Identification of RNAs Engaged in Direct RNA-RNA Interaction with a Long Non-Coding RNA
Summary
An easy-to-use RNA pull-down protocol is designed for the identification of RNAs engaged in direct RNA/RNA interaction with a long non-coding RNA. The protocol uses psoralen as a fixative to cross-link only RNA/RNA interactions and provides the whole direct RNA interactome of a long non-coding RNA when coupled with RNA sequencing.
Abstract
The growing role attributed nowadays to long non-coding RNAs (lncRNA) in physiology and pathophysiology makes it crucial to characterize their interactome by identifying their molecular partners, DNA, proteins and/or RNAs. The latter can interact with lncRNA through networks involving proteins, but they can also be engaged in direct RNA/RNA interactions. We, therefore, developed an easy-to-use RNA pull-down procedure that allowed identification of RNAs engaged in direct RNA/RNA interaction with a lncRNA using psoralen, a molecule that cross-links only RNA/RNA interactions. Bioinformatics modeling of the lncRNA secondary structure allowed the selection of several specific antisense DNA oligonucleotide probes with a strong affinity for regions displaying a low probability of internal base pairing. Since the specific probes that were designed targeted accessible regions throughout the length of the lncRNA, the RNA-interaction zones could be delineated in the sequence of the lncRNA. When coupled with a high throughput RNA sequencing, this protocol can be used for the whole direct RNA interactome studies of a lncRNA of interest.
Introduction
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts longer than 200 nucleotides in length. Their number is ever increasing, more than 58,000 in humans. Furthermore, their crucial role in physiology and pathophysiology makes it essential to characterize their molecular partners that allow them to implement their regulatory functions. Actually, one approach to understand the functions of lncRNAs is the detection of the interacting molecular partners of each lncRNA.
The molecular targets of lncRNAs can be DNA, proteins, or RNAs, and various techniques have been developed to identify them. In this regard, the identification of proteins interacting with lncRNAs is the objective of various protocols, including different RNA-immunoprecipitation (RIP) procedures in which an antibody against the protein of interest is used to specifically pull down lncRNAs (for a review1). Other techniques such as Capture Hybridization Analysis of RNA Targets2 (CHART) or Chromatin Isolation by RNA Purification3 (ChIRP) allow to pull-down lncRNAs together with the associated protein complexes. In these latter techniques, the lncRNA is used as bait. CHART and ChIRP are also powerful techniques to identify genomic maps showing lncRNA occupancy2,3. In addition, these RNA pull-down methods make it possible to identify the RNA partners of a lncRNA. A procedure that allows the capture of RNAs targeted by a lncRNA has been described. Antisense DNA biotinylated oligonucleotide probes are designed against regions of low probability of internal base pairing as determined by bioinformatics modeling of the lncRNA secondary structure4. However, this procedure does not discriminate whether the RNA partners interact indirectly via a protein network or directly via direct RNA/RNA interactions with the lncRNA. It has been shown that cross-linking with psoralen derivatives is the method of choice to select direct RNA/RNA interactions mediated by base-pairing5. Psoralen derivatives are indeed able to intercalate into double-stranded DNA or RNA and to covalently link pyrimidines after irradiation with UV light (365 nm)6. Coupling the RNA pull-down using biotinylated oligonucleotide probes with the cross-linking with psoralen is an easy-to-use procedure proposed here for the identification of the RNAs engaged in direct RNA/RNA interactions with a lncRNA. Moreover, this procedure can allow delineating the RNA-interaction zones in the sequence of the lncRNA if different oligonucleotide probes designed along the length of the lncRNA are used.
Protocol
1. Probe design
- Generate the secondary structure of the lncRNA using a specialized free web server software : RNAstructure software7 or Vienna RNA web suite8. Select regions that display a low probability of internal base pairing and design 25 bases long antisense oligonucleotide probes for different regions of this lncRNA.
- Check all the oligonucleotides designed with the free academic software, AmplifX (https://inp.univ-amu.fr/en/amplifx-manage-test-and-design-your-primers-for-pcr), in order to obtain different information on these probes and to choose those with the best parameters.
NOTE: Check for optimal parameters on the software as following. Percentage of GC: between 40 and 60%, 3' stability, complexity, self-end dimer: "good". Melting temperature (TM) is not important because the affinity is performed at room temperature. - Using an alignment search tool (i.e., Blast, etc.), ensure that the selected antisense oligonucleotide probes do not recognize nucleotide sequences in any other RNA expressed in the cells of choice.
- Design a 25 bases long non-specific DNA oligonucleotide probe. Ensure this probe neither shows an affinity for the lncRNA of interest nor any other RNA sequences in the genome of interest.
