Single Molecule Fluorescence Microscopy on Planar Supported Bilayers
Summary
Preparation of protein-functionalized planar glass-supported lipid bilayers, determination of protein mobility within and measurement of protein densities is shown here. A roadmap to building a noise-reduced Total Internal Reflection microscope is outlined, which allows visualizing single bilayer-resident fluorochromes with high spatiotemporal resolution.
Abstract
In the course of a single decade single molecule microscopy has changed from being a secluded domain shared merely by physicists with a strong background in optics and laser physics to a discipline that is now enjoying vivid attention by life-scientists of all venues 1. This is because single molecule imaging has the unique potential to reveal protein behavior in situ in living cells and uncover cellular organization with unprecedented resolution below the diffraction limit of visible light 2. Glass-supported planar lipid bilayers (SLBs) are a powerful tool to bring cells otherwise growing in suspension in close enough proximity to the glass slide so that they can be readily imaged in noise-reduced Total Internal Reflection illumination mode 3,4. They are very useful to study the protein dynamics in plasma membrane-associated events as diverse as cell-cell contact formation, endocytosis, exocytosis and immune recognition. Simple procedures are presented how to generate highly mobile protein-functionalized SLBs in a reproducible manner, how to determine protein mobility within and how to measure protein densities with the use of single molecule detection. It is shown how to construct a cost-efficient single molecule microscopy system with TIRF illumination capabilities and how to operate it in the experiment.
Introduction
Understanding the mechanisms by which membrane proteins fulfill their cellular function can often be a challenging task, because their modes of action are often defined by low affinity interactions, which are difficult to detect and evaluate by conventional biochemical methodologies. Membrane proteins may furthermore be highly enriched in specific membrane domains 5,6 or form dynamic higher order structures, which can in principle alter their biological activity 7.
Non-invasive laser-assisted single molecule fluorescence microscopy has thus become the methodology of choice to study protein behavior in complex cellular environments. This is in particular true for biochemical processes taking place at the plasma membrane, as these can in principle be monitored in noise-reduced Total Internal Reflection (TIRF) mode. Here sample illumination is restricted to an up to 200 nm-thin slice in close proximity to a glass surface, which results in a significant loss in cellular background and, due to the characteristics of the evanescent excitation light, also in a substantial increase in fluorescence signal intensity 8,9. Both aspects are important when aiming for high positional and temporal resolution in single molecule detection.
Planar SLBs are not only compatible with TIRF microscopy. When functionalized with appropriate ligands they also provide direct access to studying the molecular dynamics of cell-cell recognition as they occur in immune cells or other cells. They can also be simply utilized to position motile and otherwise non-adherent cells close enough to the glass slide so that such cells can be imaged in TIRF illumination, which is highly desirable when conduction experiments involving single molecule tracking or single molecule-based superresolution. To this end proteins have to be loaded onto a highly mobile SLB. An inherent advantage of the used lipids are that there is no unspecific binding of proteins at all. Moreover, SLBs can be regarded as two-dimensional liquids with an inherent capacity to heal themselves; irregularities within the glass support are compensated up to a certain degree. A step-by step protocol is provided to generate highly mobile bilayers charged with proteins of interest. The formation of planar SLBs is described, which contain 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) as the main ingredient (90-99%) and the synthetic and functionalized lipid 1,2-dioleoyl-sn-glycero-3-{[N(5-amino-1-carboxypentyl)iminodiacetic acid] succinyl}nickel salt (DGS NTA-Ni) in low abundance (1-10%). POPC carries one saturated fatty acid and one mono-unsaturated fatty acid and forms lipid bilayers of high fluidity even at RT. DGS NTA-Ni binds with its headgroup to poly-histidine tails and serves as anchor for poly-histidine-tagged proteins of choice (Figure 1).
Importantly, the microscopy hardware must include a number of peripherals which are described below. With the information provided and some basic knowledge in optics and laser physics, any biologist should be able to build a single molecule imaging setup. Sample excitation should ideally be performed on an inverted microscope in Total Internal Reflection (TIRF) mode as this regimen reduces spurious light and excessive background resulting otherwise from cellular auto-fluorescence 9. For this, a special TIRF-capable objective (see below) and laser illumination are needed. Some experiments will require illumination times within the millisecond range and operated in rapid succession. To save illumination time or to avoid unnecessary bleaching caused by repetitive illumination it is often helpful to split the emitted light into two or more spectral channels. This allows for simultaneous readout of two or more emission profiles, which can become a significant factor when conducting experiments involving Förster Resonance Energy Transfer (FRET). Here the emission resulting from single excitation light pulse can be recorded both from the FRET donor and FRET acceptor (as sensitized emission). The camera should capture enough photons with sufficiently low background to visualize single fluorophores during the illumination time. Computer software will have to be up to the task to synchronize excitation shutting and image acquisition. A comprehensive overview is given below and in Figure 2:
TIRF-capable objective: For objective-based TIRF illumination the numerical aperture (NA) of the objective must be equal to or larger than 1.4 using standard glass slides (n=1.515) 9. Magnification can range between 60x to 150x. The objective should be chromatically corrected to allow for confocality when imaging different fluorophores.
Lasers: The number of available laser options has significantly increased over the last years. Modern electronically modulatable continuous wave diode lasers are cost-efficient and can be operated with unprecedented frequency and speed. More expensive gas ion lasers excel in laser beam quality, but require external shutting (see below) and often extensive (water-) cooling.
Excitation shutters: Acousto-optical modulators (AOM) allow fast shutting in rapid succession 10. For tight shutting the combination of an AOM with mechanical shutters is recommended. This way extensive heating of the AOM due to continuous light exposure is avoided and light leaking from the AOM in the off-position is canceled out.
Lenses are used to widen laser beams and to focus them onto the back focal plane of the objective. This way, the excitation light leaves the objective as a parallel beam (Figure 3), which is required for TIRF illumination of the sample. Moving the focal point within the back focal plane from the center into the periphery of the objective will change the angle at which the beam leaves the objective, but not the location of the laser spot at the specimen (Figure 3), which is a function of the overall beam geometry. TIRF illumination ensues at a critical angle, which can be adjusted using a set of mirrors functioning as a periscope to translate the focal point of the laser within the focal plane of the objective. Lenses should be chromatically corrected and can be used in a set of two or three lenses. A three-lens system consists of two lenses acting as a telescope to widen the laser beam (and hence the illumination spot at the specimen) and a third lens to focus the widened beam into the back focal plane of the objective (Figure 4). Both functions (telescope and focusing) can also be achieved by a combination of two lenses only (see Figure 4).
Mirrors should be at least 95% reflective to avoid loss of laser light. For each beam adjustment a set of two mirrors is usually applied as such arrangement is sufficient to precisely adjust any angle and position of a laser beam.
Dichroic overlays reflect and transmit light of different specified wavelengths and are used to superimpose or split the beams of two lasers.
Polychroic filters are more complex than dichroic filters and are needed to reflect incoming excitation light and transmit exiting emission light from the specimen. They are placed into the filter cube between the objective and the microscope tube lens.
Clean up filters: Depending on the type of laser employed laser clean up filters with a narrow transmission bandwidth should be placed into the laser beam right after it leaves the laser.
