Plasma Lithography Surface Patterning for Creation of Cell Networks
Summary
A versatile plasma lithography technique has been developed to generate stable surface patterns for guiding cellular attachment. This technique can be applied to create cell networks including those that mimic natural tissues and has been used for studying several, distinct cell types.
Abstract
Systematic manipulation of a cell microenvironment with micro- and nanoscale resolution is often required for deciphering various cellular and molecular phenomena. To address this requirement, we have developed a plasma lithography technique to manipulate the cellular microenvironment by creating a patterned surface with feature sizes ranging from 100 nm to millimeters. The goal of this technique is to be able to study, in a controlled way, the behaviors of individual cells as well as groups of cells and their interactions.
This plasma lithography method is based on selective modification of the surface chemistry on a substrate by means of shielding the contact of low-temperature plasma with a physical mold. This selective shielding leaves a chemical pattern which can guide cell attachment and movement. This pattern, or surface template, can then be used to create networks of cells whose structure can mimic that found in nature and produces a controllable environment for experimental investigations. The technique is well suited to studying biological phenomenon as it produces stable surface patterns on transparent polymeric substrates in a biocompatible manner. The surface patterns last for weeks to months and can thus guide interaction with cells for long time periods which facilitates the study of long-term cellular processes, such as differentiation and adaption. The modification to the surface is primarily chemical in nature and thus does not introduce topographical or physical interference for interpretation of results. It also does not involve any harsh or toxic substances to achieve patterning and is compatible for tissue culture. Furthermore, it can be applied to modify various types of polymeric substrates, which due to the ability to tune their properties are ideal for and are widely used in biological applications. The resolution achievable is also beneficial, as isolation of specific processes such as migration, adhesion, or binding allows for discrete, clear observations at the single to multicell level.
This method has been employed to form diverse networks of different cell types for investigations involving migration, signaling, tissue formation, and the behavior and interactions of neurons arraigned in a network.
Protocol
1. Creation of molds used for patterning
- Conceptual design of patterns. Patterns are created which serve to form cell networks, control cell contact, isolate cells, mimic natural structures, or otherwise guide cell adhesion and placement.
- Computer-aided design or CAD based creation of pattern and creation of photomask. The desired pattern is designed in CAD software, such as AutoCAD, and is then sent to an outside company for producing a photomask.
- Photolithography to produce master pattern. Glass slides are patterned with either a thin positive resist or a thick negative resist depending upon the size of the structure being created. Glass slides are used for their benefits of being low cost, stronger than silicon wafers and not producing as much dust when broken. Alternatively, other convenient structures such as diffraction gratings can be used as the master pattern.
- The patterned slide is attached to a stack of slides which have not been patterned. All steps are performed inside a clean flow hood to minimize dust.
- Creation of master molds. A container of tinfoil slightly larger than the stack of slides is created to hold the 30 g of Polydimethylsiloxane (PDMS) which is then poured over the photolithographically patterned surface to create an initial mold. The PDMS is degassed and allowed to cure for one to two days.
- Once cured the PDMS is peeled off of the master pattern and forms a mold for the next step.
- 6 g of 2 part epoxy ( Devcon #14310(6 g) is mixed for 1 minute and then poured into the master mold, degassed and allowed to cure. Degassing the epoxy must be done quickly (<8 min) using many bubble breaking cycles and using only a thin layer before it the epoxy sets and bubble removal becomes impossible to remove the bubbles. The 6 g used produces a thin layer which facilitates quick bubble removal.
- After one hour, the 2 part epoxy will be partially cured and more epoxy can be added on top without degassing to produce a thicker structure which is easer to handle in later steps. The epoxy is then allowed to cure for one to two days.
- Once cured, the epoxy is removed from the PDMS master mold and tape is wrapped around the epoxy mold to form a container. A working mold is then created by pouring PDMS onto the epoxy mold, degassing the PDMS, and curing for one to two days.
- Once cured, the working mold is peeled off of the epoxy and stored to maintain cleanliness.
2. Patterning of surfaces with molds for cell guidance
- Small sections of the working mold are cut out and placed onto a polystyrene Petri dish. The locations of these mold sections are labeled.
- A tripod is formed from the small sections and weighted to ensure conformal contact between the working mold and the Petri dish surface. Good placement can be observed by looking at the bottom of the dish.
- The assembly is placed into the plasma chamber. Plasma treatment is initiated and continued at 150 Pa at 29.6 W (max power) for 10 minutes using air plasma.
- After plasma treatment, the mold and weight arraignment are removed.
- The Petri dish is placed under UV light for 10 minutes of sterilization inside the biosafety cabinet and stored there until seeded with cells.
3. Seeding surfaces with cells
- SH-SY5Y CRL-2266 Human Neuroblastoma or other cells are cultured using standard protocols for the cell type.
- The confluence of the SH-SY5Y CRL-2266 Human Neuroblastoma cells is measured visually before seeding onto the patterned surface.