- Order all probes with a biotin modification added to their 3'-end along with a triethyleneglycerol spacer (TEG). This spacer increases the oligo-biotin distance in order to avoid steric hindrance.
NOTE: To obtain an optimal result and assess the specificity of results of the pull-down, design a minimum of two different antisense oligonucleotide probes in the same region and then compare their efficiency experimentally.
2. Cross-linking
- Grow cells (e.g., GH4C1 cells used in this experiment) until confluence in 78.5 cm2 cell culture dishes. After removing the cell culture medium, rinse the cell layer with 5 mL of cold phosphate-buffered saline enriched with Ca2+ and Mg2+ (PBS+) and add 3 mL of psoralen-derived molecule (4′Aminomethyltrioxsalen hydrochloride) solution prepared in PBS+ at 0.1 mg/mL concentration.
- Place the culture plate for 30 min in a mammalian cell culture incubator at 37 °C with 5% CO2. After 30 min, remove the lid, place culture dishes on ice at 2.5 cm away from 365 nm UV tubes in an UV crosslinker for 10 min. Agitate the dishes delicately and restore the UV exposure for an additional 10 min period.
- Discard media by aspiration, add 1 mL of PBS+ to collect cells with a cell scraper, and transfer to a micro-tube. Centrifuge at 4 °C, 400 x g for 5 min. Remove as much supernatant as possible and stock pellets at -80 °C.
3. Cell lysis
- Prepare Proteinase K buffer by adding 100 mM NaCl, 10 mM Tris-HCl, pH 7.0, 1 mM EDTA, 0.5% SDS and 5 µL/mL RNase inhibitor.
- Resuspend the cell pellets in this buffer (approximately 200 µL per 1 x 107 cells). Add proteinase K to a final concentration of 0.1 µg/µL and incubate for 45 min at 50 °C. Then incubate for 10 min at 95 °C, to inactivate the proteinase K.
4. Sonication
- Distribute 100 µL of this lysate (corresponding to 5.106 cells) in two micro-tubes and add 200 µL of hybridization buffer (700 mM NaCl, 70 mM Tris-HCl, pH 7.0, 1 mM EDTA, 1.25% SDS, 5 µL/mL RNase inhibitor solution and 15% formamide) in each tube.
- Place the tubes in the 4 °C water bath of a sonicator and start the sonication with 2 series of 30 s pulses (high intensity) separated by 30 s off.
- Collect 20 µL of samples to serve as input control samples and store at -80 °C.
NOTE: This sonication procedure was previously optimized for the GH4C1 cell line in order to obtain RNA fragments ranging in 2,000 nucleotides9. For others cell lines, the sonication procedure must be optimized. When this technique is performed to study the RNA partners of a very long non-coding RNA, the sonication step proves to be indispensable to obtain an adequate recovery of the lncRNA. In this case, it is, therefore, necessary to design several probes with a binding site located every 4,000 nucleotides maximum along the lncRNA length. To recover the full length of the lncRNA, use a pool of several specific probes. However, if the purpose is to identify the regions of the lncRNA involved in RNA-RNA binding, the specific probes must be used separately. Furthermore, if studying RNA partners of a smaller lncRNA, experiments should be performed to determine if the sonication step is required.
5. RNA pull-down
- Day 1 – Hybridization step
- Add 150 pmol (1.5 µL of a 100 µM stock solution) of a biotinylated specific or non-specific probe to each sample tube. Incubate for 4 h under agitation on a tube rotator at room temperature (RT) (approximately 30 rpm).
- Take the necessary volume of magnetic beads (i.e., 40 µL of beads per 150 pmol of probes hybridization reaction). Use magnet support to separate beads from commercial media. Discard the supernatant and wash the beads with 900 µL of the Hybridization buffer. Once done, resuspended the magnetic beads in the same volume of the Hybridization buffer.
- Add magnetic beads in each sample tube (40 µL of beads per 150 pmol of probes hybridization reaction). Incubate overnight under agitation on a tube rotator at RT (approximately 30 rpm).
- Day 2 – RNA isolation step
- Separate beads from the cell lysate using magnet support (2-3 min). Discard the supernatant and wash the beads with 900 µL of the Wash buffer (0.5% SDS, 2x SSC) with 5 min incubation under agitation on the rotator at RT. Repeat this wash 5 times.
- After removing the last wash, add 95 µL of Proteinase K buffer on the beads. Defrost the input samples and add 75 µL of Proteinase K buffer. Add 5 µL of proteinase K to a final concentration of 1 µg/µL in all tubes (samples and input samples). Incubate at 45 °C for 45 min and then at 95 °C for 10 min.
