Time-lapse 3D Imaging of Phagocytosis by Mouse Macrophages
Summary
Here we describe methods using spinning disk confocal microscopy to image single phagocytic events by mouse resident peritoneal macrophages. The protocols can be extended to other phagocytic cells.
Abstract
Phagocytosis plays a key role in host defense, as well as in tissue development and maintenance, and involves rapid, receptor-mediated rearrangements of the actin cytoskeleton to capture, envelop and engulf large particles. Although phagocytic receptors, downstream signaling pathways, and effectors, such as Rho GTPases, have been identified, the dynamic cytoskeletal remodeling of specific receptor-mediated phagocytic events remain unclear. Four decades ago, two distinct mechanisms of phagocytosis, exemplified by Fcγ receptor (FcγR)- and complement receptor (CR)-mediated phagocytosis, were identified using scanning electron microscopy. Binding of immunoglobulin G (IgG)-opsonized particles to FcγRs triggers the protrusion of thin membrane extensions, which initially form a so-called phagocytic cup around the particle before it becomes completely enclosed and retracted into the cell. In contrast, complement opsonized particles appear to sink into the phagocyte following binding to complement receptors. These two modes of phagocytosis, phagocytic cup formation and sinking in, have become well established in the literature. However, the distinctions between the two modes have become blurred by reports that complement receptor-mediated phagocytosis may induce various membrane protrusions. With the availability of high resolution imaging techniques, phagocytosis assays are required that allow real-time 3D (three dimensional) visualization of how specific phagocytic receptors mediate the uptake of individual particles. More commonly used approaches for the study of phagocytosis, such as end-point assays, miss the opportunity to understand what is happening at the interface of particles and phagocytes. Here we describe phagocytic assays, using time-lapse spinning disk confocal microscopy, that allow 3D imaging of single phagocytic events. In addition, we describe assays to unambiguously image Fcγ receptor- or complement receptor-mediated phagocytosis.
Introduction
Twenty years before Metchnikoff's observation of phagocytic mesenchyme cells in starfish larvae, in 1882, and subsequent development of his theory of phagocytosis1, Ernest Haeckel described, in 1862, the engulfment of insoluble dye particles by blood cells of Thetis fimbris (Tethys fimbria), a species of predatory sea slug (Ernest Haeckel. Die Radiolarien (Rhizopoda radiaria): Eine Monographie; Druck und Verlag von Georg Reimer, Berlin, 1862). He explicitly described membrane protrusions enveloping the particles, which were subsequently taken up into the cytoplasm and accumulated around the cell nucleus. More than 100 years later, a pioneering study by Kaplan suggested that there were at least two morphologically distinct mechanisms of phagocytosis2. Kaplan showed by means of scanning electron microscopy that mouse peritoneal macrophages ingested an IgG-opsonized sheep red blood cell using thin membrane extensions which reached up and tightly enveloped the particle, initially giving rise to a cup-like structure. Phagocytic cup formation required actin polymerization since it was abrogated by the cell-permeable fungal toxin cytochalasin B, known to block actin dynamics3. In contrast, sheep red blood cells opsonized with complement appeared to sink directly into the macrophage without the generation of membrane extensions, although, in some images, membrane ruffles can be seen in the immediate vacinity of the sinking particles. Unlike phagocytic cup formation, complement receptor-mediated sinking in was insenstive to cytochalasin B treatment2. In the experiments described by Kaplan, complement opsonization was performed by incubating immunoglobulin M (IgM) labeled sheep red blood cells with serum from complement C5-deficient mice, which circumvents hemolysis by the complement C5-dependent terminal complement complex.
The two modes of phagocytosis, phagocytic cup formation and sinking in, identified by Kaplan have become established opinion in the field4,5,6,7,8,9. However, the ultra-high resolution images used in the original study by Kaplan2, as well as a similar study by Munthe-Kaas et al.10, only provide snapshots of phagocytic events. In a recent review, Rougerie et al.11 stressed that morphological differences between FcγR- and CR-mediated phagocytosis remain to be clarified, and, moreover, membrane ruffles have been observed during complement receptor-mediated particle uptake2. Live-cell imaging of single phagocytic events spanning from particle capture to internalization, combined with genetically modified mouse models, could greatly improve our understanding of how phagocytes capture and ingest particles. One approach could be to use fast atomic force microscopy (AFM) which allows ultra-high resolution (10–20 nm) topographical imaging of living cells. Recently, a fast AFM system12 has been developed, which is suitable for imaging cell surfaces rapidly with low noise. This technique has the advantage that high-resolution, topographical and mechanical parameters of living cells can be measured at short intervals (seconds), unlike scanning electron microscopy, which necessitates the fixation and critical point drying of cells. Another approach is time-lapse 3D confocal microscopy, which is widely available, although phototoxicity and bleaching are limiting factors during recordings. This approach is highly versatile and allows optical sectioning with high spatial resolution and enables extraordinary flexibility in labeling with a staggering range of fluorescent probes, including genetically encoded fluorescent proteins. Here we describe phagocytosis assays using time-lapse spinning disk confocal microscopy that allow high spatiotemporal resolution of specific receptor-mediated phagocytic events.
Protocol
The protocols follow the guidelines of our local human research ethics committee, as well as the animal care guidelines.
