Phloem Sap Sampling from Brassica napus for 3D-PAGE of Protein and Ribonucleoprotein Complexes
Summary
Here we present a protocol to analyze the protein composition of large native protein:protein and protein:nucleic acid complexes from oilseed rape (B. napus) phloem exudate using a 3D polyacrylamide gel electrophoresis (PAGE) approach combining blue native (BN) with two denaturing PAGEs followed by mass spectrometric identification.
Abstract
Sampling the phloem of higher plants is often laborious and significantly dependent on the plant species. However, proteome studies under denaturing conditions could be achieved in different plant species. Native protein:protein and protein:nucleic acid complexes from phloem samples have as yet scarcely been analyzed, although they might play important roles in maintenance of this specialized compartment or in long-distance signaling. Large molecular assemblies can be isolated using a blue native gel electrophoresis (BN-PAGE). Their protein components can be separated by a subsequent sodium dodecyl sulfate PAGE (SDS-PAGE). However, proteins with similar molecular weights co-migrate, what can hinder protein identification by mass spectrometry. Combining BN-PAGE with two different denaturing gel electrophoresis steps, namely Tris-Tricine-urea and SDS-PAGE, enables the additional separation of proteins according to their hydrophilicity/hydrophobicity and thus increases resolution and the success of protein identification. It even allows distinguishing proteins that only differ in their posttranslational modifications. In addition, blue native northern blotting can be applied to identify the RNA components in macromolecular complexes. We show that our protocol is suitable to unravel the protein and RNA components of native protein:protein and ribonucleoprotein (RNP) complexes occurring in phloem samples. Combining a blue native PAGE with two different denaturing PAGE steps can help to separate different kinds of large protein complexes, and also enables an increased identification rate of their components by mass spectrometry. Furthermore, the protocol is robust enough to simultaneously detect potentially bound nucleic acids within single protein complexes.
Introduction
The long-distance conduits of the phloem are essential for the allocation of organic nutrients in higher plants, but are also involved in regulating many different processes important for growth and development, pathogen defense, and the adaptation to adverse environmental conditions. Besides small effectors like ions, phytohormones, or metabolites themselves, macromolecules like proteins and RNAs have recently emerged as potential signals.
Studies have shown that phloem loading of proteins is size dependent and controlled by the size exclusion limit (SEL) of the pore-plasmodesmal units connecting companion cells (CC) and sieve elements (SE) that are typically in the range of 10 to >67 kDa1,2,3. However, despite these size limitations larger proteins and protein complexes have been found within the phloem system challenging the idea of size exclusion4, and even nonspecific loss of proteins from CC to SE has been suggested5.
Multiprotein as well as large ribonucleoprotein complexes play pivotal roles in biosynthesis and maintaining the structural integrity of the cell6. There is evidence that phloem-mobile protein-protein and ribonucleoprotein complexes exist4,7, but to date little about their function is known. In phloem sieve elements protein:protein and RNP complexes have been implicated with long-distance transport and/ or signaling8,9. To unravel the functions of the translocated complexes, studying their composition under varying environmental or stress conditions is required. To achieve this, native complexes have to be isolated and their components have to be separated with sufficient resolution.
To isolate high-molecular-weight complexes from the phloem, it is first necessary to obtain sap samples in sufficient quality and quantity. The technique that can be used for phloem sampling depends on the plant species of interest. Three main methods to obtain phloem sap exist 10: insect stylectomy11, EDTA-facilitated exudation12, and spontaneous exudation13.
Phloem samples from Arabidopsis thaliana (A. thaliana), the major model plant in laboratories worldwide, can only be obtained by EDTA-facilitated exudation or aphid stylectomy14,15. Both methods lead to either highly diluted phloem sap or very low sample amounts. EDTA-facilitated exudation is also prone to contamination and might not represent the real phloem composition16. Spontaneous phloem exudation is limited to plant species like cucurbits, lupine or yucca, where cutting off plant parts leads to a spontaneous emission of phloem sap that can be easily collected13,17,18,19. We recently established oilseed rape (Brassica napus) as a suitable model system for phloem analysis, since it is a close relative of A. thaliana and several microliters of phloem sap can be obtained from small incisions that has been shown to be of high purity4,8,20. Furthermore, the B. napus genome sequence was released in 201421.
Except for aphid stylectomy, all phloem collection methods described include damaging of surrounding tissue at the site of sampling, and the composition of phloem sap may change in response to injury. Therefore it is essential to check the quality of phloem samples, regardless of the sampling method applied. There are different methods for quality control of collected phloem sap, e.g. by determination of RNase activity22,23, RT-PCR with primers against Rubisco8,24, or the analysis of the sugar composition8.
Different methods to isolate and separate protein complexes can be applied, including size exclusion chromatography, sucrose density ultracentrifugation, or sample filtration through membranes with distinct molecular weight cut offs. However, these approaches do not allow high resolution. The technique called blue native PAGE (BN-PAGE), first described by Schägger et al.25, is superior in terms of resolution. It was successfully used to determine the molecular weights of large native protein complexes from various organelles or from cellular lysates and their relative abundances26,27.
To further elucidate the complex composition of protein complexes, it is necessary to separate the individual components under denaturing conditions, leading to complete disassembly of the complex components. This can be achieved by a second dimension of SDS-PAGE 28,29 or, to achieve higher resolution, by a second dimension of isoelectric focusing (IEF) followed by a third dimension of SDS-PAGE30 and subsequent identification of the proteins using mass spectrometry.
In this protocol, we describe sampling of pure phloem sap from B. napus plants and the analysis of protein complexes from such phloem samples using a 3D electrophoretic approach combining BN-PAGE with Tris-Tricine-urea-PAGE and SDS-PAGE. The separated protein complex components are subsequently identified using mass spectrometry. In addition, we introduce blue native northern blotting as a method allowing the detection of RNAs in large RNP complexes4, extending the applicability of the approach.
Protocol
1. Sampling and preparation of phloem sap from B. napus plants 4,8,20
- For phloem sap sampling, use well-watered 8-week-old B. napus plants that show only initial flowering to prevent pollen contaminations.
- Use a hypodermic needle (Ø 0.8 mm) and punctate the inflorescence stem several times. Repeat at several other inflorescence stems and on other plants to obtain enough phloem sap (Figure 2a).