Notch filters are designed to effectively absorb laser light with a narrow bandwidth and transmit all other light. They are placed within the emission path to filter out any spurious laser excitation light. However, notch filters work only at 0° incoming angle. If the band-width of the notch filter is very sharp, the different incoming angles may not be reflected any more. Blocking of collimated laser laser light is not affected, but back-scattered light might not be efficiently hindered from reaching the camera.
Motorized filter wheels equipped with appropriate filters wheels can be easily placed into the excitation and emission pathway and allow simple switching between different light intensities (when equipped with ND filters) or fluorescent channels (when placed in front of a xenon or mercury arc lamp for ratiometric calcium imaging or in front of the camera to select for the emitting fluorophore).
50:50 cube beam splitter can be used to split laser beams into two separate excitation beam paths and also to combine them in front of the periscope.
Beam splitter (emission path): For fast image acquisition without any need for physical filter changes, the emission beam is split into a blue-shifted and red-shifted channel. In principle, beam splitters can be built by employing a dichroic wedge or a set of mirrors and a dichroic mirror to separate the emission beam in a wavelength-dependent manner. Two emission filters are needed to clean up the emitted channels.
Spatial filter: Spatial filtering of the excitation beam is sometimes required to remove non-parallel light rays emitted from low quality laser beams. A spatial filter consists of a two-lens telescope with a small pinhole placed exactly at the focal point of both lenses. This way spurious light resulting from non-parallel portions of the laser beam becomes efficiently blocked. Such conditions must be met when establishing TIRF-based microscopy. Spatial filtering increases with smaller pinholes, which are more difficult to position into the focal point, and with smaller focal lengths of the first lens. To reduce effects caused by lens aberrations it is advantageous to employ instead of simple lenses high quality and infinity-corrected microscope objectives with low magnification (10x or 20x).
Periscope: A periscope setup is necessary to translate the focused laser beam within the back focal plane of the objective, a prerequisite for TIRF imaging. It can be easily built from two two-inch mirrors, a translational stage for adjusting the first mirror and a post for positioning the second mirror to reflect the widened and focused excitation beam into the microscope (Figure 5).
Camera: Back-illuminated Electron-Multiplying Charge Coupled Devices (EMCCD) are routinely used for the recording of single molecule signals. This is because of their high quantum efficiency (up to 95%), high acquisition speed (up to 30 MHz) and comparatively low noise. Cooling down to -80 °C reduces thermal noise and is supported by a number of currently available EMCCD cameras. A limitation of the EMCCD technology is that camera noise increases linearly with the both the camera gain and the signal being captured. This is not the case with scientific CMOS (sCMOS) cameras, which are significantly less expensive and operate due to the special architecture of the sCMOS chip also much faster than EMCCD cameras. However a problem associated with the CMOS technology is that image acquisition still lacks some degree of a quantitative readout on a single molecule level, because each pixel features different detection sensitivity. In principal this can be compensated through pixel normalization, but this procedure is by no means trivial 11. This is why we still hesitate to recommend sCMOS cameras for single molecule microscopy, but in view of the rapid development of this technology, sCMOS cameras might soon become the camera of choice. Slow scan CCD cameras cameras avoid amplification-related noise and pixel variance all together and support fastest acquisition rates if they can be operated in a so-called ‘kinetic’ mode. In this mode the entire camera chip except for a region of interest (ROI) is masked, which makes it possible to employ the chip itself as a storage device. After the ROI is exposed for the first time, the resulting charges are shifted into the masked area of the chip pixel line by pixel line, where the image is protected from further light exposure. Once the shifting of all lines of the ROI into the masked region is completed (in the sub-millisecond range), the ROI itself is ready for the next exposure. This cycle is repeated until all pixel lines of the CCD chip are charged. The chip is then read out slowly with markedly reduced readout noise. For example on a 1000 x 1300 pixel chip, 20 ROIs of 50×50 pixel can be recorded in rapid succession. Because the images remain on the chip for a considerable time before being read out, it is critical to ensure high-quality masking and to cool the camera (e.g. with liquid nitrogen) as a means to reduce excessive thermal noise. Some EMCCD cameras also support the kinetic mode.
Software: Timing of the lasers, shutters, AOMS and camera exposure as well as proper image storage are integral to any successful imaging experiment. In principal many defined operations can be programmed with available software packages that come with the camera. Commercial software packages support a great number of hardware peripherals, which can be implemented with little technical knowhow.
Pulse generator, Data Acquisition (DAQ) board (with analog and digital output channels) and oscilloscope: A pulse generator is an excellent choice to convert trigger pulses into pulses of defined time and voltage. This way lasers can be precisely controlled for output power and timed in the millisecond to sub-millisecond range. DAQ boards with analog outputs achieve the same and are easily integrated into the computer’s motherboard via PCI slots. Pulse duration, amplitude and frequency are verified with an oscilloscope.
Enclosure of the entire excitation beam path: To avoid fluctuations in the excitation profile due to air convections the entire excitation beam path should be enclosed from its lab environment. This measure is especially important when conducting TIRF microscopy. Optical components are also protected from dust and the human eye from laser light exposure. Enclosures can be easily build from black card board, which can be bought in arts supply stores.
To determine the fluidity of the SLB Fluorescence Recovery After Photobleaching (FRAP) measurements 12 are performed. For FRAP the existence of two excitation beam paths is advisable (see Figure 3). The first beam path is designed to image the fluorescence of the bilayer. This can be done in TIRF configuration and with low light intensity. The second beam path should allow for a short but intense bleach pulse and should be configured in non-TIRF mode, so that it leaves the objective along the optical axis. A round aperture can be placed into the excitation beam path (see Figure 8) to project a perfectly round bleach profile with defined edges. In order to image this aperture onto the object plane, its optimum position would be in the focal plane of lens 3 (see Figure 3). However, due to the long focal length of this lens, an aperture image of sufficient quality can also be generated, when the aperture is placed at slightly shifted positions.
After having performed the imaging experiment the raw data must be analyzed appropriately. Several step-by-step protocols are offered, which cover the adjustment of the main settings (e.g. illumination power and time, TIRF angle), acquiring and analyzing the data.
Protocol
1. Generation and Functionalization of Planar SLBs
- Mixing of the lipids
- To arrive at a 10% DGS NTA-Ni/90% POPC lipid composition dissolve 45 mg of POPC and 6.9 mg DGS NTA-Ni in chloroform in a 250 ml round bottom flask. Keep the volume of chloroform low (just several ml) to evaporate it in the next step. For a 1% DGS NTA-Ni/99% POPC lipid composition use 49.5 mg POPC and 0.69 mg DGS NTA-Ni.
- Remove the chloroform using a rotary evaporator slowly – an increase above ambient temperature is not necessary. Alternatively, blow it off inside a chemical hood in a stream of inert gas such as nitrogen or argon. While doing this constantly turn the flask to allow equal deposition of the drying lipid on the lower half of the round bottom flask.
- Once the majority of the chloroform is evaporated, attach the flask to vacuum O/N to remove the chloroform quantitatively.
NOTE: This step is critical to avoid contamination of proteins and cells with toxic chloroform at later stages and to ensure lipid bilayer fluidity.