1. Isolation of Resident Mouse Peritoneal Macrophages
- Sacrifice the mouse (3–4 months of age (either sex); e.g. C57BL/6 strain) using an overdose of the volatile anesthetic isoflurane (>5% in air) or carbon dioxide13, followed by cervical dislocation. Induction of anesthesia can be readily assessed by loss of righting reflex14, as well as by loss of paw withdrawal reflex.
- After cleaning the belly of the mouse with 80% ethanol in water, make a midline skin incision and expose the underlying abdominal wall.
- Place a 24-G plastic catheter into the peritonium and lavage the cavity with 2x 4.5 mL ice-cold Hank's buffered salt solution (HBSS), without Ca2+ and Mg2+via a 5 mL plastic syringe. While injecting, leave about 0.5 mL residual HBSS (5–4.5 mL (injected) = 0.5 mL) in the syringe in case tissue is sucked into the tip of the catheter and needs to be expelled. Transfer the aspirated suspension into a 14 mL polypropylene round-bottom tube and centrifuge at 300 x g for 6.5 min at room temperature.
NOTE: The tip of the plastic catheter is blunt, which, compared to a needle, minimizes injury to the abdominal contents. Typically, following gentle abdominal massage, 8 mL peritoneal cell suspension can be retrieved from the peritoneal cavity. - Discard the supernatant and resuspend the peritoneal cells in 1 mL bicarbonate-free RPMI 1640 medium containing 20 mM Hepes, 10% heat-inactivated fetal bovine serum and antibiotics, such as penicillin (100 units/mL) and streptomycin (100 µg/mL), prepared by diluting 100x penicillin/streptomycin 1:100. This typically gives a concentration of around 6 x 106 cells/mL.
NOTE: About 30% of isolated peritoneal cells are macrophages, whereas the other (smaller) cells are lymphocytes, predominantly B cells.
2. Seeding of Peritoneal Cells in Channel Slides
- After pipetting the cell suspension up and down to reduce clumping, pipette 100 µL suspension into the prefilled channel (volume, 100 µL) of a fibronectin-coated channel slide (a 100 µL channel has the dimensions (length x width x height): 50 mm x 5 mm 0.4 mm).
- Prefill the chamber by adding 1 mL serum-supplemented RPMI 1640 (Hepes) medium to one of the two reservoirs of the channel slide, tilting the slide, and then aspirating the medium from the downstream reservoir (Figure 1).
- Remove unwanted air bubbles, or long strips of air, in the channel by adding 1–2 mL medium to one of the two reservoirs, tightly applying a reservoir cap and then pumping out the air through rhythmic thumb pressure on the cap and, where necessary, tilting the slide. After expelling air, tilt the slide with the uncapped reservoir (and containing medium) downward before removing the cap to avoid air being sucked back into the channel.
- Prefill the chamber by adding 1 mL serum-supplemented RPMI 1640 (Hepes) medium to one of the two reservoirs of the channel slide, tilting the slide, and then aspirating the medium from the downstream reservoir (Figure 1).
- Incubate the channel slide, seeded with cells, in a moist chamber for 2 h at 37 °C in the absence of CO2 (CO2 is not required since the HCO3-/CO2 buffer system is replaced by Hepes). The channel slides can be conveniently placed on a rack, which holds eight slides. Typically, 6–8 slides are prepared from one mouse, although up to 10 or more is possible.
- Remove the rack and exchange the RPMI 1640 (Hepes) medium in each slide for RPMI 1640 medium containing bicarbonate, supplemented with 10% fetal bovine serum, as well as penicillin/streptomycin.
- Tilt the slide and aspirate medium first from the lower reservoir and then from the upper reservoir. Next, add 1 mL of the new medium to one of the reservoirs, tilt the slide and aspirate medium from the downstream reservoir, after it has flowed through the channel.
- After this wash (medium exchange) step, fill the channel slide by adding 1 mL medium to one of the reservoirs.
NOTE: Washing steps using channel slides is very simple and effective in terms of solution exchange and removing non-adherent cells. Note that the RPMI 1640 (bicarbonate) medium is normally pre-incubated with 5% CO2 overnight to ensure thermal and pH equilibrium.
- Incubate the cells overnight at 37 °C in a humidified incubator with 5% CO2. Perform experiments on the following day.
3. Isolation of Human Red Blood Cells
- On day 2 (second day of experiments; after overnight incubation of the isolated peritoneal macrophages), collect 1–2 mL peripheral venous blood from a healthy donor, into a heparinized tube. Transfer 1 mL into a round bottom 2.0 mL (polypropylene) microcentrifuge tube, centrifuge at 300 x g for 5 min at 18 °C (empirical setting), and remove the supernatant (plasma and buffy coat).
- Gently aspirate 100 µL of the sedimented red blood cells and mix this volume 1:1 with modified RPMI 1640 (Hepes) medium (described below) in a round bottom 2.0 mL microcentrifuge tube. Label the tube "1:1".
NOTE: The RPMI 1640 (Hepes) medium used on day 2 (for assays) after isolating cells contains, in addition to 10% heat-inactivated fetal bovine serum, penicillin (100 units/mL), streptomycin (100 µg/mL), and 1 mM N-(2-mercaptopropionyl)glycine (to reduce phototoxicity). Hereafter, this medium is referred to as "modified" RPMI 1640 (Hepes) medium. - Pipette 4 µL of the 1:1 diluted red blood cell suspension into 2 mL modified RPMI 1640 (Hepes) medium, pre-pipetted into a 2.0 mL microcentrifuge tube (repeat this step, i.e. prepare 2 tubes). Label each tube "4:2000". Place the "1:1" and "4:2000" tubes on ice.