NOTE: For complex analysis, it is recommended to start with at least 600 µL of phloem sap. 4 to 5 plants will yield between 200 and 700 µL of phloem sap. - Wipe off the first drop from the injured sites with filter paper. These drops mainly contain highly contaminated material from surrounding injured tissue and need to be discarded.
- Collect the following drops by pipetting and store the collected exudate in a pre-chilled conical 1.5 mL reaction tube at -20 °C using an ice-free cooling system.
- Continue collecting until no further drop formation is observable, but no longer than 1 h. This procedure can be repeated after two days without significant loss of collected phloem sap.
NOTE: The collected phloem sap can be used immediately or stored at -80 °C for future analysis. For -80 °C storage, shock-freeze the reaction tube in liquid nitrogen. - Concentrate the phloem sample to approximately 1/6th of the initial volume using centrifugal concentrators with a molecular weight cut off (MWCO) of 10,000 Daltons (Da). Centrifuge the concentrated sample at 4 °C and 20,000 x g for at least 15 min to remove dust particles.
NOTE: Phloem sap concentration does not lead to a significant loss of proteins. - For subsequent complex analysis, proceed with step 3.1.
2. Phloem sap purity control using reverse transcriptase PCR (RT-PCR) 4,8,20
- Isolate total RNA from phloem sap using a standard RNA extraction method for liquid material (e.g. TRIzol or phenol-chloroform extraction followed by isopropanol precipitation from the aqueous phase and ethanol washing steps according to the manufacturer's protocol).
Caution: Phenol-chloroform is toxic. Work under a hood with appropriate personal protection equipment. The extracted RNA can be stored in ethanol at -80 °C for up to six months. - Remove contamination of DNA by digestion with DNase I (1 u/µg) for 45 min at 37 °C and inactivate the enzyme by adding EDTA to a final concentration of 5 mM and incubate the sample at 75 °C for 10 min.
- To get pure RNA, purify and concentrate the sample by another purification step (e.g. using RNA Cleanup Kits, according to the manufacturer's instructions).
- Perform a reverse transcriptase PCR (RT-PCR) for synthesis of cDNA with a cDNA synthesis kit using 150 ng of pure RNA as described by the manufacturer's protocol.
NOTE: The cDNA can be stored at -20 °C until further use. - Use 5 µL of cDNA as a template for a standard PCR reaction as specified by the manufacturer, to amplify compartment-specific transcripts and check by agarose gel electrophoresis. Confirm the purity of phloem samples by e.g. using primers for rubisco small subunit, thioredoxin h, and the pollen coat protein (Figure 1b, Table 1).
3. Blue native PAGE 4,25,26,31
- Use either self-poured native gradient gels as described elsewhere27 or commercial Bis-Tris gradient gels.
- Load 20 to 30 µg of the concentrated protein sample per well onto the gel in at least triplicates.
- Run the gel at 4 °C and 150 V using dark blue cathode buffer B and anode buffer (Table 2) until the sample passed 1/3rd of the gel (ca. 20 to 30 min, Figure 3a).
NOTE: For northern blotting, a single gel lane is sufficient, while for the analysis of protein composition by 3D PAGE and mass spectrometry it is recommended to combine the same band from up to ten gel lanes to concentrate less abundant proteins. - Pause the run and exchange dark blue cathode buffer B with light blue cathode buffer B/10 (Table 2) and continue the run. Finish the gel run when the blue front starts to elute out of the gel (ca. 120 to 180 min, Figure 3a).
NOTE: The finished blue native gel can be stored at 4 °C for at least one week. Prolonged storage can lead to diffuse bands and should be avoided. Dark blue buffer B can be reused.
4. OPTIONAL: Detection of RNA using blue native northern blotting 4
- Cut out distinct bands or use the whole gel lane from the Blue native gel and transfer them onto a nylon membrane by semi-dry blotting at 3.5 mA/cm2 for 60 min and mark the upper and lower gel border with a pencil on the membrane (Figure 4a).
- After blotting, wash the membrane with DEPC-treated water and dry between two filter papers.
- Perform UV-crosslinking of the blotted membrane with 120,000 µJ/cm2 (85 s if using the Crosslinker in Table of Materials).
- Pre-hybridize the membrane with 6 mL of hybridization buffer (commercial or self-made) for 60 min at 68 °C in a hybridization oven (Figure 4a).
- Add an additional 1 mL of hybridization buffer containing 4 µL of 3'-biotinylated probes (100 nM).
- Incubate the membrane with the probe overnight, while cooling the oven from 68 °C to 37 °C.
- Wash the membrane with buffer containing 2x SSC and 0.1 % SDS, for 5 min at room temperature to remove probes bound unspecifically.
Caution: SDS can cause irritations. Wear gloves and lab coat when working with SDS and SDS containing solutions. - Perform the detection of the 3'-biotinylated probes on the membrane e.g. with a Streptavidin-HRP-conjugate and luminol as a horseradish peroxidase (HRP) substrate, resulting in a sensitive chemiluminescent reaction (Figure 4a).
5. Second dimension: Tris-Tricine-urea PAGE 4,28
- Excise single bands from the blue native gel. Combine bands from samples run in parallel lanes to achieve a further concentration of complex proteins (Figure 5).
NOTE: Excised bands can be stored in single reaction tubes until further use at 4 °C. - Pour a 12 % Tris-Tricine-urea gel (thickness of 1 mm, 6.8cm x 8.6 cm (width x length)). Prepare 10 mL of gel solution A for the separating gel (Table 2), pour between two glass plates and overlay with isopropanol. Remove the isopropanol after the polymerization is finished and transfer 4 mL of gel solution B (Table 2) for the stacking gel onto the gel and insert a 10 well comb.
Caution: Unpolymerized acrylamide can be neurotoxic and TEMED is harmful if inhaled. Always wear personal protection equipment and work under a hood. - Transfer the excised gel bands into a 1.5 mL reaction tube and add 1 mL of 2x SDS sample buffer for equilibration of the gel pieces (Figure 5, Table 2).
- Incubate the samples for 10 min at room temperature, prior to boiling in a heating block (95 °C) or in a microwave and incubate the samples again at room temperature for a further 15 min.
- Stack several equilibrated gel pieces representing the same complex into one single gel pocket.
- Perform the electrophoresis using anode and cathode running buffers at 150 V for 45 to 60 min and excise the whole gel lane.