- Generation of small unilamellar vesicles (SUV’s) from dried lipids
NOTE: Small unilamellar vesicles (SUVs) are between 20 nm and 100 nm in diameter and can be readily produced by bath sonication. Lipid extrusion, an alternative method, can give rise to SUVs and, depending on the pore size of the filter applied for extrusion, also large unilamellar vesicles (LUVs), which are 100 nm to 1,000 nm in size. However, SUVs are best suited for forming SLBs.- SUVs through bath sonication (shown in the video):
- Degass PBS. Suspend the dried lipid mixture with the 250 ml round bottom flask in 10 ml degassed PBS.
- Fill the flask with inert gas such as nitrogen or argon, close it with a stopper and seal the flask with autoclave tape.
- Place the sealed flask into a bath sonicator and sonicate the lipid suspension at 120 to 170 W until it has turned clear. This takes between 30 and 60 min.
- Monitor the progress of vesicle formation spectro-photometrically: Ensue that the absorption of undiluted emulsion in comparison to PBS remains constant at 234 nm (as an approximate indicator for the lipid concentration) yet should drop significantly at 550 nm (due to reduced light scattering via larger particles).
- To pellet heavy non-unilamellar vesicles, which interfere with the formation of a contiguous SLB, pellet the vesicle suspension by centrifugation, at 150,000 x g for 1 hr at 25 °C and then for 8 hr at 288,000 x g, 4 °C.
- Filter the supernatant through a syringe filter with a pore diameter of 0.2 µm.
- Measure the OD at 234 nm and 550 nm to monitor the clarity of the vesicle preparation and the amount of lipid left.
- Store the vesicles at 4 °C under argon or nitrogen. Optionally, use vesicle suspension for bilayer preparation for several months.
- SUVs through lipid extrusion:
- In brief, pass the lipid suspension several times through a filter with a pore size of 0.1 µm. Regardless of the fabrication method, centrifuge the vesicle suspension twice (1h, 150,000g , 25 °C; then 8h, 4 °C, 288,000 g) to pellet non-unilamellar vesicles which are heavier .
NOTE: When stored at 4 °C under inert gas, vesicles can be used for bilayer preparation for several months.
- In brief, pass the lipid suspension several times through a filter with a pore size of 0.1 µm. Regardless of the fabrication method, centrifuge the vesicle suspension twice (1h, 150,000g , 25 °C; then 8h, 4 °C, 288,000 g) to pellet non-unilamellar vesicles which are heavier .
- SUVs through bath sonication (shown in the video):
- Formation of an SLB and charging with protein
- Treat 24 x 50 mm #1.5 glass slides with a 3:1 mixture of concentrated sulfuric acid and 30% hydrogen peroxide for 30 min.
NOTE: Due to the aggressive nature of the mixture take special care, i.e. wear a lab coat and safety goggles as shown in the video! - Rinse glass slides under a stream of ddH2O out of a squirt bottle. Place them on a Teflon stand and let them dry for 10 to 30 min.
- Remove the bottom glass slide of 8 or 16 well chambers, fill the bottom with epoxy glue and carefully place a clean and dry glass slide on the glue-covered chamber bottom. Let the glue harden for about 10 min. Remove excess glue from the bottom.
NOTE: A tight seal between the glass slide and the chamber can also be easily achieved with the use of a dental two-component silicone modeling paste, which hardens within 10 to 30 min. This seal is not as firm as epoxy glue but withstands regular physical strain. After the experiment it can be easily removed from the chamber, which is then reusable in future experiments. - Pipette 120 µl (60µl) of a 10-fold PBS-diluted lipid suspension into each well of an 8 (16) – well chamber (prepared as described above) and let the bilayer form for 20 min. Carefully rinse the bilayer twice with 15 ml PBS. Once the bilayer has formed, it must always be submerged in buffer and never be exposed to air.
- Fill each well all the way with PBS, then take off 330µl (8-well chamber) or 165µl (16-well chamber). There are 350µl (175µl) left in the well. Add 50µl (25µl) of cocktail containing His-tagged proteins diluted in PBS to each well (400 or 200 µl final) and incubate at RT for 60 min in the dark. The surface protein density can be adjusted by varying the protein concentration during this incubation step.
- Rinse each well twice with 15 ml PBS.
NOTE: Typically, SLBs should be used in experiments no longer than 6 hr after their charging with protein. During this period, fluorophore recovery after photobleaching remains up to 95% and no loss in bilayer-associated protein can be detected. The protein density should be determined prior to the experiment (see below). - Optional step: To test for non-specific binding of the protein the DGS NTA-Ni containing SLB, wash the SLB once with PBS containing 300 mM imidazole. The protein should come off completely.
- Treat 24 x 50 mm #1.5 glass slides with a 3:1 mixture of concentrated sulfuric acid and 30% hydrogen peroxide for 30 min.
2. Microscope Setup
- For building a microscopy system capable of detecting the fluorescence of single fluorochromes refer to the recommendations stated in the introduction.
3. Power Measurements
NOTE: Fluorophores can be easily saturated with excess excitation light. This accelerates photobleaching with no further gain in emission and should thus be avoided. To optimize sample illumination the excitation power density should be measured directly at the specimen. Such measurements can also serve as a reference for future experiments. Furthermore, knowing the ratio between input and output laser power is helpful when assessing where light is lost in the imaging system.
- Move the excitation beam into the upright position so that it leaves the objective in a 90° angle.
- Place the head of the power meter on top of the beam and measure the power of the beam (=P). Document the result.
- Using the periscope, turn the beam into the TIRF position and image an intact homogenous fluorescent bilayer. To avoid bleaching and CCD signal saturation place a neutral density (ND) filter with an optical density (OD) of 2 to 3 into the excitation light path.
- Measure the integrated counts of the entire illuminated field (= Itotal). Also, measure the size of the illuminated area (= Atotal). Measure the integrated counts (= ICenter) of the maximally illuminated center spot as well as the size of the spot (= Acenter).
- Measure the background signal per pixel (=B) either outside the illuminated area or without sample illumination.
- Subtract the background signal from the results measured in 3.). Do this by multiplying the number of pixels of the area in question (whole illuminated area or center spot) with the background counts per pixel. (=Itotal – Atotal .B and Icenter – Acenter .B).
- Divide the integrated and background corrected signal of the center spot by the integrated and background-corrected signal of the entire field of illumination.
NOTE: The result indicates the fraction of total power located within the center spot (= (Icenter – Acenter .B) / (Itotal – Atotal .B) ). Multiplication of this result with the power P measured in 2.) results in the power of light illuminating the center spot. - Determine the size of the spot Acenter in µm². For this divide the camera pixel size in µm by the magnification of the objective, take the square of the result as µm² per pixel.
- Alternatively measure the pixel size within the image with the use of a micrometer slide placed on the microscope and take the square of the result as area per pixel. Multiply this value by the number of pixels located within the center spot to arrive at the illuminated spot area.
- Divide the power P illuminating the center spot by the size of the center spot Acenter,µm² to calculate the illumination intensity per µm2. An example is given in Figure 6.