4. Labeling of the Macrophage Plasma Membrane
- Remove the seeded channel slides from the CO2 incubator and exchange the RPMI 1640 (bicarbonate) medium in each slide for modified RPMI 1640 (Hepes) medium. Next, place the cells in a moist chamber at 37 °C in the absence of CO2.
NOTE: After overnight incubation and washing, most of the lymphocytes are removed from the channel. The majority of the remaining adherent cells are macrophages, which can be identified either morphologically or by immunofluorescence staining using anti-F4/80 antibody. Around 95% of mouse resident peritoneal macrophages are F4/80 positive. - Dilute fluorescently green labeled anti-mouse F4/80 antibody (stored at 4 °C) 1:40 in modified RPMI 1640 (Hepes) medium. Take a slide, tilt it, and pipette (drop by drop) 100 µL of the antibody mixture into the opening of the 100 µL channel of the slide (see Figure 1). Aspirate medium that flows into the downstream reservoir. Incubate the cells for 20 min at 37 °C (humidified environment, without CO2). During the incubation period, label the human red blood cells (see Section 5).
NOTE: As alluded to above, CO2 is not required since the HCO3-/CO2 buffer system is replaced by Hepes. - After 20 min incubation time (during which human red blood cells are stained and opsonized), wash the slide by adding 1 mL modified RPMI 1640 (Hepes) medium to one of the reservoirs and aspirating the medium after it has flowed through the channel, facilitated by tilting the slide. Add 1 mL medium for short-term storage or proceed to an experiment (i.e., pipetting in particles (opsonized red blood cells) and imaging phagocytosis by time-lapse spinning disk confocal microscopy).
5. Labeling the Plasma Membrane of Human Red Blood Cells
- Begin the labeling of human red blood cells immediately after incubating a slide of macrophages with green fluorescently labeled anti-F4/80 antibody (after Section 4.2). After gentle mixing, take 400 µL from one of the "4:2000" tubes (see Section 3.3) and dispense it into a (round bottom) 2.0 mL microcentrifuge tube. Allow time for the suspension to warm to around 37 °C (see paragraph below). Add 0.4 µL orange/red fluorescent plasma membrane stain (aliquots stored at -20 °C), mix and incubate at 37 °C for 5 min.
NOTE: A heated aluminium block, placed inside the laminar flow hood, is useful for minimizing heat loss while preparing slides. Tubes can be placed into bore wells of the heating block and a separate, or integrated, heated aluminium plate can serve as a working space. - After the 5 min incubation period, prepare the first wash step by adding 1600 µL modified RPMI 1640 (Hepes) medium (to fill the 2.0 mL microcentrifuge tube).
- Centrifuge the tube at 300 x g for 5 min at 18 °C (empirical setting). A compact red pellet should be visible on the wall (i.e., off-center) at the bottom of the tube. Rotate the tube so that the pellet is facing upwards and carefully remove all of the supernatant successively (in two steps) using a 1–1.5 mL pipette tip.
- After aspirating the supernatant, add 2,000 µL modified RPMI 1640 (Hepes) medium, mix (to resuspend the cells), and repeat the above centrifugation and supernatant aspiration steps.
- Resuspend the pellet (of plasma membrane stained and 2x washed red blood cells) with 400 µL of modified RPMI 1640 (Hepes) medium. Label the tube PMS (abbreviation for plasma membrane stain).
NOTE: Alternatively, the succinimidyl ester of a pH-sensitive rhodamine derivative, which becomes more strongly fluorescent at acidic pH, could be used to label human red blood cells. In this case, the fluorescence intensity additionally serves as a measure of phagosome maturation after particles have been internalized.
6. Opsonization (Labeling) of Human Red Blood Cells with Mouse Immunoglobulin G (IgG)
- Add 1 µL mouse (IgG2b) monoclonal (clone HIR2) anti-human CD235a (1 mg/mL) antibody (stored at -20 °C) to the tube labeled PMS (see Sections 5.2–5.5), containing plasma membrane stained human red blood cells suspended in 400 µL medium. CD235a (also known as glycophorin A) is an erythroid lineage-specific membrane sialoglycoprotein.
- Incubate at 37 °C for 8 min. Note that opsonization of human red blood cells with IgG causes agglutination (cell clumping). Although agglutination can serve as a visual indicator of opsonization, it is undesirable for the imaging of single phagocytic effects. To circumvent agglutination, repeatedly mix the cell suspension (every 1 min) using, for example, a variable 20–200 µL volume pipette set to 200 µL.
- Towards the end of the 8 min incubation period, if desired, wash the macrophage slide, incubated with green fluorescently labeled anti-F4/80 antibody, with 1 mL modified RPMI 1640 (Hepes) medium. Ensure that both reservoirs of the channel slide are free of medium after the washing steps.
7. Imaging the Phagocytosis of Plasma Membrane Stained and IgG-coated Human Red Blood Cells
- After the 8 min incubation with anti-CD235a antibody (Section 6.2), pipette 100 µL suspension containing plasma membrane-stained and IgG-opsonized human red blood cells into the channel of a slide containing macrophages labeled with green fluorescent anti-F4/80 antibody (Step 4). Thus, red fluorescent, IgG-opsonized human red blood cells are added to green fluorescent phagocytes (macrophages).