- Incubate the cut lane for 20 min in acidic buffer (100 mM Tris, 100 mM acetic acid) and equilibrate in 125 mM Tris-HCl, pH 6.8 for another 20 min (Figure 5).
6. Third dimension: SDS-PAGE 4,32
- Pour a 15 % SDS separating gel according to Laemmli32 (thickness: 1.5 mm, 6.8cm x 8.6 cm (Width x Length)) (Table 2) and add a thin layer of a 4 % stacking gel above.
Caution: SDS, acrylamide and TEMED are toxic and harmful. Work under a hood and wear personal protection equipment. - Transfer the equilibrated gel lane onto the SDS gel and cover the gel with 0.5 % (w/w) agarose in 1x SDS running buffer supplemented with a trace of bromophenol blue (Figure 5).
- Boil the SDS running buffer with agarose in the microwave prior loading onto the gel.
- For running a protein marker to estimate the molecular weights of the single components, insert a piece of filter paper on one end of the gel until the agarose solidified or use a special comb typically used for IPG gels.
- Run the gel at 150 V for 60 min and stain the gel with either colloidal Coomassie, Silver stain or any other staining method suitable for subsequent mass spectrometric analysis.
- Analyze the visible protein spots using mass spectrometric approaches like matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) or LC-MS/MS mass spectrometry.
NOTE: We recommend to use the colloidal Coomassie staining as already described33. In our hands, even less abundant proteins can be sufficiently stained and allows a mass spectrometric analysis with reasonable data quality.
Representative Results
Here we present a protocol that allows the analysis of protein:protein as well as protein:RNA complexes in phloem samples from Brassica napus. The workflow is illustrated in Figure 1. One major advantage of B. napus over other model organisms is the possibility of relatively easy phloem sample collection from small punctures in the inflorescence stem. This allows to obtain pure phloem samples in comparably large amounts. When sampling phloem, it is always advisable to check for contaminations, for example by RT-PCR8. A representative result obtained from a pure phloem sample is depicted in Figure 2. Non-contaminated phloem samples should show a visible band for thioredoxin h, whereas no signal for rubisco and the pollen coat protein should appear. The rubisco small subunit can serve as a control in samples from leaves, inflorescence stems or pollen. Thioredoxin h should be detectable in all samples, since its mRNA has been found in the phloem of different species, while the pollen coat protein transcript should only be visible in flower bud samples. Contaminations can occur when phloem sampling is performed at fully flowering plants (pollen coat protein) or when the first droplets from the phloem sampling punctures are not discarded properly (rubisco).
Prior to BN-PAGE it is mandatory to concentrate the phloem sap. Using 20 to 30 µg of protein per well, at least four major protein complex bands become visible after BN-PAGE of B. napus phloem sap, too low or too high concentrations do not allow a clear separation of the complexes (Figure 3). It is recommended to load several gel bands representing the same complex into one single pocket of the second-dimension gel to further concentrate the proteins from each single complex for downstream processing and subsequent mass spectrometric identification.
To identify the individual components of phloem macromolecular complexes, we combined the initial BN-PAGE with a second dimension Tris-Tricine-urea and a third dimension SDS-PAGE to achieve a separation of single proteins. Here, a separation with high resolution can be observed due to the different protein migration behaviors in urea and SDS-PAGE gels. Typically, proteins belonging to a single complex will be visible as a diagonal line across the gel. Proteins above this line have an increased hydrophobicity, whereas proteins below the line represent more hydrophilic ones28 (Figure 5, Figure 6).
For the characterization of RNP complexes, we used a part of a BN-PAGE gel to perform BN northern blotting. After blotting of a whole lane onto a nylon membrane and crosslinking by UV radiation, the RNA can be visualized using RNA-specific probes as illustrated in Figure 4. Here it is shown that a specific tRNA is only present in one specific complex. The protein components of this tRNA-binding complex can be further investigated using the 3D gel approach described above. Mass spectrometric analyses showed that this complex contains mainly tRNA-ligases what confirms the tRNA binding activity found by BN northern blotting.
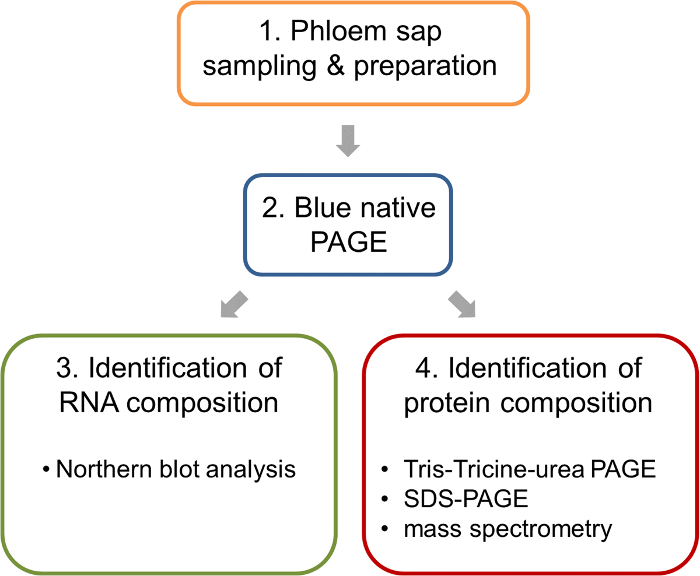
Figure 1: Work flow of the analysis of ribonucleoprotein complexes in the phloem sap of oilseed rape. After sampling and preparation of phloem samples, BN-PAGE is performed to separate native complexes. Such BN-PAGE gels allow the parallel analysis of nucleic acids by BN northern blotting and the identification of the protein components of the complexes by mass spectrometry. Please click here to view a larger version of this figure.
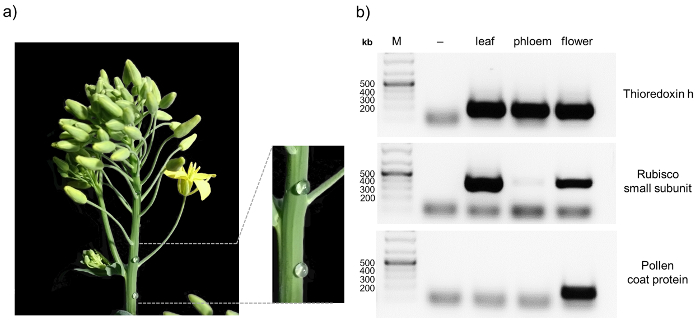
Figure 2: Sampling and purity control of phloem sap of B. napus. After puncturing the inflorescence stem of 8 – 10 week old oilseed rape plants with a sterile hypodermic needle, exudate is collected with a pipette and transferred to an ice-cold reaction tube (a) To confirm the purity of the phloem samples RT-PCR is performed, amplifying the transcripts of thioredoxin h, rubisco small subunit, and pollen coat protein in tissue-specific samples (b) RT-PCR without reverse transcriptase was performed as control (-). Please click here to view a larger version of this figure.