- Keep the sample on ice. Use magnetic support to separate beads from RNAs present in the supernatant. Purify RNAs with an RNA purification kit that includes a DNase digestion step. Store purified RNAs at -80 °C.
NOTE: Eluted RNA can be subjected to RT-qPCR or high throughput RNA sequencing to detect enriched transcripts. For RNA sequencing analysis, the reads can then be aligned to the genome of interest using free software, such as STAR10 or HISAT211, and the quantification can be done using FeatureCounts12 or HTSeq count13.
Representative Results
The elucidation of the lncRNA interactome i.e., the cellular components that interact with lncRNAs, proteins, RNA, and DNA, is of key importance for understanding the functions of lncRNAs. Various techniques have been developed to characterize the lncRNA interactome, including RIP, CHART, ChIRP, and RNA pull-down. While the latter has been shown to be powerful in identifying RNA targets of lncRNAs, these procedures do not indicate whether the RNA partners interact indirectly via a protein network or directly via direct RNA/RNA interactions. However, it is now believed, that the mechanisms through which numerous lncRNAs exert their function rely on RNA/RNA interactions with other RNAs, often mRNAs, making crucial the development of methods for genome-wide discovery for these direct RNA targets. Furthermore, these RNA/RNA interactions, mediated by base pairing, can be cross-linked with psoralen derivatives6,5. Therefore, a procedure is proposed to identify RNAs engaged in direct RNA/RNA interactions with a lncRNA (workflow is depicted in Figure 1) and is based on the psoralen cross-linking of the cells coupled with RNA pull-down protocol.
The secondary structure of lncRNAs makes certain regions of lncRNA, particularly those with a low probability of internal base pairing, more capable of being hybridized by specific oligonucleotide probes than others. Then, as a first step of the procedure, the secondary structure of the lncRNA is modeled using the RNAstructure software7 to design oligonucleotides that target accessible regions and can hybridize specifically to lncRNA. Cells were lysed after cross-linking with psoralen and extracts were sonicated to obtain RNA fragments of ~2,000 nucleotides (Figure 1). Due to the length of some lncRNAs, this step is indispensable for their effective pull-down. Sonication of lncRNA generated short fragments that were pulled down using specific antisense probe located up to 2,000 nt upstream and 2,000 nt downstream from the probe binding site (Figure 1). Consequently, fragments corresponding to around 4,000 nt of a lncRNA were pulled with a specific probe (Figure 1). Then, high-throughput RNA sequencing or qPCR analysis can be performed after the RNA pull-down method to obtain the comprehensive list of RNA targets for a lncRNA of interest.
The procedure was applied to the lncRNA, nuclear-enriched abundant transcript one (Neat1). This lncRNA is essential to the formation and structure of nuclear bodies, the paraspeckles, found in the nucleus of the most cultured cells. In addition to Neat1, the paraspeckles contain several RNA binding proteins (RBP)14, and they retain RNA targets within the nucleus15. The nuclear retention of RNA targets by these nuclear bodies, i.e., paraspeckles, may occur through their binding to RBP or directly through their binding to Neat1 utilizing RNA/RNA interactions. To identify the RNAs targeted directly by Neat1 in a rat pituitary cell line, i.e., GH4C1 cells, the procedure whose graphical representation is given in Figure 1 was applied to the pull-down of Neat1 (Figure 2). In order to ensure the pull-down of the entire Neat1 long isoform (21 kb in length in rat), six Neat1-specific biotinylated probes (a-f, Table 1) that can bind to six different Neat1 parts were designed (Figure 2A). The parts of Neat1 pulled down by each probe are shown in Figure 2A. qPCR with specific primers targeting each of the 6 parts of Neat1 (Table 1), showed that each probe (a-f) was specific to the part of Neat1 against which it was designed (see Figure 2B).
In order to pull down the entire length of Neat1, the six probes (a-f) were pooled and named as pool SP1. To assess the specificity of the Neat1 RNA pull-down experiments, another additional pool of six probes (SP2) that were each designed in adjacent corresponding regions of binding of the six a-f probes, was used (Table 1). Every six parts of Neat1 were shown to be effectively pulled down with the SP1 pool, and the SP2 pool of the probes. However, the degree of recovery of each part was not the same (Figure 3A). Furthermore, the use of a non-specific probe (NSP, Table 1) recovered very low Neat1 as compared to the two specific pools SP1 and SP2 (Figure 3A).