- As soon as human red blood cells have been added, mount the channel slide on a spinning disk confocal microscope (with the stage incubator set to 37 °C) for imaging, for example, via a 60X/1.49 oil-immersion objective lens. Start imaging as soon as possible, since particles begin to settle within 1 min.
NOTE: Using fluorescent beads with a diameter of 100 nm, we measured, by analysis of point spread functions, XY resolutions of x = 0.22 µm, y = 0.23 µm (488 nm laser) and x = 0.27 µm, y = 0.27 µm (561 nm laser). However, to reduce photobleaching and phototoxicity during time-lapse 3D imaging of living cells, we compromise spatial resolution using 2 x 2 binning. Binning increases sensitivity (improves the signal-to-noise ratio) and the allowable frame rate, but at the expense of spatial resolution. - Use a perfect focus (or similar) system to prevent focus drift during recordings. After activating the perfect focus system, use the offset control to focus onto macrophage lamellipodial protrusions immediately above the coverslip (see note below) of the channel slide. This focus level corresponds to Z = 0 µm. Obtain Z-stacks from -1 µm to +16 µm at 0.8 µm steps, which amounts to 22 Z-slices. Z-stacks can, for example, be obtained at a rate of 1 stack (for each channel) every 15 s for a total of 16 min.
NOTE: Longer recording periods suffer from losses of image quality, mainly due to photobleaching. Z-stacks are acquired for each of the two channels: macrophages are imaged using a 488 nm laser (green channel) and human red blood cells are imaged using a 561 nm laser (red channel). Using this approach, various views of macrophages ingesting IgG-opsonized human red blood cells at different timepoints can be obtained (for example, see Figure 2).
NOTE: Typical settings of the system (with 2 x 2 binning) are: green channel (20.5% laser (50 mW; 488 nm) power, 101 sensitivity (range, 0–255) and 200 ms exposure time); red channel (10.5% laser (50 mW; 561 nm) power, 101 sensitivity (range, 0-255) and 98 ms exposure time).
NOTE: The bottom of the channel slide is a polymer with the same thickness as a #1.5 glass coverslip and the same optical properties of glass, but, notably, the material has the advantage of gas permeability.
8. Imaging the Phagocytosis of Plasma Membrane Stained and Complement-coated Human Red Blood Cells
- Complement-opsonize plasma membrane stained and IgG-opsonized human red blood cells similarly to Sections 5 and 6, except, instead of pipetting 4 µL of the 1:1 diluted red blood cell suspension into 2 mL modified RPMI 1640 (Hepes) medium, as described in Section 3.3, pipette 4 µL into 1 mL modified RPMI 1640 (Hepes) medium, and accordingly label the tube 4:1,000. Thus, the human red blood cells are 2x more concentrated.
- Proceed with the steps in Sections 4 and 5, but starting out with the 4:1,000 rather than the 4:2,000 diluted stock of human red blood cells (4:1,000 and 4:2,000 refer to the subsequent dilutions of the initially 1:1 diluted human red blood cells described in Sections 3.1 and 3.2).
- Sacrifice a C5 null mouse (e.g., B10.D2-Hc0 H2d H2–T18c/oSnJ; Jackson Laboratory) by (> 5% in air) isoflurane inhalation followed by decapitation, and collect the blood. We usually drip about 0.8 mL of blood into a round bottom 14 mL plastic tube immediately following isoflurane inhalation and decapitation.
- After 1 h, when the blood is fully coagulated, transfer the residual fluid into a 2.0 mL microcentrifuge tube and centrifuge at 300 x g for 5 min at room temperature. Subsequently, carefully collect the yellowish serum. Typically, we recover at least 200 µL C5 null serum. Place the C5 null serum on ice.
- After 4 min incubation of plasma membrane stained human red blood cells with IgG, add another 1 µL of anti-CD235a antibody (giving 2x concentrated), and then take 50 µL of the cell suspension and mix it in a separate 2.0 mL microcentrifuge tube containing 50 µL C5 null serum. Continue to repeatedly mix (every 1 min) by pipetting up and down, as in Section 6.2, for another 4 min. Thus, after this step, the human red blood cells have been incubated with anti-CD235a antibody for a total of 8 min.
NOTE: The 4 min incubation period with C5 null serum is sufficient to activate the classical complement cascade and opsonize the human red blood cells with C3b, which is cleaved by Factor I to iC3b.
9. Confirmation of IgG- and C3b-opsonization of Human Red Blood Cells
- Confirm opsonization of human red blood cells with mouse anti-CD235a IgG (IgG2b; see Section 6.1) antibody by incubating the cells with goat anti-mouse (secondary) IgG antibody conjugated to a green or red fluorescent fluorophore.
- Add 1 µL mouse anti-CD235a IgG antibody and 1 µL fluorescently labeled anti-mouse secondary antibody to a 400 µL suspension of human red blood cells, and incubate at 37 °C for 8 min. Subsequently, wash twice (as in Sections 5.2–5.5).
- Resuspend the cell pellet with 400 µL modified RPMI 1640 (Hepes) medium and pipette 100 µL of the suspension into a channel slide (see Figure 1C) for confocal fluorescence imaging.
