Cost-effective Method for Microbial Source Tracking Using Specific Human and Animal Viruses
Summary
The study describes a cost-effective method for the identification of the source of fecal/urine contamination or contamination by nitrates in water using qPCR for the specific quantification of human/porcine/bovine DNA viruses, adenoviruses and polyomaviruses, proposed as MST tools.
Abstract
Microbial contamination of the environment represents a significant health risk. Classical bacterial fecal indicators have shown to have significant limitations, viruses are more resistant to many inactivation processes and standard fecal indicators do not inform on the source of contamination. The development of cost-effective methods for the concentration of viruses from water and molecular assays facilitates the applicability of viruses as indicators of fecal contamination and as microbial source tracking (MST) tools. Adenoviruses and polyomaviruses are DNA viruses infecting specific vertebrate species including humans and are persistently excreted in feces and/or urine in all geographical areas studied. In previous studies, we suggested the quantification of human adenoviruses (HAdV) and JC polyomaviruses (JCPyV) by quantitative PCR (qPCR) as an index of human fecal contamination. Recently, we have developed qPCR assays for the specific quantification of porcine adenoviruses (PAdV) and bovine polyomaviruses (BPyV) as animal fecal markers of contamination with sensitivities of 1-10 genome copies per test tube. In this study, we present the procedure to be followed to identify the source of contamination in water samples using these tools. As example of representative results, analysis of viruses in ground water presenting high levels of nitrates is shown.
Detection of viruses in low or moderately polluted waters requires the concentration of the viruses from at least several liters of water into a much smaller volume, a procedure that usually includes two concentration steps in series. This somewhat cumbersome procedure and the variability observed in viral recoveries significantly hamper the simultaneous processing of a large number of water samples.
In order to eliminate the bottleneck caused by the two-step procedures we have applied a one-step protocol developed in previous studies and applicable to a diversity of water matrices. The procedure includes: acidification of ten-liter water samples, flocculation by skimmed milk, gravity sedimentation of the flocculated materials, collection of the precipitate and centrifugation, resuspension of the precipitate in 10 ml phosphate buffer. The viral concentrate is used for the extraction of viral nucleic acids and the specific adenoviruses and polyomaviruses of interest are quantified by qPCR. High number of samples may be simultaneously analyzed using this low-cost concentration method.
The procedure has been applied to the analysis of bathing waters, seawater and river water and in this study, we present results analyzing groundwater samples. This high-throughput quantitative method is reliable, straightforward, and cost-effective.
Protocol
1. Concentration of the viral particles present in water samples
- Collection and conditioning of water samples
- Collect a minimum of 2 replicates of 10 L per sample in plastic containers with flat bottom and one extra sample as a process control. This last sample will be spiked with a known amount of viral particles and used as a control.
Note: It is recommended to have special separate material (bottles, tube, etc.) for spiked samples. - Check the calibration of the conductimeter and recalibrate if necessary. Prepare a negative control by using tap water previously adjusted to the adequate conductivity (see below 1.6) in one extra plastic 10 L container.
- For spiked samples: Add the standard volume of the control virus (approximately 105 genome copies per 10 L of water) to the sample. Mix by stirring avoiding splashing and aerosols. Positive controls could consist in an uncommon strain of adenovirus such as HAdV-35 or a bacteriophage such as MS2.
- If the sample presents high quantity of suspended material (sand or other materials), let it sediment for 15 minutes. Transfer the water into a new container.
- Adjust the pH of the water sample to 3.5 (± 0.1) by the addition of 1 N HCl. This step is important for the concentration of the viruses, so make sure the pH has been properly adjusted. Mix the water thoroughly by vigorous stirring while adding the HCl. (Note: If the pH is lower than 3.5 add 1 M NaOH).
- Adjust the conductivity. If the sample has a conductivity of 1500 μS/cm or higher, this step is not needed. If the conductivity is lower than 1500 μS/cm the formation of flocculated material (flocs) is not guaranteed so adjust the conductivity to 1500 μS/cm by the addition of artificial sea salts (Sigma). Mix vigorously by stirring while adding the sea salts.
- Record the pH of the samples before and after conditioning as well as the volume of HCl used. The conductivity should also be recorded after adjusting the pH. Always disinfect the pH-meter and conductimeter electrodes with a fresh HCl solution, dechlorinate with 10% sodium tiosulphate solution and finally rinse with distilled water.
- Collect a minimum of 2 replicates of 10 L per sample in plastic containers with flat bottom and one extra sample as a process control. This last sample will be spiked with a known amount of viral particles and used as a control.
- Preparation of pre-flocculated 1 % skimmed milk (PSM)
- Check the calibration of the pH-meter and the conductimeter and recalibrate if necessary.