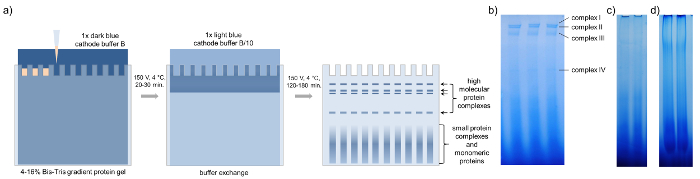
Figure 3: BN-PAGE. Schematic representation of blue native PAGE with concentrated phloem samples of B. napus. (a) After loading the gradient protein gel with 15-20 µL of concentrated phloem sap, BN-PAGE is performed with 1x dark blue cathode buffer B at 150 V for 20 – 30 min in a cold room. Then the buffer is exchanged against 1x light blue cathode buffer B/10 and the PAGE is finished at 150 V for 120 – 180 min until the dark blue running front runs out of the gel. The BN-PAGE gels show four distinct bands representing four different high-molecular-weight complexes. (b) Loading too little (c) or too much (d) phloem proteins reduces the visibility of individual complexes. Please click here to view a larger version of this figure.
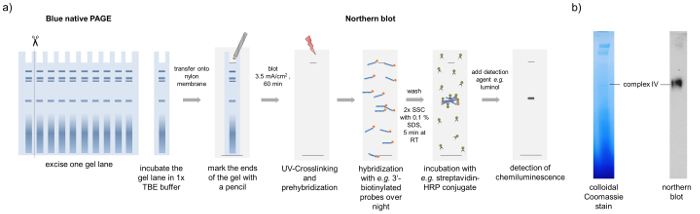
Figure 4: BN northern blotting. Schematic representation of the BN northern blotting method. (a) A lane of the BN-PAGE is transferred onto a nylon membrane by semi-dry blotting. After UV-crosslinking and pre-hybridization, the membrane is incubated with RNA specific probes (here a methionine tRNA-specific probe), which are biotinylated at the 3'-end in a hybridization oven over night. Free probes are removed by washing steps and the detection of the RNA is carried out by a streptavidin-HRP conjugate and luminol. Using this approach, we observed that complex IV from the BN-PAGE contains methionine tRNA. (b) The Coomassie stained gel and the tRNA blot resemble two different gel lanes run in parallel to avoid migration differences. Please click here to view a larger version of this figure.
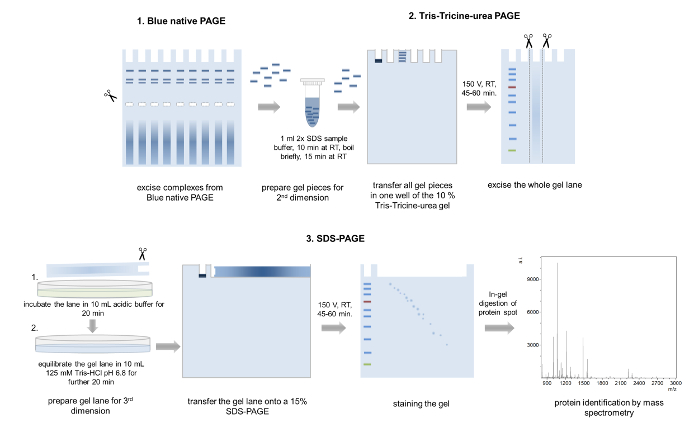
Figure 5: 3D-PAGE. Schematic representation of the 3D-PAGE approach. Several bands from the same complex are excised from a BN-PAGE gel, incubated in loading buffer and transferred into one well of the second dimension Tris-Tricine-urea-PAGE. After electrophoresis, whole gel lanes are cut out, washed in acidic acid buffer, equilibrated, and placed onto a 15% SDS-PAGE gel. After the third dimension PAGE, the separated proteins are Coomassie stained and analyzed by mass spectrometry. Please click here to view a larger version of this figure.
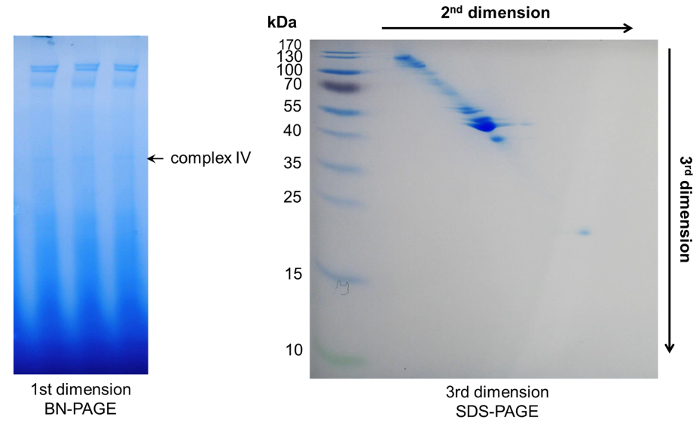
Figure 6: Representative results after 3D-PAGE. Within the first dimension BN-PAGE several complexes appeared. Their protein components could be further separated by two subsequent denaturing PAGEs, resulting in a diagonal spot pattern. Please click here to view a larger version of this figure.
| Probe | Sequence |
| Thioredoxin h | forward: CTCAAGGCAGCCAAAGAATC reverse: ATGGCCTGAACATCGAACTC |
| Rubisco small chain | forward: TTCACCGGCTTGAAGTCATC reverse: CCGGGTACTCCTTCTTGCAT |
| Pollen coat protein BRU77666 | forward: TCAGAACTGGAGCTTCAACG reverse: TTCCTTAATGGCCTCAGTGG |
Table 1: Primers used for phloem sample purity control. To confirm phloem sample purity primers specific for rubisco small chain, thioredoxin h, and pollen coat protein can be used for cDNA amplification.