NOTE: The cells will clump (agglutinate) following washing and resuspension, but washing is necessary to reduce background fluorescence due to unbound secondary antibody.
- Confirm opsonization of human red blood cells, pre-labeled with mouse IgG and incubated with C5 null mouse serum, with C3b/iC3b using, for example, rat anti-mouse C3b/iC3b/C3c antibody and a secondary antibody, for example, goat anti-rat IgG antibody conjugated to a green fluorescent fluorophore.
- Repeat the steps in Section 8 using unstained human red blood cells.
- Add 0.25 µL fluorescently labeled anti-mouse C3b/iC3b/C3c antibody to the 100 µL mixture (50 µL IgG-labeled human red blood cells + 50 µL C5 null mouse serum) and incubate at 37 °C for 8 min.
- Wash twice, resuspend the pellet in 100 µL modified RPMI 1640 (Hepes) medium and pipette the suspension into a channel slide (see Figure 1C) for confocal fluorescence imaging.
Representative Results
A schematic diagram of the channel slide used for the imaging of phagocytosis by time-lapse spinning confocal microscopy is shown in Figure 1. Human red blood cells (hRBCs) are stained with the red fluorescent plasma membrane marker CellMask Orange, whereas isolated mouse resident peritoneal macrophages (Ms) are labeled with green fluorescent Alexa Fluor 488-conjugated anti-F4/80 antibody (Figure 2), which serves both as a specific marker of mouse macrophages and as a plasma membrane label. Human red blood cells can be opsonized with mouse IgG (mIgG), or mouse IgM (mIgM), by incubating the cells with IgG (or IgM) anti-CD235a antibody (CD235a, also known as glycophorin A, is a protein specifically expressed on human red blood cells). Time-lapse 3D imaging of green fluorescent macrophages presented with red fluorescent human red blood cells enables visualization of single particle phagocytic events (Figure 2). Close observation of single phagocytic events allows details of particle capture and ingestion to be delineated. For example, the capture of a mIgG-opsonized human red blood cell by a macrophage filopodium, a thin, finger-like projection, can be observed (Figure 3A; see also Horsthemke et al.15). Moreover, the squeezing of a human red blood cell during phagocytic cup formation can be observed (Figure 3A). Upon introduction of fresh mouse serum, mIgG-opsonized, or mIgM-opsonized, human red blood cells trigger the classical complement cascade, which culminates in the formation of a hemolytic membrane attack complex. The kinetics of complement-mediated hemolysis can be measured by imaging cells dual-stained with CellMask Orange and Calcein. Green fluorescent cytosolic Calcein is rapidly released from cells during hemolysis (Figure 3B).
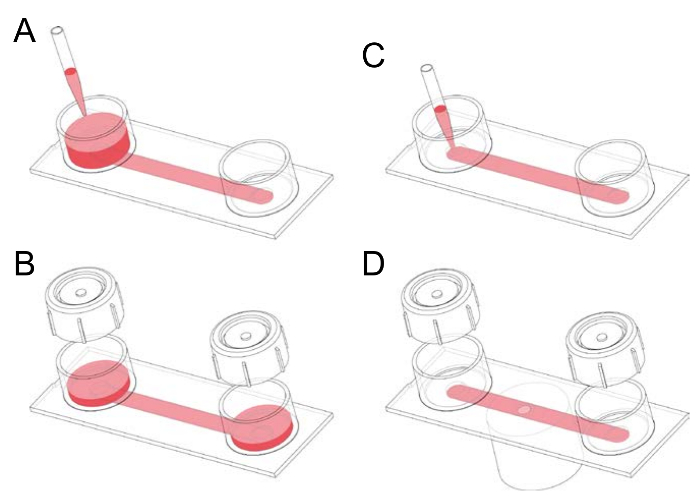
Figure 1: Handling of fibronectin-coated channel slides. (A) A channel slide consists of two reservoirs connected by a channel with the dimensions 50 mm x 5 mm 0.4 mm. Channel slides are initially prefilled by applying 1-2 mL medium to one of the two reservoirs and tilting the slide. (B) Caps can be placed onto the reservoirs prior to incubation. The caps can be conveniently used to pump out unwanted air bubbles prior to seeding the channel with cells. (C) The air bubble-free 100 µL channel can be filled by directly pipetting medium into the mouth of a channel. This step is used, for example, to seed macrophages into a slide or to add gfluorophore (green fluorescent)-conjugated anti-F4/80 antibody, which serves as a membrane label, as well as a mouse macrophage marker. (D) After pipetting particles, such as opsonized human red blood cells, into a channel seeded with fluorescently stained macrophages, the slide can be placed on the stage of an inverted microscope, and time-lapse spinning disk confocal microscopy can be performed. Please click here to view a larger version of this figure.

Figure 2: Time-lapse 3D imaging of phagocytosis. (A) Schematic diagram showing the opsonization of plasma membrane stained (red fluorescent) human red blood cells (hRBCs) with mouse (m) anti-CD235a immunoglobulin G (mIgG) antibody, and presentation of labeled hRBCs to mouse macrophages (Ms), labeled (green fluorescent) with green fluorescent fluorophore-conjugated anti-F4/80 antibody. (B)Time-lapse images (XZ views), obtained by spinning disk confocal microscopy, showing phagocytic cup formation and ingestion of mIgG-opsonized hRBCs. Scale bar = 10 µm. (C) 3D reconstructions showing macrophages ingesting mIgG-opsonized hRBCs. Corresponding XZ views (for 3 of the timepoints) are shown in B. Grid spacings represent 5.07 µm. Please click here to view a larger version of this figure.