- Prepare pre-flocculated skimmed milk solution (1 % PSM, w/v) by dissolving 10 g skimmed milk powder (Difco) in 1 L artificial seawater (dissolve 33.3 g of artificial sea salts in 1 L of dechlorinated tap water and autoclave) and carefully adjusting the pH to 3.5 with 1N HCl. The flocs should be visible. Prepare the solution just before to be used or store at 4°C for 24 h. For dechlorination use 1 ml of 10% tiosulphate solution per 100 ml of water.
- Flocculation of viral particles present in water samples
- Add 100 ml of 1 % PSM to the 10-L water sample.
- Stir the samples for 8-10 h to allow the viruses to adsorb to the flocs. Use a timer to switch-off the stirring after 8-10 h.
- Stop the stirring and let the flocs sediment by gravity for 8-10 h.
- Collecting and re-dissolving the flocs. Centrifugation
- Remove the supernatant using a peristaltic pump and a plastic pipette connected to a plastic tube. For spiked samples the supernatant should be collected into a bottle and disinfected according to internal procedures. In all cases TAKE CARE not to collect the pellet.
- Collect the sediment with the flocs (approximately 500 ml) into a centrifuge bottle.
- Balance the pots by the addition of PSM pH 3.5.
- Centrifuge the pots in a high-speed centrifuge at 8,000 x g for 30 min at 4°C. As soon as the centrifuge stops, carefully remove the centrifuge pots from the centrifuge.
- Very gently pour off and discard the supernatant. Follow appropriate measures for infectious material.
- Add 7 ml of phosphate buffer to dissolve the pellet in each centrifuge bottle.
- Once the flocs have been dissolved, measure and add phosphate buffer to reach a total volume of 10 ml.
- Homogenize the viral concentrate by vortexing and distribute the 10 ml into clean microtubes which should be frozen at -80°C until needed in further analysis.
2. Nucleic acid extraction
- Perform a nucleic acid extraction with the QIAamp Viral RNA Mini Kit following manufacturer instructions. This kit enables the use of an automated platform (such as Qiacube, Qiagen).
3. Quantitative PCR of human adenoviruses (HAdV), JC polyomaviruses (JCPyV), porcine adenoviruses (PAdV) and bovine polyomaviruses (BPyV)
- Quantification of genome copies in the samples
- Prepare the qPCR mix in a clean separate area by using TaqMan Environmental PCR Master Mix 2x (Applied Biosystems) . The reaction takes place in a 96-well optical reaction plate covered with optical adhesive covers. Concentrations of the Master Mix, primers and probes are described in Table 1.
- Once the mix has been prepared, aliquot 15μl into each well including the controls (see 3.2). The total volume for one reaction after addition of target will be 25μl (15μl mix + 10μl sample or standard).
- Add the nucleic acid extractions from the samples (10μl) in a separate area. Run direct and a ten-fold dilution in purified water of each sample in duplicate. Cover the wells containing the samples with part of an adhesive cover, keep the other part of the cover for the following step.
- Add dilutions of the DNA standard suspensions (10μl) from 100 to 106 GC/10μl by triplicate and using a micropipette exclusively used for the standard DNA. It is advisable to add standards in an area equipped with UV radiation for destroying plasmid DNA and to clean the micropipette after each use. Cover the wells containing the standards with the pre-cut adhesive cover.
Note: To prepare standard suspensions to be used in the quantification of genome copies, the target DNA region should be cloned into a plasmid and linearized. In the following address you will find a detailed procedure on how to create standard curves with plasmid DNA templates to be used in qPCR:
http://www.appliedbiosystems.com/support/tutorials/pdf/quant_pcr.pdf - Perform the qPCR into an adequate system selecting the appropriate parameters (considering the use of adhesive cover and the total volume in each well, etc). Following activation of the AmpliTaq Gold for 10 min at 95°C, 40 cycles of amplification are performed as follows: 15 s at 95°C and 1 min at 60°C for HAdV, JCPyV and BPyV, and 15 s at 95°C, 20s at 55°C and 20s at 60°C for PAdV.
- Once the reactions are completed, store data and results as described in the user’s manual of the equipment used. The amount of DNA will be defined as the median of the data obtained after correcting the dilution factor when needed.
- Controls
- Use positive and negative controls. The assay must include more than one non-template control (NTC) to prove mix does not produce fluorescence. It is advisable to run the positive process control in order to evaluate potential enzymatic inhibition due to inhibitors present in the studied samples.
- Record results of qPCR assays of two different dilutions of the standard DNA and from the process control. Use results to prepare control charts for quality control (QC) programs related to the sensitivity and efficiency of the assays.
- Confirmation of results
- Positive results may be further confirmed using nested-PCR and nucleotide sequencing of the amplicons, producing additional data on the nucleotide sequences of the strains detected1,5,6,7,9,12.