To obtain a comprehensive list of direct RNA targets of Neat1 in GH4C1 cells, RNA-sequencing analysis was performed after Neat1 pull-down using the two specific probe pools described above16. It should be mentioned that low concentrations of RNAs recovered after Neat1 pull-down with NSP did not allow for the construction of libraries. Therefore, the lists obtained with the two specific probe pools directed to Neat1 (Table 1) were cross checked to assess the specificity of the results. 1791 RNAs were common in the two pools and corresponds to 70% of RNAs obtained with pool SP1 and 75% of RNAs obtained with pool SP2, respectively16. Some of these mRNAs were assessed with qPCR analysis using specific primers (Table 1). Consistent with results obtained in RNA-sequencing analysis, the transcripts Cntn1, Malat1, Pou1f1 and Rplp0 were found to be directly associated with Neat1 in RT-qPCR experiments (Figure 3B). By contrast, Tapbp a transcript not included in the list of direct RNA targets as determined by RNA-Seq, was not found enriched after RT-qPCR analysis (Figure 3B). The pull-down procedure described here has, therefore, proven to be an effective tool to explore the direct base-paired interactions between Neat1 and its RNA targets.
In addition, by using the 6 probes a-f separately, it was possible to assess the unique regions of Neat1 that were involved in RNA/RNA binding. Indeed, qPCR was performed for the five transcripts mentioned above using each of the 6 probes separately and compared to the non-specific probe (Figure 4). Each value of enrichment was normalized with the corresponding Neat 1 fragment percentage recovery. This was done to ensure that results obtained were independent of the degree of recovery for each Neat1 fragment by its corresponding probe sequences. The 4 mRNAs of Cntn1, Malat1, Pou1f1 and Rplp0 were shown to bind to the Neat1 5' end (Neat1_part1 bound by probe a). On the other hand, Tapbp was never enriched by any of the 6 probes (Figure 4). The procedure allows for the identification of the 5' region of Neat1 as the preferred location for base-paired RNA interactions.
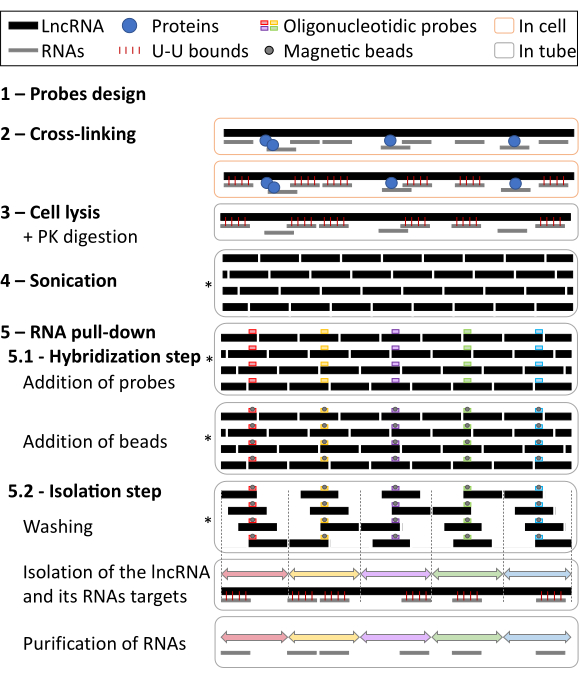
Figure 1: Graphical representation of the procedure. Cells are cross-linked with psoralen derivatives that intercalate into double-stranded RNA and, after irradiation with long-wavelength (365-nm), UV light covalently links pyrimidines on adjacent strands. Cells are then lysed, and proteins digested by Proteinase K. Sonication fragments RNAs to around 2000 nt fragments. Biotinylated specific probes are added for the hybridization with the lncRNA. Magnetic streptavidin beads are then added to separate specific material from the rest of the cell lysate. Beads are then isolated by a magnet and washed several times. After recovery RNAs are purified and used for RT-qPCR or RNA-seq analysis. In the four steps marked with (*), RNAs linked by U-U bounds ( ) are omitted to simplify the graphical representation. Please click here to view a larger version of this figure.
) are omitted to simplify the graphical representation. Please click here to view a larger version of this figure.
