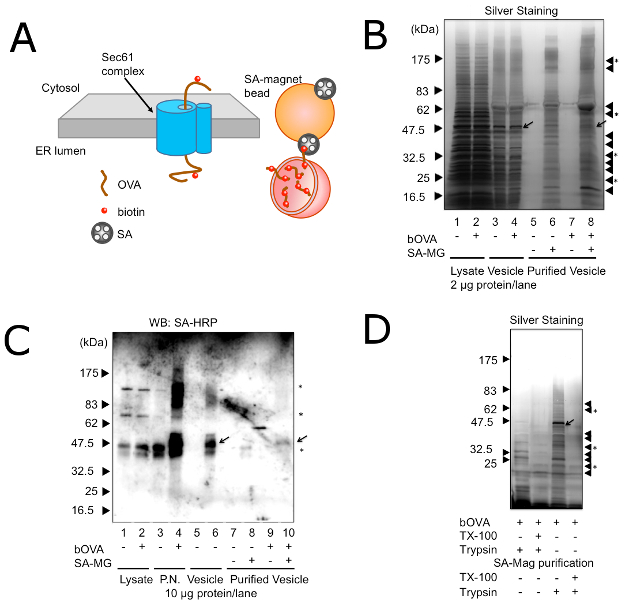
Figure 2: Purification of Microsomes with bOVA Undergoing ERAD. (A) A schematic model of purification of microsomes with bOVA undergoing ERAD. bOVA is associated with the membrane through the Sec61 translocon and targeted with SA-magnetic beads. (B) Microsomes with (+) or without (-) prior addition of bOVA were purified with (+) or without (-) SA-magnetic beads. Proteins (2 µg) or corresponding volumes of purified proteins were resolved on 7.5 – 15% SDS-PAGE, and silver staining was used to visualize protein bands. Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Triangles with asterisks indicate unique proteins found only in the presence of exogenously added bOVA and SA-magnetic beads. The arrow shows bOVA. (C) Microsomes with (+) or without (-) prior addition of bOVA were purified with (+) or without (-) SA-magnetic beads. Proteins (10 µg) or corresponding volumes of purified proteins were resolved on 7.5 – 15% SDS-PAGE, and subjected to Western blotting with SA-HRP. P.N.: post-nuclear fraction. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained by at least three independent assays. (D) Microsomes with prior addition of bOVA were purified with SA-magnetic beads. Microsomes were treated with (+) or without (-) trypsin and TX-100 before purification (left two lanes) or after purification (right two lanes). Proteins (2 µg) or corresponding volumes of purified proteins were resolved on 7.5 – 15% SDS-PAGE, and silver staining was used to visualize protein bands. Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Triangles with asterisks indicate unique proteins found only in the presence of exogenously added bOVA and SA-magnetic beads. Equivalent results were attained by at least three independent assays. Reprinted with permission from reference28. Please click here to view a larger version of this figure.
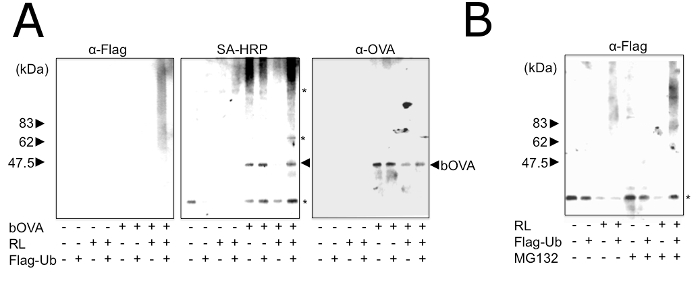
Figure 3: In Vitro Reconstitution of Processing and Ubiquitination using OVA in Purified Microsomes. (A) Purified microsomes with (+) or without (-) prior addition of bOVA were treated with (+) or without (-) RL and Flag-Ub for 1 h and were solubilized using TNE. bOVA was purified with SA-magnetic beads and subjected to Western blotting with the indicated antibodies. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained by at least three independent assays. (B) Purified microsomes were treated with (+) or without (-) RL, Flag-Ub, and MG132 for 1 h and were then solubilized using TNE. bOVA was purified with SA-magnetic beads and subjected to Western blotting with SA-Flag. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained by at least three independent assays. Reprinted with permission from reference28. Please click here to view a larger version of this figure.