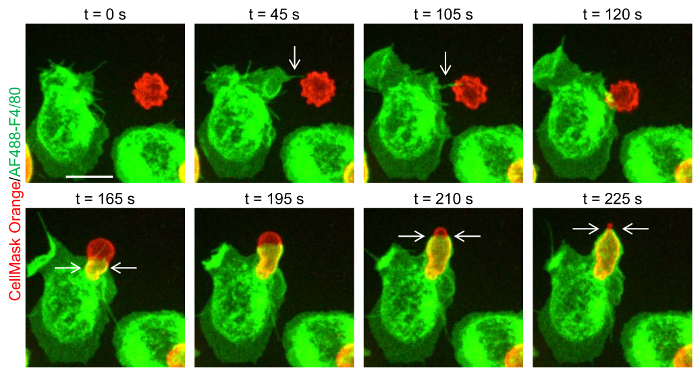
Figure 3: Capture of a particle by a filopodium. Time-lapse images, obtained by spinning disk confocal microscopy, showing a mouse macrophage capturing a mouse immunoglobulin G (mIgG)-opsonized human red blood cell (hRBC) via a filopodium (arrows in upper panel), a finger-like projection. Note that the red blood cell loses its crenations early during phagocytic cup formation. Furthermore, the phagocytic cup appears to squeeze the enveloped red blood cell (indicated by arrows in lower panel). Scale bar = 10 µm. Please click here to view a larger version of this figure.
Discussion
The vast majority of phagocytosis assays, especially end-point and high-throughput assays, do not provide visualization of how particles are actually captured, enveloped and ingested. Pioneering studies by Munthe-Kaas et al.10 and Kaplan2 in the 1970s suggested that strikingly different cytoskeletal reorganizations were involved in the phagocytosis of IgG-opsonized versus complement-opsonized particles (sheep red blood cells). Here we describe phagocytosis assays using spinning disk confocal microscopy which allow high-resolution, real-time imaging of single phagocytic events. Our model phagocyte is the the mouse resident peritoneal macrophage, which can be isolated with minimal handling, and we use freshly isolated human red blood cells as particles. However, the phagocytosis assays could be applied to other phagocytes, such as mouse bone marrow-derived macrophages or neutrophils, mouse macrophage cell lines, human monocyte-derived macrophages or human peripheral blood neutrophils. In the case of human phagocytes or mouse neutrophils, alternative fluorescently labeled antibodies would be required, such as fluorescently labeled anti-CD14 antibodies (human monocytes/macrophages)16 or anti-Gr-1 (Ly-6G) antibodies (mouse neutrophils).
Unopsonized human red blood cells, like traditionally-used sheep red blood cells, are inert in the sense that these cells are not (or, at least, very rarely) ingested by mouse peritoneal macrophages. This ensures, in contrast to polystyrene beads, low background activity. Human red blood cells can be conveniently opsonized with immunoglobulins using mouse IgG or IgM monoclonal antibodies against CD235a (glycophorin A), a specific marker of human erythrocytes (red blood cells) and erythroid precursors17,18. In parallel assays, fluorescently labeled anti-mouse IgG or IgM secondary antibodies can be applied to confirm opsonization. The IgG and IgM antibody classes are hemagglutinins, substances (antibodies) that cause red blood cells to agglutinate. To avoid agglutination, we intermittently mix the cell suspension during the 8 min incubation period with anti-CD235a antibody, and then we add the mixed suspension directly to a macrophage-containing channel slide (fibronectin-coated slide) without a wash step. Wash steps involve sedimentation of red blood cells by centrifugation, which strongly promotes agglutination. Before opsonizing human red blood cells, we label the plasma membrane with a lipophilic orange/red fluorescent probe. This probe is brightly fluorescent at the beginning of time-lapse recordings, but the signal gradually fades, probably largely due to photobleaching19. In addition, macrophages and the fibronectin coating of the slide may become weakly orange/red fluorescent during recordings. This problem is presumably due to insufficient washing of human red blood cells after labeling. Instead of using a lipophilic fluorescent plasma membrane marker, human red blood cells could be labeled with a pH-sensitive rhodamine derivative using its amine reactive succinimidyl ester15,20. This has the advantage of allowing visualization of phagosome maturation since fluorescence intensity increases with decreasing pH15,20, but this approach has the disadvantage that reactive ester preparations are currently expensive and unstable after reconstitution in aqueous medium.