| Buffers and Solutions | Content | Comment | |
| dark blue cathode buffer B | 15 mM Bis-Tris pH 7.0 50 mM Tricine 0.02% (w/v) Coomassie blue G250 |
recipe is adapted from Fiala et al. 17 pH should be adjusted to 7.0 Buffer can be made as a 10x stock |
|
| light blue cathode buffer B/10 | 15 mM Bis-Tris pH 7.0 50 mM Tricine 0.002% (w/v) Coomassie blue G250 |
Buffer can be prepared by diluting cathode buffer B with cathode buffer without Coomassie | |
| anode buffer | 50 mM Bis-Tris pH 7.0 |
Buffer can be prepared as a 10x stock solution | |
| 2x SDS sample buffer | 125 mM Tris-HCl pH 6.8 4% SDS 20% Glycerol 0.2 M DTT 0.02% (w/v) bromophenol blue |
||
| Transfer buffer 1x TBE buffer | 89 mM Tris pH 7.6 89 mM boric acid 2 mM EDTA |
TBE buffer can be made and stored as stocks of 5x or 10x. It is important to use RNase-free deionized water. | |
| 2x SSC buffer | 300 mM NaCl 30 mM Na3citrate pH 7.0 |
||
| 3x gel buffer | 3 M Tris 1 M HCl 0.3% SDS pH 8.45 |
recipe adapted from Schägger 19 | |
| Tris-Tricine gel solution A | For 10 mL: 30% acrylamide-bisacrylamide (29:1) 3.34 mL 6 M Urea 3x gel buffer 3.34 mL 70% glycerol 1.4 mL add ddH2O to 10 mL 10% APS 40 µL TEMED 4 µL |
Weight 6 M Urea and add 3x gel buffer, the acrylamide-bisacrylamide solution and 70% gycerol. Heat to 60 °C in a water bath to solubilize the urea. Add ddH2O to 10 mL, 10% APS and finally TEMED for polymerization. | |
| Tris-Tricine gel solution B | For 6 mL: 30% acrylamide-bisacrylamide (29:1) 0.8 mL 6 M Urea 3x gel buffer 1.5 mL add ddH2O to 6 mL 10 % APS 45 µL TEMED 4.5 µL |
Weight 6 M Urea and add 3x gel buffer and acrylamide-bisacrylamide solution. Heat to 60 °C in a water bath to solubilize the urea. Add ddH2O to 10 mL, 10% APS and finally TEMED for polymerization. | |
| 1x Tris-Tricine running buffer (anode) | 100 mM Tris 22.5 mM HCl pH 8.9 |
recipe adapted from Schägger 19 | |
| 1x Tris-Tricine running buffer (cathode) | 100 mM Tris 100 mM Tricine 1% SDS pH 8.25 |
recipe adapted from Schägger 19 | |
| Acidic buffer | 100 mM Tris 100 mM Acetic acid |
||
| 4% SDS stacking gel | For 2 mL: ddH2O 1.4 mL 30% acrylamide-bisacrylamide (37.5:1) 0.33 mL 1 M Tris pH 6.8 0.25 mL 10% SDS 20 µL 10% APS 20 µL TEMED 2 µL |
||
| 15% SDS separating gel | For 10 mL: ddH2O 2.3 mL 30% acrylamide-bisacrylamide (37.5:1) 5 mL 1 .5 M Tris pH 8.8 2.5 mL 10% SDS 100 µL 10% APS 100 µL TEMED 4 µL |
||
| 1x SDS running buffer | 25 mM Tris 192 mM glycine 0.1% SDS |
||
| Coomassie staining solution | 5% (w/v) aluminum sulfate-(14-18)-hydrate 10% (v/v) ethanol (96%) 2% (v/v) orthophosphoric acid (85%) 0.02% (w/v) Coomassie Brilliant Blue G-250 |
||
| ULTRAhyb Ultrasensitive Hybridization Buffer | Ambion, Life Technologies | ||
Table 2: Solutions and buffer recipes. List of all buffers and solutions necessary for 3D-PAGE analysis.
| Spot no. | MW obs. [kDa] | Identification | Organism | Accession no. | MW [kDa] | MASCOT score | |
| CIV_1 | 130 | Putative disease resistance protein At4g19050 | B. napus | CDX78917 | 131.1 | 110 | |
| CIV_2 | 130 | Putative disease resistance protein At4g19050 | B. napus | CDX78917 | 131.1 | 89 | |
| CIV_3 | 100 | Heat shock 70 kDa protein 14-like | B. napus | XP_013748865 | 89.9 | 113 | |
| CIV_4 | 90 | Eukaryotic elongation factor 2 Cell division control protein 48 homolog A |
B. napus B. napus |
CDX90241 CDY41316.1 |
89.5 89.5 |
111 197 |
|
| CIV_5 | 85 | Heat shock protein 90-2-like | B. rapa | XP_009132342 | 79.8 | 172 | |
| CIV_6 | 80 | Threonine–tRNA ligase Glycine–tRNA ligase |
B. oleracea B. napus |
XP_013599115 CDY43195 |
81.5 80.8 |
110 88 |
|
| CIV_7 | 70 | Heat shock protein 70 kDa Lysine–tRNA ligase-like |
B. oleracea B. napus |
XP_013685267 XP_013747530 |
67.8 69.9 |
230 150 |
|
| CIV_8 | 65 | Myrosinase Aspartate–tRNA ligase 2 Asparagine–tRNA ligase 1 |
B. napus B. napus B. napus |
ABQ42337 XP_013670803 XP_013729126 |
60.6 61.6 63.5 |
183 141 153 |
|
| CIV_9 | 60 | Myrosinase | B. napus | ABQ42337 | 60.6 | 164 | |
| CIV_10 | 55 | Adenosylhomocysteinase 2-like | B. rapa | XP_009135865 | 53.1 | 139 | |
| CIV_11 | 55 | Elongation factor 1-alpha 1-like | B. oleracea | XP_013586115 | 51.9 | 161 | |
| CIV_12 | 50 | Cystine lyase CORI3-like | B. napus | XP_013653143 | 48.1 | 237 | |
| CIV_13 | 44 | Cystine lyase CORI3-like | B. rapa | XP_009108611 | 48.3 | 213 | |
| CIV_14 | 38 | Fructose-bisphosphate aldolase | B. rapa | XP_009115993 | 38.4 | 226 | |
| CIV_15 | 45 | Cystine lyase CORI3-like | B. rapa | XP_009108611 | 48.3 | 219 | |
| CIV_16 | 45 | Cystine lyase CORI3 | B. rapa | CDY69765 | 46.9 | 209 | |
| CIV_17 | 38 | Fructose-bisphosphate aldolase | B. rapa | XP_009115993 | 38.4 | 124 | |
| CIV_18 | 18 | Peptidyl-prolyl cis-trans isomerase CYP18-3-like | B. napus | XP_009101932 | 18.3 | 128 | |
| CIV_19 | 45 | Cystine lyase CORI3-like | B. rapa | XP_009108611 | 48.3 | 177 | |
Table 3: Identified protein components from complex IV. Several tRNA ligases and further proteins have been found within the tRNA binding complex using MALDI-TOF mass spectrometric analysis after 3D-PAGE.