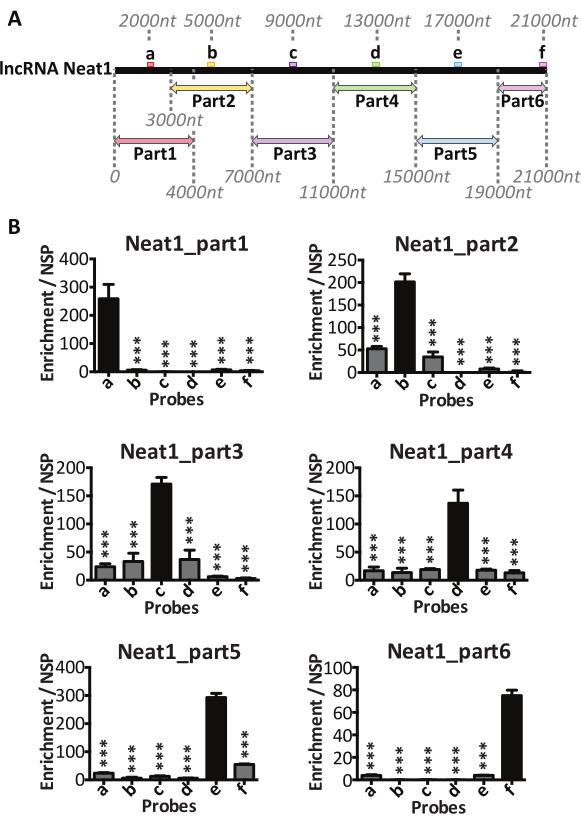
Figure 2: Pull-down of the different parts of Neat1. (A) Schematic representation of the pull-down of 6 parts of Neat1 by 6 specific probes (a-f). Schematic localization of the binding sites for 6 specific antisense oligonucleotides (a-f) designed along the length of Neat1. The different fragments of Neat1 generated by the sonication step, as well as the corresponding parts of Neat1 pulled-down by the different probes (a-f) are shown. (B) This figure has been modified from Jacq et al, 20219 reprinted by permission of Taylor & Francis Ltd, http://www.tandfonline.com. Specific parts of Neat1 pulled-down by each specific probe (a-f). The specific enrichment of the different parts of Neat1 was determined by RT-qPCR after pull-down with each specific probe (a-f) compared to a non-specific probe (NSP).*** p<0.0001 compared to the probe that specifically binds the part of Neat1 pull-down. Please click here to view a larger version of this figure.
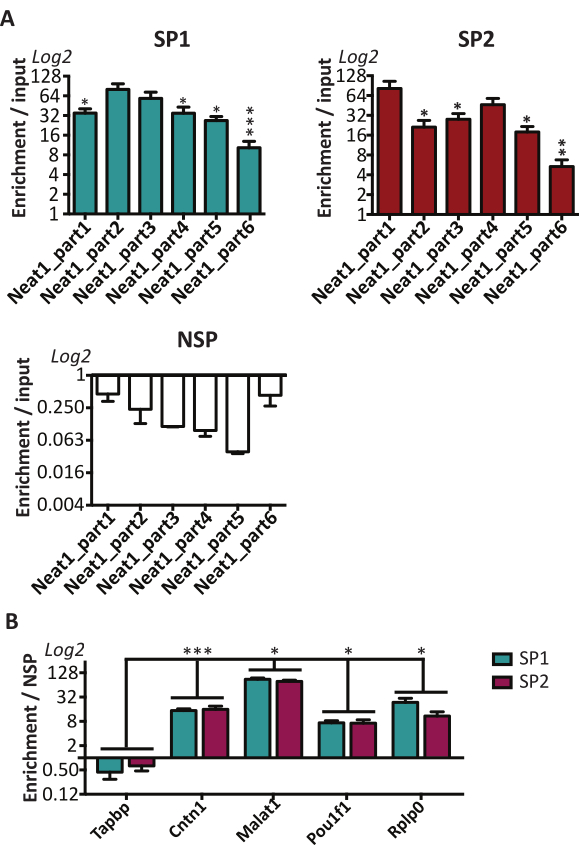
Figure 3: RNAs in direct RNA/RNA interaction with Neat1 identified by RT-qPCR. (A) This figure has been modified from Jacq et al, 20219 reprinted by permission of Taylor & Francis Ltd, http://www.tandfonline.com. The two pools of 6 specific probes, SP1 and SP2, were shown to induce a specific enrichment in every part of Neat1 as compared to a non-specific probe (NSP). However, while every 6 parts of Neat1 were efficiently pulled down both with the pool SP1 and the pool SP2 probes, the efficacy of both pools could differ depending on the part of Neat1 considered. SP1: *p<0.05 ***p<0.0001 compared to the Neat1_part2. SP2: *p<0.05 **p<0.01 compared to the Neat1_part1. (B) Four RNAs selected from the list of direct RNA targets established by RNA-seq were shown to be significantly enriched after Neat1 RNA pull-down with the pool SP1 and SP2 of specific probes relative to a non-specific probe (NSP) in contrast to a non-target RNA. *p<0.05 ***p<0.0001 compared to the non-target RNA (Tapbp). Please click here to view a larger version of this figure.