IgG-opsonized human red blood cells are ingested via FcγRs, which can be confirmed using peritoneal macrophages isolated from NOTAM21 or Fcer1g-/- (Fcer1g knockout) mice. NOTAM macrophages bind IgG-opsonized human red blood cells, but lack ITAM (immunoreceptor tyrosine-based activation motif)-mediated signaling required to induce phagocytosis, whereas Fcer1g knockout macrophages do not express surface FcγRs. IgG- or IgM-opsonized human red blood cells can be additionally opsonized with C3b (which is cleaved to iC3b) by incubating the cells with freshly isolated serum from a complement C5 null mouse (wild-type serum causes hemolysis). The opsonins IgG and IgM activate the classical complement cascade, which leads to formation of pores (membrane attack complexes) and cell lysis. In mice lacking complement C5, the complement cascade proceeds to complement C3 cleavage, but C5 convertase lacks the substrate required to catalyze the terminal pathway. We developed simple assays to measure the kinetics of the complement cascade. In short, human red blood cells can be co-labeled with a red fluorescent, plasma membrane marker and the green fluorescent, cytosolic fluorophore. Upon formation of the membrane attack complex, formed by complement components C5-C9, the cytosolic fluorophore is rapidly (in seconds) lost from the cytosol. Visualization of the end-effector (cytolysis) function of the complement cascade indicated that 4 min incubation time is sufficient for C3b/iC3b opsonization of human red blood cells. In parallel assays, C3b/iC3b coating of human red blood cells can be easily assessed after applying a mixture of anti-mouse C3b and fluorescently labeled secondary antibodies. In this case, a wash step is required to remove unbound fluorescent antibodies. Although the wash step involves cell sedimentaion by centrifugation, which promotes agglutination, successful opsonization can be readily assessed by confocal microscopy. Complement receptor-mediated phagocytosis can be imaged by either applying IgG-/iC3b-opsonized human red blood cells to NOTAM or Fcer1g-/- macrophages or by introducing IgM-/iC3b-opsonized human red blood cells to wild-type macrophages. IgM-opsonized blood cells are not recognized by the ITAM-containing FcγRs (FcγRI, FcγRIII and FcγRIV) required for the phagocytosis of IgG-opsonized particles22.
To image phagocytic events, the plasma membrane of macrophages can be labeled with green fluorescent fluorophore conjugated anti-F4/80 antibody, which also serves as a specific marker of mouse macrophages. Human red blood cells can be rendered red fluorescent by incubation with an orange/red fluorescent plasma membrane marker, as discussed above. This lipophilic plasma membrane marker avoids potential confounding effects of antibody-based labels. Red fluorescent human red blood cells, with or without opsonization, can be directly pipetted into the 100 µL channel of a channel slide and 3D time-lapse imaged via a 60X/1.49 oil immersion (or similar) objective lens performed using the 488 nm and 561 nm laser lines, respectively, of a spinning disk confocal (or similar) microscope. It is tempting to optimize the system for high-resolution imaging, but the acquisition of repeated Z-stacks over 16 min or so may cause considerable photobleaching and phototoxicity. We chose to use 2×2 binning to promote good signal-to-noise ratios and allow reductions in excitation intensity and/or exposure times, but at the expense of optical resolution. In addition, to reduce phototoxicity, we add a scavenger of reactive oxygen species to the medium. In future studies, the assays could be modified to image the phagocytosis of apoptotic human red blood cells. Application of a Ca2+ ionophore, such as A23187, can be used to induce phosphatidylserine externalization23, an "eat me" signal and hallmark of early apoptosis24,25.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the grants HA 3271/4-1 and HA 3271/4-2 from the DFG (Deutsche Forschungsgemeinschaft), and the grant FF-2016-05 from EXC 1003 (Cluster of Excellence 1003), Cells in Motion (CiM), DFG.
Materials
| 24-G plastic catheter | B Braun Mesungen AG, Germany | 4254503-01 | Used for peritoneal lavage |
| Hank’s buffered salt solution without Ca2+ and Mg2+ | Thermo Fisher Scientific | 14170120 | Used for peritoneal lavage |
| 14 ml polypropylene round bottom tubes | BD Falcon | 352059 | Used to collect peritoneal cells |
| RPMI 1640 medium containing 20 mM Hepes | Sigma-Aldrich | R7388 | Basis medium for assays |
| Heat-inactivated fetal bovine serum | Thermo Fisher Scientific | 10082139 | Used as supplement for RPMI 1640 media |
| 100x penicillin/streptomycin | Thermo Fisher Scientific | 15140122 | Used as supplement for RPMI 1640 media |
| Fibronectin-coated µ-Slide I chambers | Ibidi, Martinsried, Germany | 80102 | Channel slides used for assays |
| µ-Slide (anodized aluminum) rack | Ibidi, Martinsried, Germany | 80003 | Autoclavable stackable rack for channel slides |
| RPMI 1640 medium containing bicarbonate | Sigma-Aldrich | R8758 | Medium for overnight culture |
| N-(2-mercaptopropionyl)glycine | Sigma-Aldrich | M6635 | Scavenger of reactive oxygen species |
| Alexa Fluor 488-conjugated rat (IgG2a) monoclonal (clone BM8) anti-mouse F4/80 antibody | Thermo Fisher Scientific | MF48020 | Mouse macrophage marker and plasma membrane label |
| CellMask Orange | Thermo Fisher Scientific | C10045 | Red fluorescent plasma membrane stain |
| Succinimidyl ester of pHrodo | Thermo Fisher Scientific | P36600 | Amine-reactive succinimidyl ester of pHrodo |
| Mouse (IgG2b) monoclonal (clone HIR2) anti-human CD235a | Thermo Fisher Scientific | MA1-20893 | Used to opsonize human red blood cells with IgG |
| Alexa Fluor 594-conjugated goat anti-mouse (secondary) IgG antibody | Abcam | Ab150116 | Used to confirm opsonization of human red blood cells with mouse IgG |
| Rat anti-mouse C3b/iC3b/C3c antibody | Hycult Biotech | HM1065 | Used to confirm C3b/iC3b opsonization of human red blood cells |
| Alexa Fluor 488-conjugated goat anti-rat IgG antibody | Thermo Fisher Scientific | A-11006 | Used as secondary antibody to confirm C3b/iC3b opsonization |
| Calcein/AM | Thermo Fisher Scientific | C3100MP | Used to load human red blood cells with Calcein |
| UltraVIEW Vox 3D live cell imaging system | Perkin Elmer, Rodgau, Germany | Spinning disk confocal microscope system | |
| Nikon Eclipse Ti inverse microscope | Nikon, Japan | Inverted microscope | |
| CSU-X1 spinning disk scanner | Yokogawa Electric Corporation, Japan | Nipkow spinning disk unit | |
| 14-bit Hamamatsu C9100-50 Electron Multiplying-Charged Couple Device (EM-CCD) peltier-cooled camera | Hamamatsu Photonics Inc., Japan | EM-CCD camera of the spinning disk confocal microscope system | |
| 488 nm solid state laser, 50 mW | Perkin Elmer, Rodgau, Germany | Laser (488 nm) source of spinning disk confocal microscope system | |
| 561 nm solid state laser, 50 mW | Perkin Elmer, Rodgau, Germany | Laser (561 nm) source of spinning disk confocal microscope system |
References
- Tauber, A. I. Metchnikoff and the phagocytosis theory. Nature reviews. Molecular cell biology. 4, 897-901 (2003).