Discussion
The phloem is a compartment of high interest, since it constitutes the major transport route for photoassimilates, and is also essential for long-distance signaling between different plant parts9,20,34,35. In addition to small molecules, macromolecules like proteins and RNAs have been implicated with phloem maintenance and signaling4,7,8. The analysis of phloem composition is restricted by the limited accessibility to phloem samples in most plant species. The insect stylet technique is widely applicable, but experimentally demanding and results in very small amounts of phloem samples11. One easy technique used for phloem sampling is EDTA-facilitated exudation12. EDTA chelates divalent ions like Calcium and thus prevents closing of sieve plates by P-proteins and callose. It is easy to use, but is prone to contamination by damaged cells and samples are diluted. Depleting divalent ions by EDTA could also lead to a breakdown of existing complexes. However, it allows sampling from a wide range of species including the model plant A. thaliana15. Spontaneous exudation of phloem sap only occurs in a small number of plant species. It is an invasive method that involves significant injury of plant tissues10. We recently established B. napus as a model species for studying long-distance transport4,8,20. Here, phloem sap can be sampled from small incisions, which reduces wounding effects and sources of contamination.
Using different sampling techniques, the proteome of phloem samples from different plant species has been investigated in recent years7,8,17,36,37,38,39. For these proteome studies, proteins were extracted and precipitated under denaturing conditions and therefore do not allow conclusions about protein:protein and protein:nucleic acid interactions present under native conditions. Co-immunoprecipitation (CoIP) and overlay assays indicated that a major ribonucleoprotein complex might exist in phloem sap from pumpkin40, but it was not demonstrated that any macromolecular complexes exist under native conditions within the phloem system.
Blue native PAGE, introduced by Schägger et al.26, in combination with subsequent high-resolution gel electrophoresis steps enable the separation of the individual components of large protein complexes. Using 3D-PAGE analyses of native B. napus phloem exudate, we could recently demonstrate that several high-molecular-weight protein:protein and RNP complexes exist in phloem samples and are enzymatically active4. Alternative methods to isolate protein complexes as well as RNPs like size exclusion chromatography might exist, but fail in terms of resolution when compared to BN-PAGE. Furthermore, for a reasonable quality of complex separation, size exclusion column matrices have to be adapted according to the overall complex molecular weights being investigated. Analyzing complexes of different size will therefore demand different types of columns which is cost intensive and does not allow as high focusing power as a BN-PAGE.
Critical steps to analyze complexes from B. napus include the total amount of phloem sap used as starting material. At least 600 µL of phloem sample are necessary and can be obtained from five well-watered plants. For 3D-PAGE and BN northern blotting it is necessary to start the initial BN gel with a sufficient amount of phloem samples. To achieve this, phloem samples are concentrated until 20 to 30 µg of protein can be loaded into each well and at least triplicates of the same sample are run in parallel. Too low or too high concentrations reduce the detection of complexes (Figure 3). Phloem samples need be collected into reaction tubes on ice and should be stored frozen for later use to avoid protein degradation and turnover. Protease inhibitors have been found in all phloem proteomes analyzed, but also a fully active proteasome was described in the phloem of B. napus4. Furthermore, volume and composition of phloem samples depend on the sampling time-point and environmental conditions10. Therefore care must be taken to collect at the same time of the day under similar conditions. Stress treatments (e.g. drought) can reduce the amount of phloem that can be collected. To identify single proteins by mass spectrometry, it is mandatory to limit possible keratin contaminations by wearing gloves all the time. Keratin leads to additional signals during mass spec analysis and hampers protein identification. Running the BN-PAGE at a higher voltage or at ambient temperature can lead to heating of the gel that might result in disassembly of fragile complexes.
The protocol presented here is suitable for the analysis of protein:protein complexes. We also show that it can be used to detect specific RNAs contained in the complexes in parallel. To achieve this, we recently introduced a protocol for BN northern blotting4. RNAs are prone to degradation by RNAse contaminations. To limit such degradation DEPC-treated, water should be used.
Main limitations of the presented method include the estimation of the molecular weight of protein complexes separated by BN-PAGE, since migration behavior depends on size, shape and even on posttranslational modifications of the complexes31,41. Size estimation of RNP complexes is even more complicated due to the influence of nucleic acids on the migration behavior. It can also not be prevented that complexes of the same size co-migrate in one single band. This can lead to the erroneous prediction of complex compositions. Therefore verifying the observed interactions with complementary techniques, e.g. by functional assays4, might be necessary. Also proteins only weakly or transiently associated to a macromolecular complex might be lost in the BN buffer used. To avoid this, samples could be crosslinked, what can also lead to artifacts or can prevent protein identification, or the buffer conditions can be optimized.
In contrast to classical 2D-PAGE, the combination of urea- and SDS-PAGE allows the separation according to hydrophobicity and molecular weight instead of isoelectric point (pI) and molecular weight 28, and does not limit the separation to proteins of a specific pI range. Similar to classical 2D-PAGE, separated proteins are visible as single spots, but on a diagonal line 4,28 (Figure 5, Figure 6). More hydrophobic proteins appear in spots above this diagonal, whereas more hydrophilic proteins are found below the line28. The polyacrylamide concentrations used in this protocol will separate proteins between 25 to 80 kDa well. To allow a separation of smaller or larger proteins the polyacrylamide concentration can be varied or gradient gels can be used in the third dimension. The components of complex IV (Figure 3) separated by 3D-PAGE (Figure 6) were cut out, trypsin digested, and analyzed by mass spectrometry. This resulted in the identification of several tRNA ligases. The components of the other complexes have been published recently 4. Our 3D-PAGE enabled the separation of different ligases, namely aspartate, asparagine, threonine, lysine and glycine ligases, with similar molecular weights (Table 3). Using a methionine tRNA-specific probe, we were able to detect potentially bound tRNA within complex IV, but if full-length tRNA or tRNA fragments are in the complex cannot be discriminated with the probes used. To check if the nucleic acids are really bound within the complex and not co-migrating, protease digested phloem sap samples might serve as suitable migration controls.