- Standard cell splitting is carried out to remove the cells from the surface of their Petri dish.
- The cells, once free floating, are seeded onto the surface of the patterned Petri dish after being diluted to achieve the desired confluence when placed on the patterned surface.
- The cells are allowed to settle and attach to the surface.
- The patterned Petri dish is incubated for several hours to several days while the cells assemble onto the pattern created by the plasma.
- When culturing the neuroblastoma cells, retinoic acid (10 μM) is added daily in order to induce the cells to differentiate into a neuron-like state. The addition of the retinoic acid is done in the dark.
- Retinoic acid is added daily for several days, after which differentiation will be observable.
- Media is replaced every 3-4 days.
4. Observation and Analysis
- Periodic images of the live cells or videos are taken in order to observe the cell behavior over time. For live images, the cells are placed in a microscope stage top incubator which keeps the cells at 37 °C, 100% humidity, and 5% CO2. Cells can also be fixed and stained for observation under fluorescence
- The images or videos of the cells that have been captured are imported to an image analysis system and measurements are carried out including growth rate, cell size, migration speed, differentiation, alignment, location of specific proteins, and others.
5. Representative Results:
A typical result of implementing the plasma lithography technique is the formation of a pattern of cells which resembles some arbitrary or natural structure. This is seen in Figure 1a-b where lines and networks of neurons have been created. Other cell types can be used as well as seen in Figure 1c-d, which shows Human umbilical vein endothelial cells (HUVEC) and C2C12 skeletal muscle cells forming grids. Materials such as poly-l-lysine can also be patterned, Figure 1e to facilitate attachment of certain cell types and for other uses. In the case of the neurons shown, what was created are networks of cells which have connections between them. This replicates what occurs naturally in the brain where neurons have discrete connections between neighboring cells which influences the operation of the brain. With the creation of such a structure in the lab, the number, position, frequency and other factors of the connections can be systematically controlled. The result of these arraignments can be measured visually and additional inputs such as chemical or electrical stimulation can be implemented to probe the network behavior.
A negative result would be an overgrown or incomplete pattern or a contaminated sample. An incomplete or overgrown pattern would result from seeding the substrate with either too few or too many cells which would not supply the pattern with an ideal amount of cells. Additionally, if the pattern is not designed correctly (e.g., lines too narrow) the cells will not be able to attach and grow successfully on it. In the case of a contaminated sample, proper cleanliness would not have been maintained during one of the steps though this is rare when following proper cell culture protocol due to the fact that the plasma patterning also sterilizes the substrate.
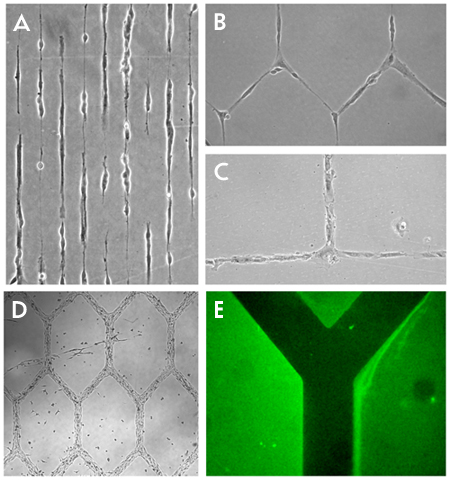
Figure 1. Patterning of cells and protein.
Discussion
The cell investigation method presented here enables creation of complex patterns of multicellular networks which mimic biological structures as well as provides a method to produce stimuli that are subcellular in nature which then facilitates investigations of both grouped cell behavior and single cell responses to environmental factors. The use of this method is simple yet robust as it can be quickly performed with low cost equipment in the same lab as the cell culture. It is also strongly cell sensitive, allows easy observation of the resultant behavior and is by nature stable for long time periods, which allows investigation of long term cell behavior. It additionally allows for a diverse range of experiments to be performed as it is compatible with many cell types and can create arbitrary patterns. The long term stability of the technique derives from the fact that the surface functionalization imparted by the plasma is part of the surface and is not a coating or other layer which can be removed or degraded. If kept under liquid, this type of modification can retain its cell guiding ability for months.
The most critical part of the experiment necessary to ensure meaningful results is that of creating the master pattern which will ultimately be used for cell guidance. If this pattern is not properly designed, cells will not appropriately respond to the pattern and may not produce useful behavior. Parameters such as line width, pattern spacing, and others can greatly influence the cell compatibility with a particular pattern, and typically a range of such parameters can be created on the initial photomask to screen for the most appropriate design.