- Kaplan, G. Differences in the mode of phagocytosis with Fc and C3 receptors in macrophages. Scandinavian journal of immunology. 6, 797-807 (1977).
- Cooper, J. A. Effects of cytochalasin and phalloidin on actin. The journal of cell biology. 105, 1473-1478 (1987).
- Caron, E., Hall, A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 282, 1717-1721 (1998).
- Aderem, A., Underhill, D. M. Mechanisms of phagocytosis in macrophages. Annual review of immunology. 17, 593-623 (1999).
- Chimini, G., Chavrier, P. Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nature cell biology. 2, E191-E196 (2000).
- Castellano, F., Chavrier, P., Caron, E. Actin dynamics during phagocytosis. Seminars in immunology. 13, 347-355 (2001).
- Swanson, J. A. Shaping cups into phagosomes and macropinosomes. Nature reviews. Molecular cell biology. 9, 639-649 (2008).
- Underhill, D. M., Goodridge, H. S. Information processing during phagocytosis. Nature reviews. Immunology. 12, 492-502 (2012).
- Munthe-Kaas, A. C., Kaplan, G., Seljelid, R. On the mechanism of internalization of opsonized particles by rat Kupffer cells in vitro. Experimental cell research. , 201-212 (1976).
- Rougerie, P., Miskolci, V., Cox, D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: role and regulation of the actin cytoskeleton. Immunological reviews. , 222-239 (2013).
- Liu, Z., et al. Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nature. 13, 143-146 (2016).
- Valentim, A. M., Guedes, S. R., Pereira, A. M., Antunes, L. M. Euthanasia using gaseous agents in laboratory rodents. Laboratory animals. 50, 241-253 (2016).
- McCarren, H. S., Moore, J. T., Kelz, M. B. Assessing changes in volatile general anesthetic sensitivity of mice after local or systemic pharmacological intervention. Journal of visualized experiments. (80), e51079 (2013).
- Horsthemke, M., et al. Multiple roles of filopodial dynamics in particle capture and phagocytosis and phenotypes of Cdc42 and Myo10 deletion. The Journal of biological chemistry. 292, 7258-7273 (2017).
- Bzymek, R., et al. Real-time two- and three-dimensional imaging of monocyte motility and navigation on planar surfaces and in collagen matrices: Roles of Rho. Scientific reports. 6, 25016 (2016).
- Poole, J. Red cell antigens on band 3 and glycophorin A. Blood reviews. 14, 31-43 (2000).
- Aoki, T. A Comprehensive review of our current understanding of red blood cell (RBC) glycoproteins. Membranes. 7, (2017).
- Song, L., Hennink, E. J., Young, I. T., Tanke, H. J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophysical journal. 68, 2588-2600 (1995).
- Miksa, M., Komura, H., Wu, R., Shah, K. G., Wang, P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. Journal of immunological. 342, 71-77 (2009).
- Boross, P., et al. FcRgamma-chain ITAM signaling is critically required for cross-presentation of soluble antibody-antigen complexes by dendritic cells. Journal of immunology. , 5506-5514 (2014).
- Ehrenstein, M. R., Notley, C. A. The importance of natural IgM: scavenger, protector and regulator. Nature reviews. Immunology. 10, 778-786 (2010).
- Closse, C., Dachary-Prigent, J., Boisseau, M. R. Phosphatidylserine-related adhesion of human erythrocytes to vascular endothelium. British journal of haematology. , 300-302 (1999).
- Barth, N. D., Marwick, J. A., Vendrell, M., Rossi, A. G., Dransfield, I. The "phagocytic synapse" and clearance of apoptotic cells. Frontiers in immunology. 8, 1708 (2017).
- Sivagnanam, U., Palanirajan, S. K., Gummadi, S. N. The role of human phospholipid scramblases in apoptosis: An overview. Biochimica et biophysica acta. 1864, 2261-2271 (2017).