It has been speculated earlier that protein translation can occur within phloem sieve tubes, since almost most components of the entire ribosome as well as elongation factors have been found in phloem samples4,7. Our finding of tRNA and tRNA ligases seems to support this idea. However, tRNA fragments present in phloem samples from pumpkin have been shown to be potent inhibitors of translation42 and functional ribosomes seem to be absent4, suggesting that translation cannot take place.
Taken together, the protocol presented here allows the separation of native protein:protein and even RNP complexes from phloem samples and the separation and identification of their individual components with high resolution. The application of this method enables the discovery of novel protein and RNP complexes in phloem samples and can therefore elucidate new functions in this highly specialized plant compartment.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Jennifer Deke and Urszula Zarzenska for their scientific support. This work was financially supported by a Career Integration Grant (CIG, PCIG14-GA-2013-63 0734) by the European Commission within the 7th framework program, the grant LFF-GK06 'DELIGRAH' (Landesforschungsförderung Hamburg), and the DFG grant (DFG KE 856_6-1) awarded to JK.
Materials
| 1.5 mL reaction tubes | Eppendorf | 30120086 | |
| 10 well comb for SDS PAGE, thickness 1 mm | BioRad | 1653359 | |
| 2-Propanol | AppliChem | 131090 | |
| 30 % Acrylamide-bisacrylamide solution (29:1) solution | BioRad | 1610156 | |
| 30 % Acrylamide-bisacrylamide solution (37.5:1) solution | BioRad | 1610158 | |
| 96 % Ethanol | Carl Roth | P075 | |
| Acetic acid | Carl Roth | 6755 | |
| Agarose | Lonza | 50004 | |
| Aluminum sulfate-(14-18)-hydrate | AppliChem | 141101 | |
| Ammonium peroxodisulfate (APS) | Carl Roth | 9592.3 | |
| Bis-Tris | AppliChem | A3992 | |
| Boric acid | AppliChem | A2940 | |
| Bromophenol blue | AppliChem | A2331 | |
| Centrifugal concentrators e.g. Vivaspin 500 (Mwco 10 kDa) | Sartorius | VS0102 | |
| Chemiluminescent Nucleic Acid Detection Module Kit | Thermo Fisher Scientific | 89880 | |
| Chromatography papers e.g. 3 mm CHR | Whatman | 3030-153 | |
| CoolBox M30 System | Biocision | BCS-133 | |
| Coomassie brilliant Blue G-250 | AppliChem | A3480 | |
| Diethyl pyrocarbonate (DEPC) | AppliChem | A0881 | |
| Dithiothreitol (DTT) | AppliChem | A1101 | |
| DNase I | AppliChem | A3778 | |
| Ethanol abs. | AppliChem | A3693 | |
| Ethylenediaminetetraacetic acid (EDTA) | AppliChem | A1103 | |
| Gel Imaging system e.g. Chemidoc Touch | BioRad | 1708370 | |
| GeneRuler 1kb plus | Thermo Fisher Scientific | SM1331 | |
| Glycerol | AppliChem | 141339 | |
| Glycine | AppliChem | 131340 | |
| Heating block TS1 | Biometra | 846-051-500 | |
| Hybridization oven e.g. GFL Hybridization Incubator 7601 | GFL | 7601 | |
| Hypodermic needle (0.8 mm) | B Braun | 4658309 | |
| IPG gel comb, thickness 1 mm | BioRad | 1653367 | |
| Labeling of RNA e.g.Biotin 3'end DNA Labeling Kit | Thermo Fisher Scientific | 89818 | |
| Native gradient gel e.g. Novex NativePAGE 4–16% Bis-Tris protein gel | Invitrogen | BN1002BOX | |
| Nylon membrane e.g. Hybond-N+ | Amersham Pharmacia | RPN203B | |
| Ortho-phosphoric acid | Carl Roth | 6366.1 | |
| PageRuler prestained protein ladder | Thermo Fisher Scientific | 26616 | |
| Power supply for Gel electrophoresis EPS | Amersham Pharmacia | 18-1130-01 | available from GE Healthcare |
| RNA Cleanup Kit | Qiagen | 74204 | |
| Semi dry blot e.g. Fastblot 43B semi dry Blot | Biometra | 846-015-100 | |
| Sodium chloride (NaCl) | AppliChem | A1149 | |
| Sodium dodecyl sulfate (SDS) | AppliChem | 142363 | |
| Synthesis of cDNA e.g. RevertAid First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | K1621 | |
| Tetramethylethylenediamine (TEMED) | AppliChem | A1148 | |
| Tricine | AppliChem | A3954 | |
| Tris | AppliChem | A1086 | |
| Tri-sodium citrate dihydrate | AppliChem | A3901 | |
| TRIzol Reagent | Thermo Fisher Scientific | 15596026 | |
| ULTRAhyb Ultrasensitive Hybridization Buffer | Thermo Fisher Scientific | AM8670 | |
| Urea | AppliChem | A1360 | |
| UV Crosslinker e.g. Stratalinker 2400 | Agilent | NC0477671 | from Fisher Scientific |
| Vertical electrophoresis apparatus e.g.The XCell SureLock Mini-Cell or Mini-Protean III System | Thermo Fisher Scientific or BioRad | EI0001 or 1658004 | |
| Wipers e.g. KIMWIPES Delicate Task Wipers | Kimberly-Clark | 34120 |
References
- Kempers, R., van Bel, A. J. E. Symplasmic connections between sieve element and companion cell in the stem phloem ofVicia faba L. have a molecular exclusion limit of at least 10 kDa. Planta. 201 (2), 195-201 (1997).
- Stadler, R., et al. Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J. 41 (2), 319-331 (2005).
- Imlau, A., Truernit, E., Sauer, N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 11 (3), 309-322 (1999).