- Power density values measured in non-TIRF illumination are typically lower than those present in TIRF illumination 13, which also depend on the exact angle at which the laser light is totally reflected. To arrive at power densities present under TIRF illumination, image calibrated fluorescent beads of 0.1µm diameter both in non-TIRF and TIRF. The ratio of bead fluorescence intensities measured in TIRF and non-TIRF mode amounts to the conversion factor C, which allows conversion of measured power density values into those effective under TIRF illumination (equation 1).
power density TIRF = C • power density non-TIRF (equation 1)
with C = bead intensity TIRF / bead intensity non-TIRF
4. Density Measurements
- Determine the average signal of individual fluorophores located on a bilayer. Bilayer-attached MHC molecules loaded with fluorescent peptides are ideal for this purpose since the protein:label stoichiometry is exactly one. If necessary bleach all labels on the bilayer in the field of view and let the fluorescence recover to visualize single fluorophores.
- Record images in rapid succession (e.g. apply a streaming acquisition protocol) and monitor single molecules mobility over several frames. For more information refer to Figure 7.
- Mark the central illumination spot within the bilayer as ROI. Measure the ROI, the variance in illumination power density (which is proportional to the fluorescence intensity) should be ideally less than 15%.
- Determine single molecule and bulk fluorescence only within this ROI. Integrate the signal of a ROI, which is large enough to capture 99% or more of the single emitter’s point spread function. When employing a camera with a pixel size of 16-20 µm (stated in the manufacturer’s camera specifications), an objective with 100-fold magnification and a numerical aperture equal to or larger than 1.45, take a region of 7 by 7 pixels with the intensity peak located in the center of the square.
- To determine the background signal, integrate the counts of a neighboring 7 by 7 pixel square, which does not contain a fluorescence signal. Subtract the background from the single molecule signal to determine the corrected value for single molecule fluorescence. Select only signals, which are still visible in the following frame.
- Repeat step 4.2 fifty to several hundred times. Average the background-corrected signal. If desired, plot single molecule intensity values as a histogram. Optional: take advantage of a suitable plugin of open-source software packages (e.g. ImageJ) or process the data using commercial software (e.g. Metamorph, Molecular Devices). Experienced users may write their own analysis program in Matlab or similar software packages.
- Place a Neutral Density filter of 2 or higher into the excitation path and take images of a fluorescent bilayer containing proteins labeled with the same fluorophore. Record the average pixel intensity within the same ROI used for single molecule intensity measurements. Record the exposure time of the bulk fluorescence measurements. Measure the same spot without illumination for background subtraction.
- To determine the protein density within the bilayer, multiply the background-corrected bulk fluorescence average pixel intensity determined in step 4 by the number of pixels per square micron (e.g. 41.5), by 100 (if an ND filter with an OD of 2 was employed) or 1,000 (if an ND filter with an OD of 3 was used) and the exposure time used in step 2 and divide it by the average single molecule signal determined in step 3, the exposure time used in step 2 and the number of fluorophores per protein.
5. Testing the Integrity of the Bilayer
- Prepare a fluorescent bilayer and place it on the microscope stage as described above.
- Adjust the focus and take an image of the bilayer in TIRF mode. Apply a short but intense bleach pulse in non-TIRF mode to ablate the fluorescence within a small disk-shaped ROI.
- Take an image of the bilayer in low intensity TIRF mode immediately after bleaching and then every five to ten sec to monitor the speed of fluorescence recovery.
- Move to a different area on the bilayer and follow step 2 and 3. Take another image in low intensity TIRF mode 5 min after the bleach pulse. Determine the intensity Ipost bleach in the bleach area and compare it to the intensity prior to bleaching I0 at the beginning of the experiment (no bleaching should have occurred) to calculate the immobile fraction (IF) as follows:
Immobile Fraction IF (%) = (I0 – Ipost bleach ) / (I0– background) x 100,
with background = measured intensity within the ROI with no excitation light applied
The IF should be less than 10% (ideally less than 5%).
Representative Results
The architecture of the single molecule fluorescence microscopy system is outlined in considerable detail in Figure 2. Individual parts such as optical components and other hardware components are explained in the introduction. The optical excitation beam paths, which give rise in a defined manner to TIRF and non-TIRF illumination, are shown and explained in Figures 3 to 5. Note the location of the 50:50 beam splitter in front of the first periscope mirror positioned on a micrometer translatable stage (Figure 5). This beam splitter superimposes the TIRF beam used for imaging (indicated in green) and the non-TIRF beam (indicated in red) used for bleaching or local aperture-defined light-mediated sample activation (as outlined in Figure 3). The combined light paths (Figure 5, indicated in orange) are reflected via the second periscope mirror into the back port of the inverted microscope. As explained in Figure 4 and outlined in Figure 2, the implementation of two or three convex lenses widens the laser beam profiles and focuses the laser beams onto the back focal plane of the objective to give rise to two collimated beams illuminating the sample in TIRF and, respectively, in non-TIRF.
An example for measured intensity values required for calculating the power density on the specimen is shown in Figure 6. The average background signal to be subtracted from intensities measured in the illuminated and central area (used for imaging), is determined in a region within the field of view, which is not illuminated by the laser beam (here 7089 counts). The background-corrected average intensities of the illuminated (1684 counts) and the central area (4830 counts) are then multiplied by the corresponding region sizes to give rise to integrated intensities (1.471 x 108 counts for the illuminated area and 3.424 x 107 counts for the central area). The ratio of the integrated intensities within the central and illuminated regions yields the fraction of the overall power illuminating the central area (0.23 or 23%). As shown in the movie, the overall power has to be determined at the objective with the use of a power meter in non-TIRF illumination mode. In our example it is adjusted to 5 mW, thus giving rise to a power of 5 mW x 0.23 = 1.15 mW within the central area. Since 41.5 pixels amount in our example to 1 µm2, the central area has a size of 7089 pixels /41.5 pixels x µm2 = 170.8 µm2. Dividing the power of the light illuminating the central area (1.15 mW) by this area (170.8 µm2) yields an average power density of 0.7 kW/cm2 within the central area.
Figure 7 features images and corresponding intensity results of single fluorophores acquired in rapid succession (100 frames per second, i.e. with a time frame of 10 msec). For intensity quantitation the average signal of a rectangular region of seven by seven (=49) pixels, which covers the fluorescence signal, is determined. Furthermore the average signal of the background is determined within a rectangular region of seven by seven pixels in close proximity of the single molecule signal. The background-corrected average signal is multiplied by the number of pixels within the region (= 49) to give rise to single molecule signal (marked in bold).
A representative FRAP experiment designed to assess the mobility of SLB-associated fluorescence-labeled proteins is shown in Figure 8. Note in Figure 8A the repetitive use of a widened low intensity imaging beam and the single use of a second more narrow and aperture-defined high intensity beam for rapid fluorochrome ablation at the zero second time point. Background-subtracted average intensities within the yellow circle, which have been normalized by the initial average intensity value prior to the bleach pulse, are plotted in Figure 8B against the time to record the speed and extent to which fluorescence has recovered. As shown, more than 90% (indicated by the green line) of the initial bilayer intensity has recovered within 75 sec following fluorochrome ablation. As a consequence less than 10% of the proteins embedded in the shown SLB are immobile.