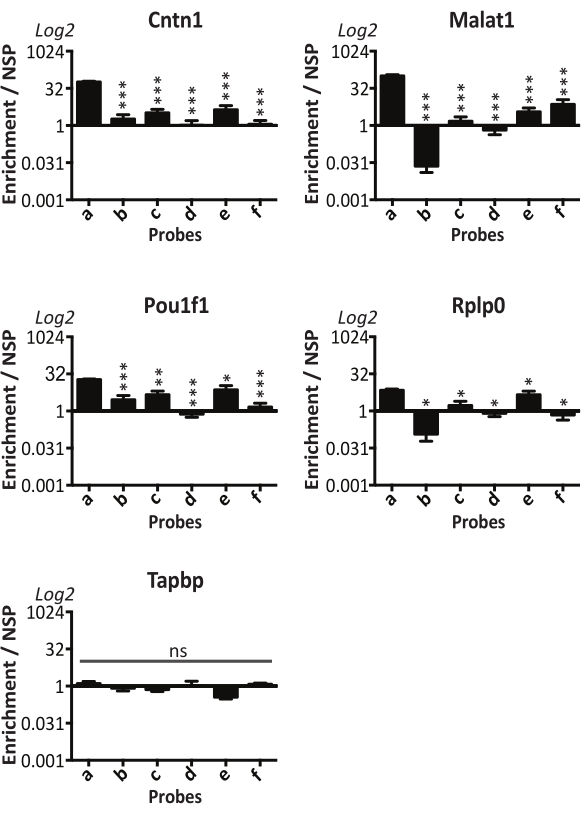
Figure 4: 5' region of Neat1 as the preferential location of base-paired RNA interactions. Each Neat1 specific probe (a-f) is used separately in RNA pull-down experiment and compared to a non-specific probe (NSP). Four Neat1 target RNAs (Cntn1, Malat1, Pou1f1 and Rplp0), as well as a non-target RNA (Tapbp) were analyzed by RT-qPCR. Enrichments relative to the NSP were normalized to the relative amount of corresponding Neat1_part captured. *p<0.05, **p<0.01, ***p<0.0001 compared to the probe a. Please click here to view a larger version of this figure.
| PROBE NAMES | SEQUENCES | |
| Specific Pool 1 (SP1) | ||
| a | CTCCACCATCATCAATCCTCTGGAC | |
| b | CATATAGCGGATGCCCAGGAACAAA | |
| c | ACAAAACAGAGCCCGAGAGTCAGTC | |
| d | ATGTACGTGACACGCTGACAACTGC | |
| e | ACTCCAAGATCTGACACCCTCACAC | |
| f | ACAGGGTCAGATGCAGTAAGACCTA | |
| Specific Pool 2 (SP2) | ||
| a | GCCTTCCCACATTTAAAAACACAAC | |
| b | TCCCCACAAGCATCTAAGAC | |
| c | TTACAATTACATACAGCCCTGTCCA | |
| d | AGTTGGTGAGTCCTAGCTCT | |
| e | TTAACCTTCCCTGGCAGTGT | |
| f | CAGGGTACTGCCTTGGTTTGGAAAT | |
| Specific Pool 2 (SP2) | ATAATTTCAAACATCAAATGGTATTTTA | |
| qPCR PRIMERS | FWD PRIMER | REV PRIMER |
| Neat1 Part_1 | AAGGCACGAGTTAGCCGCAAAT | TGTGCACAGTCAGACCTGTCATTC |
| Neat1 Part_2 | GCCTGCTTTCAGCTGTTGGTTT | TCTGGACAGCAACTGAGCAATACG |
| Neat1 Part_3 | TGCCAGTACTTCTGTCCCTAGGACAT | CTTCAGGCCTGGCTTCATTAGTGTTC |
| Neat1 Part_4 | GCTTTGGTGTATGGCGTGAGGTAA | CAGCCGACTGGGCAAAGCAATTAT |
| Neat1 Part_5 | GCTGACGTAGACTTTGAGGACCTACA | GAGATCGCTTGGGACCAGTTGGATAA |
| Neat1 Part_6 | GCATGAAGTTGGAAGCTGAGGGAAA | CTCTGAGACAGGGTCAGATGCAGTAA |
| Tapbp | GCAATTCCTGGGCTGCTTGAGAAA | TGTCCTTGCAGATAGGGCAGATGT |
| Cntn1 | GAGGTCTGGCTCCCGATACATAATCA | TGGGGACTTCTATGGAGTGCTTGT |
| Malat1 | GAAGGCGTGTACTGCTATGCTGTT | TCTCCTGAGGTGACTGTGAACCAA |
| Pou1f1 | TTCCAGAGGATGTGGGTTCCATCA | CTGTGCTCCATTTTCAGGCCAAGT |
| Rplp0 | CTTCCCACTGGCTGAAAAGGTCAA | AAGAGACCGAATCCCATGTCCTCA |
Table 1: Sequences of DNA oligonucleotide probes and qPCR primers
Discussion
Numerous lncRNAs carry out their function through complementary base pairing to mRNAs. It is, therefore, important to develop procedures that allow characterizing the direct RNA interactome of the lncRNAs. Therefore, a procedure was developed that combines the use of psoralen as cross-linking reagent with RNA pull-down technique.