- Ostendorp, A., et al. Functional analysis of Brassica napus phloem protein and ribonucleoprotein complexes. New Phytol. 214 (3), 1188-1197 (2017).
- Paultre, D. S. G., Gustin, M. -. P., Molnar, A., Oparka, K. J. Lost in Transit: Long-Distance Trafficking and Phloem Unloading of Protein Signals in Arabidopsis Homografts. Plant Cell. 28 (9), 2016-2025 (2016).
- Sali, A., Glaeser, R., Earnest, T., Baumeister, W. From words to literature in structural proteomics. Nature. 422 (6928), 216-225 (2003).
- Lin, M. -. K., Lee, Y. -. J., Lough, T. J., Phinney, B. S., Lucas, W. J. Analysis of the Pumpkin Phloem Proteome Provides Insights into Angiosperm Sieve Tube Function. Mol. Cell. Proteomics. 8 (2), 343-356 (2008).
- Giavalisco, P., Kapitza, K., Kolasa, A., Buhtz, A., Kehr, J. Towards the proteome of Brassica napus phloem sap. Proteomics. 6 (3), 896-909 (2006).
- Lucas, W. J., Yoo, B. C., Kragler, F. RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell. Bio. 2 (11), 849-857 (2001).
- Dinant, S., Kehr, J. Sampling and Analysis of Phloem Sap. Plant Mineral Nutrients. 953, 185-194 (2013).
- Kennedy, J. S., Mittler, T. E. A Method of obtaining Phloem Sap via the Mouth-parts of Aphids. Nature. 171 (4351), 528 (1953).
- King, R. W., Zeevaart, J. A. Enhancement of Phloem exudation from cut petioles by chelating agents. Plant Physiol. 53 (1), 96-103 (1974).
- Alosi, M. C., Melroy, D. L., Park, R. B. The regulation of gelation of Phloem exudate from cucurbita fruit by dilution, glutathione, and glutathione reductase. Plant Physiol. 86 (4), 1089-1094 (1988).
- Dinant, S., Bonnemain, J. -. L., Girousse, C., Kehr, J. Phloem sap intricacy and interplay with aphid feeding. Comptes rendus biologies. 333 (6-7), 504-515 (2010).
- Tetyuk, O., Benning, U. F., Hoffmann-Benning, S. Collection and analysis of Arabidopsis phloem exudates using the EDTA-facilitated Method. J Vis Exp. (80), e51111 (2013).
- Kovalskaya, N., Owens, R., Baker, C. J., Deahl, K., Hammond, R. W. Application of a modified EDTA-mediated exudation technique and guttation fluid analysis for Potato spindle tuber viroid RNA detection in tomato plants (Solanum lycopersicum). J. Virol. Methods. 198, 75-81 (2014).
- Rodriguez-Medina, C., Atkins, C. A., Mann, A. J., Jordan, M. E., Smith, P. M. Macromolecular composition of phloem exudate from white lupin (Lupinus albus L). BMC Plant Biol. 11 (1), 36 (2011).
- Taylor, J. S., Thompson, B., Pate, J. S., Atkins, C. A., Pharis, R. P. Cytokinins in the Phloem Sap of White Lupin (Lupinus albus L.). Plant Physiol. 94 (4), 1714-1720 (1990).
- Yoo, B., et al. A Systemic Small RNA Signaling System in Plants. Plant Cell. 16 (8), 1979-2000 (2004).
- Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C., Kehr, J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53 (5), 739-749 (2008).
- Chalhoub, B., et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 345 (6199), 950-953 (2014).
- Sasaki, T., Chino, M., Hayashi, H., Fujiwara, T. Detection of several mRNA species in rice phloem sap. Plant Cell Phys. 39 (8), 895-897 (1998).
- Doering-Saad, C., Newbury, H. J., Bale, J. S., Pritchard, J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J Exp. Bot. 53 (369), 631-637 (2002).
- Ruiz-Medrano, R., Xoconostle-Cázares, B., Lucas, W. J. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Dev. 126 (20), 4405-4419 (1999).
- Schägger, H., Cramer, W. A., von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217 (2), 220-230 (1994).
- Schägger, H., von Jagow, G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199 (2), 223-231 (1991).
- Fiala, G. J., Schamel, W. W. A., Blumenthal, B. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for analysis of multiprotein complexes from cellular lysates. J Vis Exp. (48), e2164 (2011).
- Rais, I., Karas, M., Schägger, H. Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics. 4 (9), 2567-2571 (2004).
- Schägger, H. Tricine-SDS-PAGE. Nature protocols. 1 (1), 16-22 (2006).
- Wittig, I., Braun, H. -. P., Schägger, H. Blue native PAGE. Nat Protoc. 1 (1), 418-428 (2006).
- Eubel, H., Braun, H. -. P., Millar, A. H. Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods. 1 (1), 11 (2005).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227 (5259), 680-685 (1970).
- Dyballa, N., Metzger, S. Fast and sensitive colloidal coomassie G-250 staining for proteins in polyacrylamide gels. J Vis Exp. (30), (2009).
- Turnbull, C. G. N., Lopez-Cobollo, R. M. Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytol. 198 (1), 33-51 (2013).
- Hall, S. M., Baker, D. A. The chemical composition of Ricinus phloem exudate. Planta. 106 (2), 131-140 (1972).
- Barnes, A., Bale, J., Constantinidou, C., Ashton, P., Jones, A., Pritchard, J. Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. J. Exp. Bot. 55 (402), 1473-1481 (2004).
- Walz, C., Giavalisco, P., Schad, M., Juenger, M., Klose, J., Kehr, J. Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry. 65 (12), 1795-1804 (2004).
- Walz, C., Juenger, M., Schad, M., Kehr, J. Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J. 31 (2), 189-197 (2002).
- Kehr, J. Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J Exp. Bot. 57 (4), 767-774 (2006).
- Ham, B. -. K., Brandom, J. L., Xoconostle-Cázares, B., Ringgold, V., Lough, T. J., Lucas, W. J. A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell. 21 (1), 197-215 (2009).
- Schamel, W. W. Biotinylation of protein complexes may lead to aggregation as well as to loss of subunits as revealed by Blue Native PAGE. J. Immunol. Methods. 252 (1-2), 171-174 (2001).
- Zhang, S., Sun, L., Kragler, F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150 (1), 378-387 (2009).