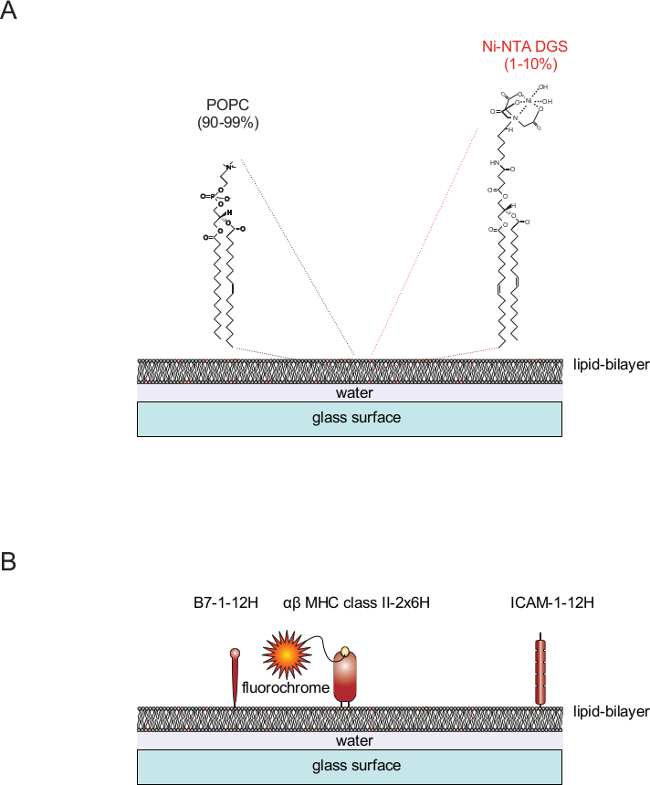
Figure 1. Schematic outline of a SLB system. (A) SLBs are made of POPC (90-99%) and the synthetic lipid DGS NTA-Ni (1-10%). They form spontaneously when clean glass surfaces are charged with SUVs consisting of the corresponding lipids. (B) Once formed, these SLBs can be easily decorated with soluble proteins extended either with one C- or N-terminal tag containing twelve histidines (12H, for example the co-stimulatory molecule B7-1 and the adhesion molecule ICAM-1) or protein dimers extended with two tags containing six histidines each (2x6H, for example a αβMHC class II dimer). Please click here to view a larger version of this figure.
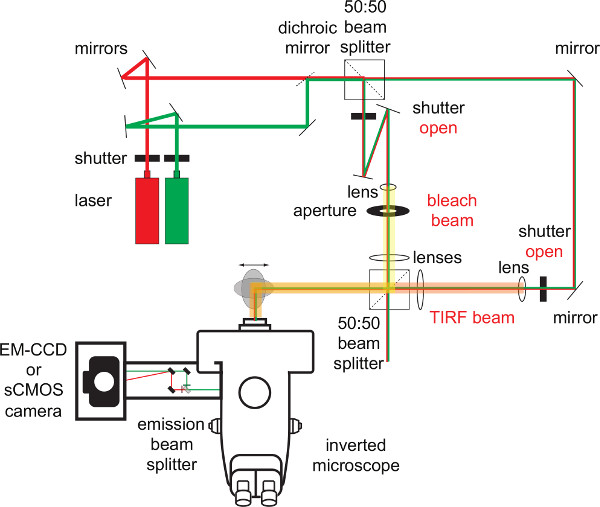
Figure 2. Overview of the excitation and emission beam paths. Gas ion lasers (e.g. Ar+ or Kr+) may be operated for fast shutting with the use of acousto optical modulators. Lasers diodes are nowadays available in many wavelengths and represent an excellent alternative as they can be electronically modulated in the microsecond-rage. Note that laser beams can be easily adjusted with two (dichroic) mirrors and split and combined with the use of simple beam splitters to give rise to two or more beam paths. In this example a TIRF imaging beam (to monitor events) and a bleach beam (to either bleach fluorophores or photo-activate caged molecules in an aperture-defined manner) are used. Both excitation paths can be operated independently of one another through the use of additional shutters. The periscope allows for translating the focused laser beams within the back focal plane of the objective to generate TIRF illumination of the sample. The fluorescent image can be spectrally split with the use of an emission beam splitter prior to acquisition with the use of an ultra-sensitive camera (e.g. EM-CCD or scientific CMOS camera). Please click here to view a larger version of this figure.
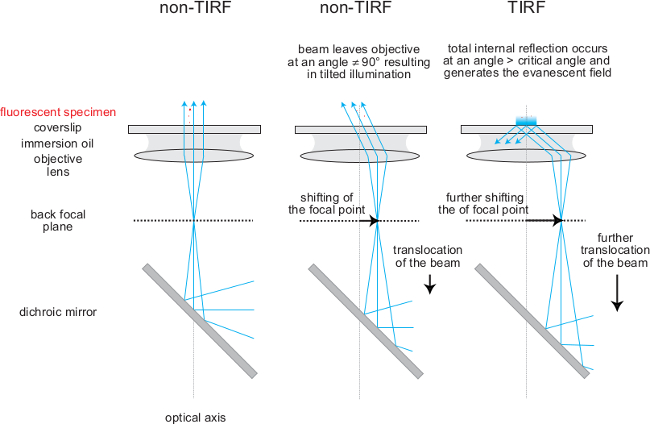
Figure 3. Adjusting TIRF illumination by translating the focused beam within the back focal plane of the objective. As shown the focal point of the laser beam is shifted within the back focal plane of the objective from the center to the periphery through translocation of the beam (using a periscope mirror arrangement). As a result a parallel beam leaves the objective in a tilted illumination until a critical angle is reached at which total internal reflection happens. The evanescent field is generated at the glass-media interface where total reflection occurs. Please click here to view a larger version of this figure.
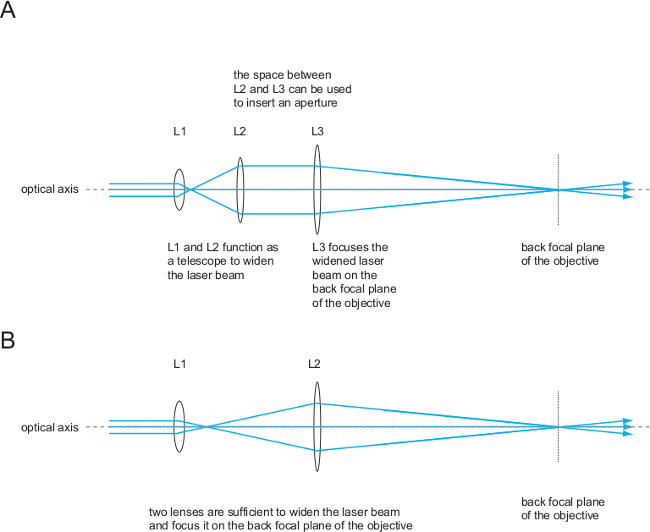
Figure 4. Simple optical systems to focus the laser beam into the back focal plane of the objective. Either three or two lenses are needed to widen the laser beam and to focus it on the back focal plane of the TIRF objective. Two lens systems contain fewer lens-associated errors than three-lens systems and might be better suited for generating the TIRF imaging beam. Three lenses might be preferable when aiming for defined beam widening and an illumination spot with sharp edges, as would be needed for studies employing photo-activation or -bleaching. Please click here to view a larger version of this figure.