In the RNA pull-down protocol described, the design and the selection of the antisense DNA biotinylated oligonucleotide probes are based on bioinformatics modeling of lncRNA secondary structure and subsequent selection of probes that hybridize against regions of the desired lncRNA that display a low probability of internal base pairing. Given that the different algorithms available predict different secondary structures, the best probes to be selected should be those that meet the above criteria in the greatest number of predicted secondary structures. Moreover, different other parameters such as the percentage of GC, 3' stability, complexity, as well as self-end dimer have to be taken into account for the selection of the probes. As we have already discussed in a previous paper4, this strategy of probe design remains less expensive and less time consuming than other strategies used in published methods such as CHART3 or ChIRP2.
The procedure described here combines the use of psoralen as a cross-linker with RNA pull-down experiments. Indeed, psoralen derivatives6,5 are known to cross-link RNA/RNA interactions mediated by base pairing and explains why several strategies have been developed to study RNA/RNA interactions using psoralen as a cross-linking reagent. This is the case of PARIS "Psoralen Analysis of RNA Interactions and Structures"17 and of SPLASH "Sequencing of Psoralen cross-linked, Ligated, And Selected Hybrids"18. The RNA pull-down procedure presented here overcomes some limitations of these previous techniques. While in SPLASH procedure, one of the limitations is that biotinylated psoralen used is less efficient at penetrating into cells than psoralen or 4'-aminomethyl trioxsalen (AMT) and high quantity of cross-linked RNA (20 µg) are necessary to have adequate material for downstream processes, in PARIS procedure four critical techniques are combined leading to a heavy-to-implement protocol. The procedure described here overcomes the heavy-to-implement aspect and the duration of the already developed procedures to provide a very easy-to-implement protocol that is less time consuming.
It should be noticed that since psoralen preferentially intercalate in double-stranded RNA, into adjacent opposite pyrimidine bases, mainly uracil, base pairing in GC-rich or U poor regions might have been not cross-linked and this can lead to an underestimation of direct RNA/RNA interactions in such regions. Consequently, direct RNA/RNA interactions that are not cross-linked by psoralen may be missed with the procedure.
Probably because of the extended length of lncRNAs, sonication of cross-linked extracts has proven to be indispensable for an efficient pull-down. In our hands, the optimal size of fragments generated by sonication is around 2000 nucleotides. As a consequence of this fragmentation several specific probes along the entire length of the lncRNA have to be designed. Since a specific antisense probe could pull-down fragments of the lncRNA that are located up to 2000 nt upstream and 2000 nt downstream from the site of probe binding and consequently can pull-down fragments of 4000 nt length, the number of probes to be designed corresponds to the number of nucleotides in the lncRNA divided by 4000. Use of a pool of these specific probes that bind along the entire length of the lncRNA allows the recovery of all RNAs engaged in direct RNA/RNA interaction with the lncRNA, whereas the use of each single probe separately allows determining which part of the lncRNA is involved in these RNA/RNA interactions.
Samples obtained with the RNA pull-down procedure can be submitted either to RT-qPCR analysis or to RNA sequencing when the objective is to establish the comprehensive list of RNAs engaged in direct RNA/RNA interaction. However, in the latter case, the main concern is to determine the non-specific background. Indeed, the experiments using a non-specific probe generally give rise to very low recovery of RNA compared to the specific probe, which makes it impossible to create a library. To overcome this problem, we suggest evaluating the specificity of RNA pull-down experiments by crossing the results obtained with two different pools of probes. In our hands, after crossing the results obtained with two pools of probes, each list of RNAs obtained with each pool of probes was cut by approximately 30% of RNAs which were not common to the two lists and were not then considered to be specifically pulled down. Actually, the use of two pools of probes increases confidence in the specificity of the identified RNAs.