Following the procedure described, human and animal viruses have been detected and quantified in bathing waters, seawater and river water2,3. As a representative example, ground water samples from areas presenting high levels of nitrates were evaluated to define the sources of the contamination. Ten-liter water samples were collected from 4 different wells in rural areas of a Northeast region in Spain. Five replicates were collected in each well being one replicate seeded with human adenovirus 2 used as process control. Samples were processed according to the protocol represented in Figure 1. The four replicates analyzed in one of the four sites studied showed positive results for PAdV (mean value 7.74×102 GC/L) which would be related to the presence of pig slurries in the areas surrounding the sampling site and would support fecal porcine contamination as the source of nitrates in groundwater (Table 2).
4. Representative Results:
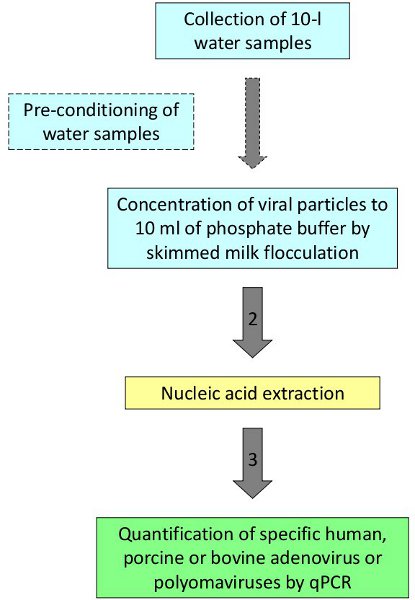
Figure 1. Procedure for the detection and quantification of viruses in water.
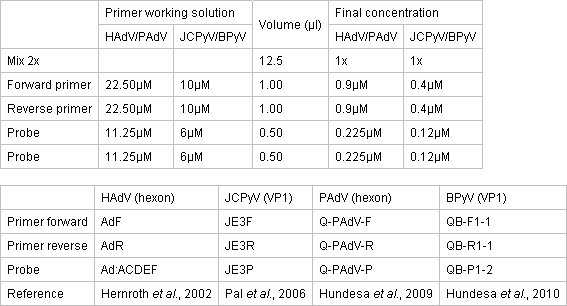
Table 1. Concentration of primers and probes for qPCR assays.

Table 2. Detection and quantification of animal and human adenoviruses and polyomaviruses in ground water samples.
n number of replicates analyzed
% percentage of positive replicates
(-) non detected
Discussion
The procedure described would fulfill the conditions for a fitting method for routine environmental and public health laboratories: reproducible, reliable, straightforward and cost-effective. The protocol is simple; however it must be followed carefully. Low conductivity in the samples without adding the requested concentration of artificial seawater salts would dramatically reduce the recovery of viruses as would be the case if the stirring time for flocculation is significantly reduced (less than 5 hours for example).
Currently applied MST tools are generally based on molecular techniques. Studies developed by different groups have shown that out of the currently used parameters (i.e. bacterial genes) none is as specific as needed. Thus the use of a combination of these parameters has been suggested as the best approximation for an efficient tracking of the origin of fecal contamination in water bodies8,11.
In recent years, the study of the selected DNA viruses that produce in many cases persistent infections in absence of clinical symptoms, has emerged as a method for source-tracking fecal contamination in the environment. Quantitative PCR techniques and the concentration method proposed provide reliable values of the concentration of these viruses and very valuable data for the development of risk-assessment studies and remediation actions. These techniques are also highly sensitive and specific, which is an absolute requirement for tracking the source of contamination. Also, they will be a very useful tool for the generation of larger data bases for the quantitative characterization of the excretion and dissemination of the specific viruses proposed as microbial source-tracking tools in diverse geographical areas.
The procedure described allows the analysis of multiple samples simultaneously in about 48 hours without intensive labor requirements. The availability of an efficient low-cost concentration method support the applicability of HAdV and JCPyV, and PAdV and BPyV in water as cost-effective assays for quantitative microbial source-tracking studies and identification of the origin of contaminants in ground water.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was partially supported by the Spanish Government “Ministerio de Educación y Ciencia” (project AGL2008-05275-C01/ALI), by the European Union Research Framework 7 funded projects VIROBATHE (Contract No. 513648), VIROCLIME (Contract No. 243923) and by the Catalan Agency of Water, Agència Catalana de l’Aigua (ACA), Departament de Control i Millora dels Ecosistemes Aquàtics. During the developed study Marta Rusiñol was a fellow of the Catalan Government “AGAUR” (FI-DGR).