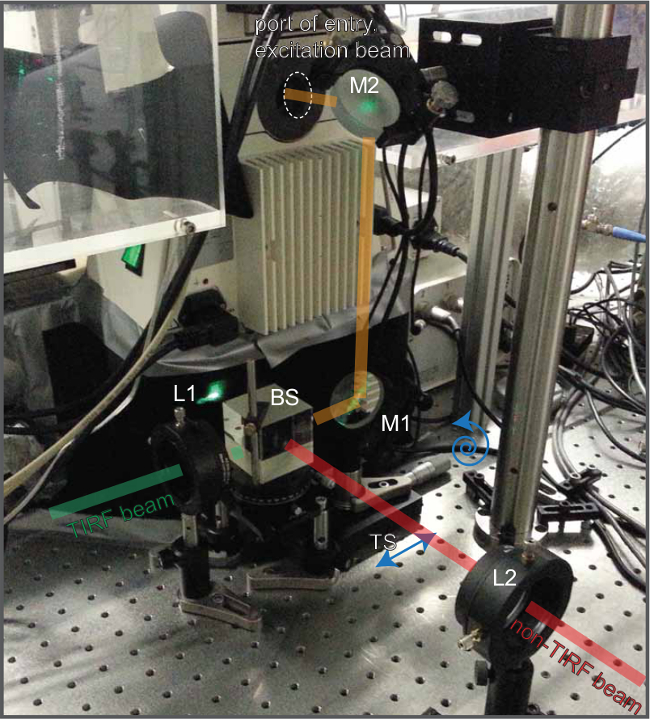
Figure 5. Annotated photograph of the beam path at the back of the fluorescence microscope. As is already outlined in Figure 2 one cane easily implement two excitation beams, one in TIRF (indicated in green) the other in non-TIRF mode (indicated in red). These become overlaid with the use of a 50:50 beam splitter (BS). Convex lenses (L1 and L2) are positioned into the respective beams to focus them onto the back focal plane of the objective (not shown). A periscope consisting of two mirrors (M1 and M2) guides both beams through the port of entry into the microscope. The first mirror (M1) can be moved with the use of a translation stage (TS) (by turning a micrometer screw as indicated) to move one of beams into TIRF position. Once TIRF has been established, the second beam (used for photo-bleaching or -activation, here indicated in red) can be adjusted independently of the TIRF beam to leave the objective in non-TIRF mode. Please click here to view a larger version of this figure.
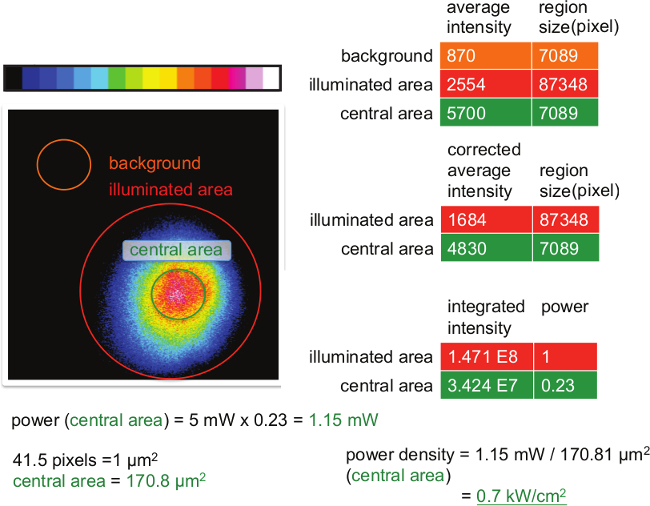
Figure 6. Measuring the power within the central area of the illumination spot. As is indicated chose appropriate regions of interest, which reflect background, the entire illuminated area (IA) and the central area (CA), where events will later be recorded. Subtract background values from those recorded for IA and CA and integrate the intensities by multiplying corrected average intensities with the number of pixels present within each area (= integrated intensities). Divide the integrated intensity of CA by that of IA to arrive at the proportion of the beam power illuminating the CA (in this example: 23%). Determine the power of the light illuminating the CA by multiplying the overall beam power (here: 5 mW) with the proportion illuminating the CA (here: 0.23). To determine the size of the central area multiply the area (here: 7089 pixel) with a pixel-to-area size conversion factor (here: 1/41.5 µm2 pixel-1). To arrive at the average power density of the excitation light illuminating the CA, divide the power (here: 1.15 mW) by the size of the CA (here: 170.8 µm2). Please click here to view a larger version of this figure.
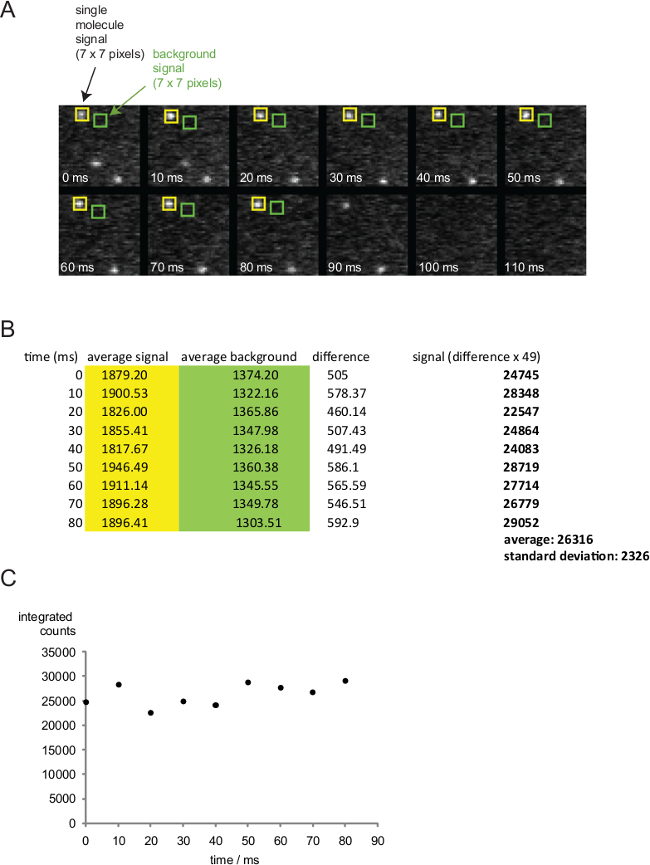
Figure 7. Quantitation of single molecule signals. (A) Time course of single fluorophores on an SLB. A 7 x 7 pixel sized region of interest (roi, yellow) is placed around the signal for quantitation. Background is determined from the a neighboring 7 x 7 pixel roi (green). (B) Quantified average pixel intensities are listed. To determine the single molecule signal, background values (green roi) are subtracted from signal values (yellow roi) and multiplied by 49. (C) Single molecule intensities of one fluorophore are plotted against time. Note, that the 90 msec time point is omitted as the fluorophore is in the process of bleaching. Please click here to view a larger version of this figure.