The aim of the proposed procedure is to allow the capture of RNAs engaged in direct RNA/RNA interaction with a lncRNA in an easy-to-implement way. This protocol, when combined with high throughput RNA sequencing, can provide the whole direct RNA interactome of a lncRNA of interest. Furthermore, by designing numerous probes specific for different parts of the lncRNA throughout its length and performing pull-downs of RNA with a single probe, it is possible to differentiate the parts of the lncRNA involved in the direct interaction with the target RNAs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by Aix-Marseille University and Centre National Recherche Scientifique and funded by a grant from Sandoz Laboratories.
Funding for open access charge: Aix-Marseille University and Centre National Recherche Scientifique
Materials
| 4′-Aminomethyltrioxsalen hydrochloride | Sigma | A4330 | Crosslinker reagent |
| Bioruptor Plus | Diagenode | B01020001 | Sonicator |
| Biotynilated probes | IDT | Oligonucleotide probes | |
| CFX96 Real Time System | BioRad | 4351107 | qPCR apparatus |
| DNA Olignucleotides | IDT | Primers for qPCR | |
| Dynabeads My One | Thermo-Fisher | 65001 | Magnetic streptavidin beads |
| Formamide | Thermo-Fisher | 15515-026 | Formamide |
| iTaq Universal SYBR Green Supermix | BioRad | 1725124 | qPCR reagent |
| Proteinase K | Sigma | P2308 | Proteinase K |
| RNA to DNA | Thermo-Fisher | 4387405 | Reverse transcription kit |
| RNA XS purification kit | Macherey-Nagel | 740902 | RNA purificationkit |
| RNAseOUT | Thermo-Fisher | 10777-019 | RNAse inhibitor |
| Tube Rotator | Stuart | SB2 | Eppendorf tube rotator |
| UV Stratalinker 1800 | Stratagene | #400072 | UV crosslinker |
References
- Chen, L. -. L., Zhao, J. C. functional analysis of long non-coding RNAs in development and disease. Systems Biology of RNA Binding Proteins. 825, 129-158 (2014).
- Simon, M. D. capture hybridization analysis of rna targets (CHART). Current Protocols in Molecular Biology. 101 (1), 2125 (2013).
- Chu, C., Quinn, J., Chang, H. Y. Chromatin Isolation by RNA Purification (ChIRP). Journal of Visualized Experiments. (61), e3912 (2012).
- Torres, M., et al. RNA pull-down procedure to identify RNA targets of a long non-coding RNA. Journal of Visualized Experiments. (134), e57379 (2018).
- Engreitz, J. M., et al. RNA-RNA interactions enable specific targeting of non-coding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 159 (1), 188-199 (2014).
- Cimino, G. D., Gamper, H. B., Isaacs, S. T., Hearst, J. E. Psoralens as photoactive probes of nucleic acid structure and function: Organic chemistry, photochemistry, and biochemistry. Annual Review of Biochemistry. 54 (1), 1151-1193 (1985).
- Reuter, J. S., Mathews, D. H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 11 (1), 129 (2010).
- Gruber, A. R., Lorenz, R., Bernhart, S. H., Neuböck, R., Hofacker, I. L. The Vienna RNA websuite. Nucleic Acids Research. 36, 70-74 (2008).
- Jacq, A., et al. Direct RNA-RNA interaction between Neat1 and RNA targets, as a mechanism for RNAs paraspeckle retention. RNA Biology. , 1-12 (2021).
- Dobin, A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29 (1), 15-21 (2013).
- Kim, D., Langmead, B., Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 12 (4), 357-360 (2015).
- Liao, Y., Smyth, G. K., Shi, W. FeatureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics. 30 (7), 923-930 (2014).
- Anders, S., Pyl, P. T., Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 31 (2), 166-169 (2015).
- Fox, A. H., et al. Paraspeckles: a novel nuclear domain. Current Biology. 12 (1), 13-25 (2002).
- Torres, M., et al. Circadian RNA expression elicited by 3′-UTR IRAlu-paraspeckle associated elements. eLife. 5, 14837 (2016).
- Jacq, A., et al. Direct RNA-RNA interaction between Neat1 and RNA targets, as a mechanism for RNAs paraspeckle retention. BioRxiv. , 354712 (2020).
- Lu, Z., et al. Psoralen Analysis of RNA Interactions and Structures with High Throughput and Resolution. Methods in Molecular Biology. 1649, 59-84 (2018).
- Aw, J. G. A., Shen, Y., Nagarajan, N., Wan, Y. Mapping RNA-RNA interactions globally using biotinylated psoralen. Journal of Visualized Experiments. (123), e55255 (2017).

.