List of Materials
| RPMI 1640 | gibco by life technologies | 11875-093 | |
| Fetal bovine serum | Equitech bio | SFB30 | |
| Sodium pyruvate | gibco by life technologies | 11360-070 | |
| MEM non-essential amino acids | gibco by life technologies | 11140-050 | |
| HEPES | gibco by life technologies | 15630-080 | |
| 2-mercaptoethanol | gibco by life technologies | 21985-023 | |
| L-glutamine | gibco by life technologies | 25030-164 | |
| Penisicillin-Sreptomycin | gibco by life technologies | 15140-122 | |
| DMEM | gibco by life technologies | 12100-46 | |
| OVA | SIGMA | A5503 | |
| Biotin-protein labelling kit | Thermo Fisher Scientific | F6347 | |
| MG-132 | Santa Cruz Biotechnology | 201270 | |
| lactacystin | SIGMA | L6785 | |
| Dounce homogenizer | IUCHI | 131703 | |
| protease inhibitor cocktails | SIGMA | P8340 | |
| iodixanol | Cosmo bio | 1114542 | |
| SA-magnetic beads | New England Biolabs | 201270 | |
| control magnetic beads | Chemagen | M-PVA012 | |
| magnetic stand | BD Biosciences | 552311 | |
| BCA protein assay kit | Thermo Fisher Scientific | 23225 | |
| silver staining kits | Cosmo bio | 423413 | |
| Reticulocyte Lysate | Promega | 1730714 | |
| Flag-tagged ubiquitin | SIGMA | U5382 | |
| anti-ovalbumin (OVA,mouse) | Antibody Shop | HYB 094-06 | |
| ant-multi-ubiquitin (mouse) | MBL | D058−3 | |
| anti-Flag (mouse) | SIGMA | F3165 | |
| trypsin | SIGMA | 85450C |
Preparação do Laboratório
Dendritic cells (DCs) are highly capable of processing and presenting internalized exogenous antigens upon major histocompatibility class (MHC) I molecules also known as cross-presentation (CP). CP plays an important role not only in the stimulation of naïve CD8+ T cells and memory CD8+ T cells for infectious and tumor immunity but also in the inactivation of self-acting naïve T cells by T cell anergy or T cell deletion. Although the critical molecular mechanism of CP remains to be elucidated, accumulating evidence indicates that exogenous antigens are processed through endoplasmic reticulum-associated degradation (ERAD) after export from non-classical endocytic compartments. Until recently, characterizations of these endocytic compartments were limited because there were no specific molecular markers other than exogenous antigens. The method described here is a new vesicle isolation protocol, which allows for the purification of these endocytic compartments. Using this purified microsome, we reconstituted the ERAD-like transport, ubiquitination, and processing of the exogenous antigen in vitro, suggesting that the ubiquitin-proteasome system processed the exogenous antigen after export from this cellular compartment. This protocol can be further applied to other cell types to clarify the molecular mechanism of CP.
Dendritic cells (DCs) are highly capable of processing and presenting internalized exogenous antigens upon major histocompatibility class (MHC) I molecules also known as cross-presentation (CP). CP plays an important role not only in the stimulation of naïve CD8+ T cells and memory CD8+ T cells for infectious and tumor immunity but also in the inactivation of self-acting naïve T cells by T cell anergy or T cell deletion. Although the critical molecular mechanism of CP remains to be elucidated, accumulating evidence indicates that exogenous antigens are processed through endoplasmic reticulum-associated degradation (ERAD) after export from non-classical endocytic compartments. Until recently, characterizations of these endocytic compartments were limited because there were no specific molecular markers other than exogenous antigens. The method described here is a new vesicle isolation protocol, which allows for the purification of these endocytic compartments. Using this purified microsome, we reconstituted the ERAD-like transport, ubiquitination, and processing of the exogenous antigen in vitro, suggesting that the ubiquitin-proteasome system processed the exogenous antigen after export from this cellular compartment. This protocol can be further applied to other cell types to clarify the molecular mechanism of CP.
Procedimento
Dendritic cells (DCs) are highly capable of processing and presenting internalized exogenous antigens upon major histocompatibility class (MHC) I molecules also known as cross-presentation (CP). CP plays an important role not only in the stimulation of naïve CD8+ T cells and memory CD8+ T cells for infectious and tumor immunity but also in the inactivation of self-acting naïve T cells by T cell anergy or T cell deletion. Although the critical molecular mechanism of CP remains to be elucidated, accumulating evidence indicates that exogenous antigens are processed through endoplasmic reticulum-associated degradation (ERAD) after export from non-classical endocytic compartments. Until recently, characterizations of these endocytic compartments were limited because there were no specific molecular markers other than exogenous antigens. The method described here is a new vesicle isolation protocol, which allows for the purification of these endocytic compartments. Using this purified microsome, we reconstituted the ERAD-like transport, ubiquitination, and processing of the exogenous antigen in vitro, suggesting that the ubiquitin-proteasome system processed the exogenous antigen after export from this cellular compartment. This protocol can be further applied to other cell types to clarify the molecular mechanism of CP.