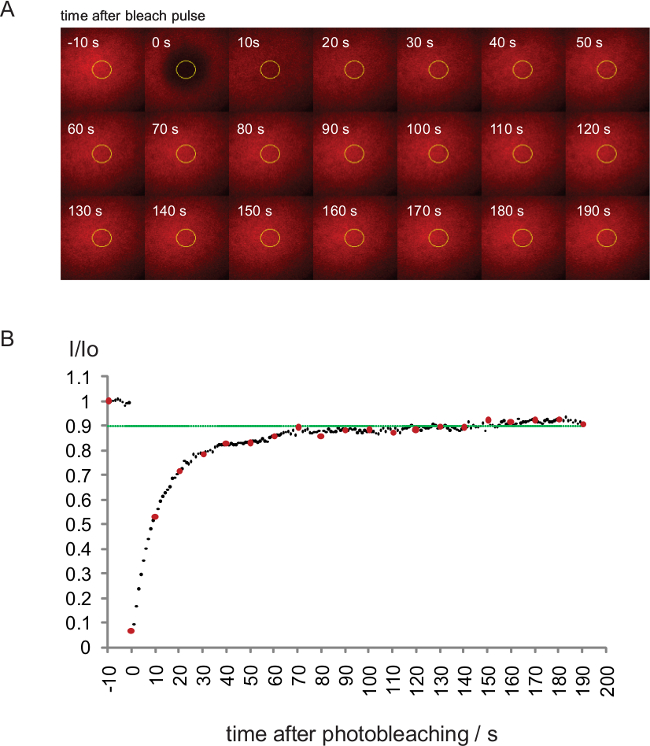
Figure 8. Fluorescence Recovery After Photobleaching (FRAP) analysis to determine the integrity of the SLB. (A) FRAP was performed on a bilayer carrying IEk/MCC-Alexa Fluor 647. Every 10th image of the experiment is shown. (B) FRAP quantitation of the experiment shown in (A). Values indicate average intensities (I) within the yellow ROI shown in (A) divided by the initial intensity (Io) before bleaching. Red data points are displayed in (A), the green line denotes 0.9 recovery of original intensities. Please click here to view a larger version of this figure.
Discussion
Single molecule microscopy provides the unique opportunity to study the behavior of proteins within their native cellular environment. Molecular imaging becomes therefore increasingly attractive to a broad scientific community, yet many life scientists still shy away from the initial investment in technology and expertise. The main benefit arising from assembling one’s own imaging system is that it can be readily adjusted to anyone’s specific need.
As suggested herein a non-TIRF excitation beam can be easily implemented in addition to an existing TIRF light path, if rapid and defined photo-bleaching (or -activation) of fluorophores and ligands is desired, e.g. when performing FRAP or ligand uncaging experiments 14. The introduction of an emission beam splitter allows for simultaneous recording of at least two and in some cases up to four fluorescent channels. The list of extensions is in essence only limited by one’s own imagination.
For example, implementing a motorized emission filter wheel in the emission path allows for fast switching between up to ten different fluorescent channels. When combining two motorized emission filter wheels (each equipped with 10 filter slots) in series, up to 18 fluorescent channels can be read out. TIRF-based imaging can be complemented with calcium mobilization measurements and Interference Reflection Microscopy (IRM) after integration of a Xenon or Mercury excitation lamp path. IRM is the method of choice to visualize the degree to which cells are attached to the glass surface or an SLB and can hence provide critical information when interpreting TIRF-based imaging data. Establishing a widened excitation beam with the use of a simple dichroic mirror and an additional laser of 405 nm allows conducting photoactivation localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM), i.e. two superresolution methodologies with a positional accuracy below the diffraction limit of visible light 15,16.
However, experimental success is not only a matter of appropriate hardware. To take full advantage of noise-reduced TIRF-based imaging, cells should make contact to a surface in a fashion that does not impose constraints on the cell surface or that interferes with the physiology of their plasma membrane. SLBs functionalized with appropriate proteins for cell adhesion are exceeding well suited for this purpose because all SLB-embedded ligands are laterally mobile and adjust their lateral position within the bilayer in response to receptor binding and segregation dynamics.
To manufacture SLBs in a reproducible fashion, it is best to start out with SUVs of high purity. As described herein, it is thus critical to remove multilamellar vesicles, which are also produced during sonication, in two ultracentrifugation steps as these interfere with the formation of contiguous SLBs featuring high fluidity. SLBs of high quality form only on clean glass surfaces. Once cleaned glass slides should be used immediately or stored in vacuum. Importantly, SLBs must never be exposed to air as this would lead to their disruption. SLB washing involves, as shown, buffer rinsing with the use of a serological pipette, as this is not only a save but also time-saving procedure.
During harvesting and purification of SLB-resident proteins the use of detergents should be avoided. This is because detergent will reduce protein mobility significantly even when present in trace amounts. To circumvent the need for detergent all together, soluble expression of secreted polyhistidine tag-equipped ligands is recommended in mammalian or insect cells. If refolding from E.coli inclusion bodies is required, caution must be applied to remove detergents effectively from the inclusion bodies prior to their un- and refolding.
A major advantage of employing SLBs results from their modular and reconstitutive nature, which allows to specifically dissect the role of a given receptor-ligand interaction for cell activation and cell adhesion. In this regard it is important to mention that DGS NTA-Ni should be present at 10% within the SLB in order to prevent proteins competing for NTA-NI binding sites. When employing SLBs harboring only 1% or 2% DGS NTA-Ni, a reduction in the association of the a given polyhistidine-tagged protein species can be noticed, especially when co-incubating increasing amounts of a second polyhistidine-tagged protein species (unpublished observation). This phenomenon is not observed when working with SLBs featuring 10% DGS NTA-Ni.
While we have never observed non-specific binding of any polyhistidine-tagged protein tested to SLBs containing DGS NTA-Ni, this possibility should be tested when introducing a protein for the first time, To this end we recommend the use of SLBs devoid of DGS NTA-Ni (the protein should not bind). Secondly, when using an SLB containing DGS NTA-Ni, the protein should come off completely after washing the SLB with PBS containing 300 mM imidazole.
Another significant advantage of employing SLBs is that transient interactions and signaling events can be monitored with enhanced spatiotemporal resolution 17-19. This is at least in part because a three-dimensional binding process is essentially reduced to two imaging dimensions, especially when recording in noise-attenuated TIRF mode. The use of SLBs is compatible with single molecule signal detection, a prerequisite for photo-activated localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM), i.e. superresolution microscopy with a resolution below the diffraction limit 15,16. These special imaging modalities enable single molecule Förster Resonance Energy Transfer experiments designed to visualize individual protein-protein interactions in a synaptic environment 20. This approach is explained in considerable detail in a JoVE publication termed “Measuring TCR-pMHC binding using a FRET-based microscopy assay” 21.
