An Ultrahigh-throughput Microfluidic Platform for Single-cell Genome Sequencing
Summary
Single-cell sequencing reveals genotypic heterogeneity in biological systems, but current technologies lack the throughput necessary for the deep profiling of community composition and function. Here, we describe a microfluidic workflow for sequencing >50,000 single-cell genomes from diverse cell populations.
Abstract
Sequencing technologies have undergone a paradigm shift from bulk to single-cell resolution in response to an evolving understanding of the role of cellular heterogeneity in biological systems. However, single-cell sequencing of large populations has been hampered by limitations in processing genomes for sequencing. In this paper, we describe a method for single-cell genome sequencing (SiC-seq) which uses droplet microfluidics to isolate, amplify, and barcode the genomes of single cells. Cell encapsulation in microgels allows the compartmentalized purification and tagmentation of DNA, while a microfluidic merger efficiently pairs each genome with a unique single-cell oligonucleotide barcode, allowing >50,000 single cells to be sequenced per run. The sequencing data is demultiplexed by barcode, generating groups of reads originating from single cells. As a high-throughput and low-bias method of single-cell sequencing, SiC-seq will enable a broader range of genomic studies targeted at diverse cell populations.
Introduction
The genome serves as a blueprint of cellular identity and function, containing the entirety of an organism's coding potential. An understanding of cellular biology at the genome level can explain the observed phenotypic diversity within heterogeneous cell populations. This heterogeneity is apparent in biological systems and has broad implications for human health and disease. For example, gene copy number variations among tumor cells are linked to the evolution and spread of cancer1,2. In bacterial infections, pathogenicity islands present in a small fraction of genomes can be horizontally transferred and lead to the proliferation of antibiotic-resistant bacteria3,4. A primary challenge in studying genomes at the single-cell level is the low quantities of DNA available, as well as the need to analyze thousands of cells to sample the full diversity of genotypes. For these reasons, limitations in experimental throughput have hindered the effectiveness of single-cell studies, biasing results towards the most abundant cells. Single-cell isolation techniques such as flow sorting5,6, optical tweezers7, embedment in bulk gels8, and microfluidics9 are capable of processing hundreds of cells for sequencing; however, this represents only a small fraction of most samples. A method for single-cell genome sequencing with substantially higher throughput would allow deeper and more complete profiling of cell populations, thereby elucidating the role of genotypic diversity within these communities.
Droplet microfluidics enables the high-throughput manipulation of cells and biological reagents within millions of picoliter-scale reactors. To date, microdroplet technologies have been used to study differential expression patterns among cells from heterogeneous tissues10,11,12, deeply sequence long molecules13,14,15, and conduct chromatin immunoprecipitation sequencing (ChiP-seq) analyses on single cells16. Indeed, microdroplets are capable of high-throughput, compartmentalized operations, making them amenable to applications in single-cell genomics. The development of this technology presents its own unique technological challenges, however. Cells must be lysed, purified, and amplified with minimal bias, to uniformly sample cell populations. Additionally, unlike polyadenylated mRNA transcripts in mammalian cells, there is no comparable molecular motif in the genome to facilitate the capture of the target nucleic acid. For these reasons, single-cell genome sequencing has been difficult to implement in microdroplet platforms.
In this work, we provide a detailed protocol of our previously reported single-cell microfluidic approach capable of sequencing the genomes of tens of thousands of cells in a single experiment17. With this technology, called SiC-seq, bacterial cells are encapsulated in micron-scale hydrogels and individually lysed, tagmented, and merged with a microdroplet containing a unique oligonucleotide barcode, which is spliced onto the cell's genomic DNA via a single overlap extension polymerase chain reaction (PCR). The hydrogels serve as isolated containers in which high-molecular-weight genomic DNA is sterically encased, allowing smaller molecules such as detergents and lytic enzymes to access and purify DNA prior to barcoding18. This protocol processes >50,000 single cells in a matter of hours, resulting in a barcoded library ready for sequencing. Following the sequencing, the reads are demultiplexed according to their single-cell barcode sequence, resulting in a dataset comprised of millions of reads, each with a cellular index.
Protocol
1. Microfluidic Device Fabrication
- Prepare the microfluidic mask designs using computer-aided design (CAD) software (provided as .DWG; see Supplementary Files). Have these designs printed by the vendor with a 10 µm resolution on a circuit board film.