Other important parameters related to carrying out patterning include mold creation, maintaining a dust free environment, cell seeding and general sterility of cell culture. Mold creation may be effected by the various transfer steps which are undertaken to allow for repeated PDMS casting off an epoxy mold. The epoxy mold is created because it will not degrade in the same way as a resist mold with small, high aspect ratio structures under repeated casting. If the transfer molding is done correctly, the dimensions and yield will not be affected but if done incorrectly such as by poorly degassing, incomplete curing, or excessive heating, bubbles, roughness and deformation of a pattern can occur which affects the final outcome. In relation to maintaining a dust free environment, the molds must be kept as clean as possible as any dust can interfere with the proper contact between the working PDMS mold and the surface and thus proper plasma shielding and chemical patterning. The seeding density of the cells also must be optimized to ensure that neither too few or too many cells inhabit the pattern area and sterility must be maintained to avoid bacterial and other contamination of the cells being used.
The technique can also be incorporated with other elements such as microfluidics, microelectrodes, and mechanical probes. This provides additional stimuli to the cells in order to better replicate various physiological conditions during experimentation and future work is focused on studying the effects of these combinations
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank D. D. Zhang for insightful discussion and generously providing reagents. M. J. is supported by the NIH Cardiovascular Training Grant, the Arizona Technology Research Initiative Fund, and Achievement Rewards for College Scientists. This work is supported by the NIH Director’s New Innovator Award (1DP2OD007161-01), the National Science Foundation (0855890), and the James S. McDonnell Foundation.
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| Plasma Cleaner And Vacuum Chamber | Harrick Plasma | PDC-001 | |
| Inverted fluorescence and phase contrast microscope | Nikon | TE2000-U | |
| Microscope stage top incubator | AmScope | Model TCS-100 | Modified to include an enclosure that supplies an appropriate atmosphere |
| Polystyrene Petri dish | VWR | 25384-090 | |
| Polydimethylsiloxane (PDMS) | Dow Corning | Sylgard 184 | |
| Epoxy | Devcon | 2 ton clear epoxy #14310 | |
| Cell culture media | Various | Media follows standard formulations for cell type | |
| Retinoic acid | Sigma | R2625 |
References
- Junkin, M., Watson, J., Vande Geest, J. P., Wong, P. K. Template-Guided Self-Assembly of Colloidal Quantum Dots Using Plasma Lithography. Adv Mater. 21, 1247-1251 (2009).
- Junkin, M., Wong, P. K. Probing cell migration in confined environments by plasma lithography. Biomaterials. 32, 1848-1855 (2011).
- Keyes, J., Junkin, M., Cappello, J., Wu, X., Wong, P. K. Evaporation-induced assembly of biomimetic polypeptides. Appl Phys Lett. 93, 023120-023 (2008).
- Langowski, B. A., Uhrich, K. E. Microscale Plasma-Initiated Patterning (μPIP). Langmuir. 21, 10509-10514 (2005).
- Tourovskaia, A., Barber, T., Wickes, B. T., Hirdes, D., Grin, B., Castner, D. G. Micropatterns of Chemisorbed Cell Adhesion-Repellent Films Using Oxygen Plasma Etching and Elastomeric Masks. Langmuir. 19, 4754-4764 (2003).
- Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M., Ingber, D. E. Geometric Control of Cell Life and Death. Science. 276, 1425-1428 (1997).
- Johansson, B. -. L., Larsson, A., Ocklind, A., Ohrlund, A. Characterization of Air Plasma-Treated Polymer Surfaces by ESCA and Contact Angle Measurements for Optimization of Surface Stability and Cell Growth. Journal of Applied Polymer Science. 86, 26185-26185 (2002).
- Murakami, T., Kuroda, S. -. i., Osawa, Z. Dynamics of Polymeric Solid Surfaces Treated with Oxygen Plasma: Effect of Aging Media after Plasma Treatment. Journal of Colloid and Interface Science. 202, 37-44 (1998).
- Strobel, M., Lyons, C. S., Mittal, K. L. . Plasma surface modification of polymers: Relevance to adhesion. , (1994).
- Xia, Y., Whitesides, G. M. Soft Lithography. Annual Review of Materials Science. 37, 550-575 (1998).
- Song, M., Uhrich, K. R. Optimal Micropattern Dimensions Enhance Neurite Outgrowth Rates, Lengths, and Orientations. Annals of Biomedical Engineering. 35, 1812-1820 (2007).
- Zhao, F., Wu, T., Lau, A., Jiang, T., Huang, Z., Wang, X. -. J. Nrf2 promotes neuronal cell differentiation. Free Radical Biology and Medicine. 47, 867-879 (2009).
- Ohl, A., Schroder, K. Plasma-induced chemical micropatterning for cell culturing applications: a brief review. Surface and Coatings Technology. 116-119, 820-830 (1999).
- van Kooten, T. G., Spijker, H. T., Busscher, H. J. Plasma-treated polystyrene surfaces: model surfaces for studying cell-biomaterial interactions. Biomaterials. 25, 1735-1747 (2004).
- Loesberg, W. A., te Riet, J., van Delft, F., Schön, P., Figdor, C. G., Speller, S. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials. 28, 3944-3951 (2007).