When interpreting SLB-based experiments one should always keep in mind that not all properties of a plasma membrane of a living cell are featured by SLBs, and that some of the missing qualities might affect the physiology under investigation. After all, SLB-embedded proteins are freely diffusing and not organized in membrane microdomains as most of their cellular counterparts are 5,6,22. Immobilized plasma membrane sheets derived from adherent cells, which conserve the plasma membrane architecture of living cells to some degree, have been succesfully employed to study the uptake from membrane-embedded antigens by B-lymphocytes 23. However, even such membranes do not interact with a highly dynamic cytoskeleton and feature no flexibility as they are supported by a rigid glass surface. In view of these discrepancies there is clearly a need for engineering SLBs, which offer means to compartmentalize proteins in a defined fashion and which are supported by surfaces of adjustable flexibility or by surfaces which change their rigidity in response to locally applied light pulses.
Disclosures
The authors have nothing to disclose.
Acknowledgements
M.A. was supported by a Schrödinger fellowship of the Austrian Science Fund (FWF, J3086-B11) and thanks the Max-Planck-Society for financial and administrative support. G.S. was supported by the Vienna Science and Technology Fund (WWTF, LS13-030). J.H. was supported by the Vienna Science and Technology Fund (WWTF, LS14-031).
Materials
| #1.5 glass slides | VWR | 631-0853 | |
| 250ml round bottom flask | VWR | 201-1357 | |
| 50:50 beam splitter cubes | Thorlabs | BS013 | |
| Acousto-opitcal modulator C1205-2 | Isomet | – | only necessary if CW lasers, which cannot be modulated, are used |
| Alexa Fluor 488-NHS | Life technologies | A-20000 | |
| Alexa Fluor 555-NHS | Life technologies | A-20009 | |
| Alexa Fluor 647-NHS | Life technologies | A-20006 | |
| autoclave tape | VWR | 489-1312 | or any other heat-stable sticky tabe |
| Avanti Mini-Extruder | Avanti Lipids | 610000 | alternative to the bath sonicator |
| bath sonicator QSONICA Q700 | QSONICA | Q700 | |
| beam splitter | Cairn | P280/210/MLS | |
| Büchi rotary evaporator | VWR | 531-0837 | |
| camera | Andor | iXon Ultra 897 | see manuscript text for alternatives |
| concentarted hydrogenperoxide | Roth | 9683.1 | |
| concentrated sulfuric acid | Roth | X944.2 | |
| epoxy glue | Uhu | 45705 | Uhu plus sofortfest 2min |
| filter wheel | Sutter instruments | Lambda 10-2 | |
| LabTek chambers | VWR | 734-2062 | |
| laser | Toptica | – | different variants (wavelengths 375 – 785 nm) are available |
| lens | Thorlabs, Newport | – | |
| DGS NTA-Ni (lipid) | Avanti Lipids | 790404C | already in Choroform-solved delivered version of the nicekl salt variant is recommended |
| POPC (lipid) | Avanti Lipids | 850457C | already in Choroform-solved delivered version is recommended |
| microscope Zeiss Axiovert 200 | Zeiss | – | |
| mirrors | Thorlabs | BB1-E02 | |
| optical filters | AHF, Chroma, Semrock | – | filter sets are available for many different combinations of dyes |
| oscilloscope | BK Precision | 2120C | |
| phosphate buffered saline (PBS) | Life technologies | 14190-136 | degas before usage |
| periscope | Thorlabs | RS99/M | |
| picodent twinsil 22 | Picodent | 13001000 | alternative to the epoxy glue |
| power meter Lasermate-Q | Coherent | – | any powermeter sensitive in the used spectral range will do |
| pulse generator | Stanford Research Systems | SRS DG535 | |
| spatial filter | Thorlabs | KT310/M | |
| syringe filter 0.2µm | Millipore | GVWP04700 | |
| TIRF-capable apochromat objective 100x, NA 1.46 | Zeiss | 440782-9800-000 | |
| trichloromethane/chloroform | Roth | 3313.1 | |
| tubes forultracentrifugation polycarbonate 1.00 mm, 11 mm x 32 mm | Thermo Fisher | 45237 | |
| Sorvall ultracentrifuge | Thermo Fisher | RC M150GX | |
| ultracentrifuge rotor | Thermo Fisher | S120-AT2 | |
| UV spectrophotometer Nanodrop 2000c | Thermo Fisher | – | |
| vacuum pump | VWR | 181-0248 |
References
- Walter, N. G., Huang, C. Y., Manzo, A. J., Sobhy, M. A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 5, 475-489 (2008).
- Sahl, S. J., Moerner, W. E. Super-resolution fluorescence imaging with single molecules. Current opinion in structural biology. 23, 778-787 (2013).
- Varma, R., Campi, G., Yokosuka, T., Saito, T., Dustin, M. L. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 25, 117-127 (2006).
- Kaizuka, Y., Douglass, A. D., Varma, R., Dustin, M. L., Vale, R. D. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc Natl Acad Sci U.S.A.. 104, 20296-20301 (2007).
- Wilson, B. S., Pfeiffer, J. R., Surviladze, Z., Gaudet, E. A., Oliver, J. M. High resolution mapping of mast cell membranes reveals primary and secondary domains of Fc(epsilon)RI and LAT. The Journal of cell biology. 154, 645-658 (2001).
- Lillemeier, B. F. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 11, 90-96 (2010).
- Woof, J. M., Burton, D. R. Human antibody-Fc receptor interactions illuminated by crystal structures. Nature reviews. Immunology. 4, 89-99 (2004).
- Axelrod, D. Cell-substrate contacts illuminated by total internal reflection fluorescence. The Journal of cell biology. 89, 141-145 (1981).
- Axelrod, D. Chapter 7: Total internal reflection fluorescence microscopy. Methods in cell biology. 89, 169-221 (2008).
- Wieser, S., Moertelmaier, M., Fuertbauer, E., Stockinger, H., Schutz, G. J. (Un)confined diffusion of CD59 in the plasma membrane determined by high-resolution single molecule microscopy. Biophys J. 92, 3719-3728 (2007).
- Huang, F. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods. 10, 653-658 (2013).
- Axelrod, D., Koppel, D. E., Schlessinger, J., Elson, E., Webb, W. W. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 16, 1055-1069 (1976).
- Axelrod, D., Burghardt, T. P., Thompson, N. L. Total Internal Reflection Fluorescence. Ann. Rev. Biophys. Bioeng.. 13, 247-268 (1984).
- Huse, M., Quann, E. J., Shouts Davis, M. M. Whispers and the kiss of death: directional secretion in T cells. Nat Immunol. 9, 1105-1111 (2008).
- Betzig, E., et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313, 1642-1645 (2006).
- Rust, M. J., Bates, M., Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods. 3, 793-795 (2006).
- Grakoui, A., et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 285, 221-227 (1999).
- Bunnell, S. C. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 158, 1263-1275 (2002).
- Sherman, E. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 35, 705-720 (2011).
- Huppa, J. B., et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 463, 963-967 (2010).
- Axmann, M., Schuetz, G. J., Huppa, J. B. Measuring TCR-pMHC binding using a FRET-based microscopy assay. Journal of Visualized Experiments. , (2015).
- Lillemeier, B. F., Schuetz, G. J., Pfeiffer, J. R., Surviladze, Z., Wilson, B. S., Davis, M. M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U.S.A.. 103, 18992-18997 (2006).
- Natkanski, E., et al. B cells use mechanical energy to discriminate antigen affinities. Science. 340, 1587-1590 (2013).