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| High speed centrifuge (8,000xg) | Berckman Coulter | Avanti J-20XP | |
| pH-meter, thermometer and conductimeter | Afora | LPPC3003 | |
| Plastic tubes 100-200 cm length | Deltalab | 350059 | |
| Sterile graduated disposable pipettes | Labclinics | PN10E1 | |
| Sterile plastic tubes of 1.5 and 10-15 mL (Eppendorf, Falcons, etc.) | Afora | KA298/00 | |
| Centrifuge pots (500 mL) | Fisher Scientific | SE5753512 | |
| Magnetic stirrers and magnets (one per sample) | Fisher Scientific | 10510 | |
| Glass or plastic containers having flat bottoms to allow the use of magnetic stirrers | Deltalab | 191642 | |
| A peristaltic pump for removing the supernatant (or a water-jet vacuum pump) | Watson-Marlow | 323E/D | |
| Timer to switch-off the stirring after 8-10 hours | Deltalab | 900400 | |
| Hydrochloric acid (1N and 0.1N) | Panreac | 141020.1611 | |
| Sodium hydroxide (1N) | Panreac | 131687.1211 | |
| Artificial seawater sea salts | Sigma | S9883 | |
| Skimmed milk (SM) | Difco | 232100 | |
| Phosphate buffer pH 7,5 | 1:2 v/v of sterile Na2HPO4 0,2M and NaH2PO4 0,2M at pH 7.5 | ||
| Thiosulphate | Panreac | 121879.1209 | Make a 10% solution in water |
| QIAamp Viral RNA Mini Kit | Qiagen | 52904 | |
| 96-well optical reaction plate (500 units) | Applied Biosystems | 43426659 | |
| Optical adhesive covers (100 units) | Applied Biosystems | 4311971 | |
| TaqMan Environmental PCR Master Mix 2x | Applied Biosystems | 4396838 |
References
- Bofill-Mas, S., Clemente-Casares, P., Major, E. O., Curfman, B., Girones, R. Analysis of the excreted JC virus strains and their potential oral transmission. J. Neurovirol. 9 (4), 498-507 (2003).
- Bofill-Mas, S., Albinana-Gimenez, N., Clemente-Casares, P., Hundesa, A., Rodriguez-Manzano, J., Allard, A., Calvo, M., Girones, R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72 (12), 7894-7896 (2006).
- Calgua, B., Mengewein, A., Grunert, A., Bofill-Mas, S., Clemente-Casares, P., Hundesa, A., Wyn-Jones, A. P., López-Pila, J. M., Girones, R. Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. J. Virol. Methods. 153 (2), 79-83 (2008).
- Hernroth, B. E., Conden-Hansson, A. C., Rehnstam-Holm, A. S., Girones, R., Allard, A. K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Appl. Environ. Microbiol. 68, 4523-4533 (2002).
- Hundesa, A., Bofill-Mas, S., de Motes, M. a. l. u. q. u. e. r., Rodriguez-Manzano, C., Bach, J., Casas, A., M, ., Girones, R. Development of a quantitative PCR assay for the quantitation of bovine polyomavirus as a microbial source-tracking tool. J. Virol. Methods. 163 (2), 385-389 (2010).
- Hundesa, A., de Motes, M. a. l. u. q. u. e. r., Albinana-Gimenez, C., Rodriguez-Manzano, N., Bofill-Mas, J., Suñen, S., E, ., Girones, R. Development of a qPCR assay for the quantification of porcine adenoviruses as an MST tool for swine fecal contamination in the environment. J. Virol. Methods. 158 (1-2), 130-135 (2009).
- Hundesa, A., de Motes, M. a. l. u. q. u. e. r., Bofill-Mas, C., Albinana-Gimenez, S., N, ., Girones, R. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl. Environ. Microbiol. 72 (12), 7886-7893 (2006).
- Layton, B. A., Walters, S. P., Lam, L. H., Boehm, A. B. Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109, 539-547 (2010).
- de Motes, M. a. l. u. q. u. e. r., Clemente-Casares, C., Hundesa, P., Martín, M., Girones, R. Detection of bovine and porcine adenoviruses for tracing the source of fecal contamination. Appl. Environ. Microbiol. 70 (3), 1448-1454 (2004).
- Pal, A., Sirota, L., Maudru, T., Peden, K., Lewis, A. M. Real-time PCR assays for the detection of virus-specific DNA in simples with mixed populations of polyomaviruses. J. Virol. Methods. 135 (1), 32-42 (2006).
- Stapleton, C. M., Kay, D., Wyer, D. a. v. i. e. s., Watkins, C., Kay, J., McDonald, C., Porter, A. T., J, ., Gawler, A. Evaluating the operational utility of a Bacteroidales quantitative PCR-based MST approach in determining the source of faecal indicator organisms at a UK bathing water. Water Res. 43, 4888-4899 (2010).
- Wang, J., Horner, G. W., Keef, O., S, J. Detection and molecular characterization of bovine polyomavirus in bovine sera in New Zealand. N. Z. Vet. J. 53, 26-30 (2005).