NOTE: For multi-layered microfluidic devices, the corresponding masks contain alignment marks. - For each device, fabricate the SU-8 master mold (Figure 1A) as follows.
- Prepare a 3-in diameter silicon wafer by pouring approximately 1 mL of SU-8 3025 photoresist onto the center of the wafer. Secure the wafer on the spin coater chuck by applying suction.
- See Table 1 for a listing of the layer thicknesses and spin speeds for each device. For all devices, begin the spin coating with 30 s at 500 rpm, followed by 30 s at the indicated speed.
- Remove the SU-8-silicon wafer from the spin coater and soft bake it on a hotplate set to 135 °C for 30 min. Allow the wafer to cool to room temperature after baking.
- Expose the SU-8-silicon wafer with the appropriate microfluidic mask under a collimated 190-mW, 365-nm UV LED for 3 min.
- After the exposure, hard bake the wafer on a hotplate set to 135 °C for 1 min. Following this baking step, allow the wafer to cool to room temperature.
- For a single-layered microfluidic device, skip to step 1.2.7. For a multi-layered microfluidic device, repeat steps 1.2.1 – 1.2.5 for the second layer of the photoresist (Figure 1B).
- Following the first hard bake for a single layer device (or the second hard bake for a multi-layered device), develop the wafer by immersing it into a bath of propylene glycol monomethyl ether acetate (PGMEA) for 30 min.
- After the wafer's development, use a squirt bottle containing PGMEA to rinse the wafer. Then rinse the wafer with a squirt bottle containing isopropanol before placing it on a 135 °C hotplate for 1 min to dry.
- Place the wafer (henceforth referred to as the master) into a Petri dish for casting with polydimethylsiloxane (PDMS).
- With the master prepared in step 1.2, proceed to perform the device fabrication with a PDMS casting.
- Prepare the PDMS by combining a silicone base with a curing agent in an 11:1 ratio by mass. Mix the silicone base and curing agent by hand with a stir stick.
- Degas the PDMS by placing it into a degassing chamber and applying a vacuum. Allow the PDMS to degas until air bubbles are no longer visible (typically 30 min).
- Carefully pour the degassed PDMS over the master, to a final PDMS-layer thickness of approximately 5 mm. Degas the PDMS again to ensure the removal of any air bubbles.
- After the degassing, bake the PDMS and the master at 80 °C for 80 min.
- Carefully excise the cured PDMS slab from the baked master using a razor blade. Ensure that all cuts are on top of the silicon wafer.
NOTE: Any cuts made off the silicon wafer may result in a lip preventing a uniform bonding. - Punch the inlets and outlets using a 0.75 mm biopsy punch. Remove any dust and stray PDMS using a packaging tape on the feature side of the device.
- Prior to plasma treating the device, clean a 50 mm x 75 mm glass slide by rinsing it with isopropanol and drying it.
- For the plasma treatment, place the PDMS slab and glass slide into the plasma bonder with the features facing up. Perform the plasma treatment using 1 mbar O2 plasma for 1 min. Bond the device to the glass slide by bringing the exposed, or face up, sides together.
- Following the plasma treatment, bake the device at 80 °C for 40 min.
- Finally, inject glass surface treatment fluid into one of the inlets to render the microfluidic channels hydrophobic. Ensure all channels are completely flooded with the solution and repeat the injection for each dropmaker. Bake the treated device at 80 °C for 10 min to evaporate excess solvent.
2. Encapsulation of Cells in Agarose Microgels
NOTE: See Figure 2A.
- Prepare 1 mL of 3% w/v low-melting temperature agarose in 1x Tris-EDTA (TE) buffer. Keep the agarose solution on a 90 °C heat block until immediately prior to the syringe loading.
- Prepare the cell suspension.
NOTE: This protocol and its associated microfluidic devices have been validated to work with bacterial cells, either from a frozen stock or fresh preparation. Mammalian cells, depending on the cell type, may require an adjustment of the microfluidic channel dimensions to accommodate the larger cell sizes.- Resuspend the cells in 1 mL of phosphate-buffered saline (PBS).
- Count the cells in a hemocytometer or by flow sorting. For 25 µm microgels with a target cell encapsulation rate of 1 in 10, prepare 1 mL of cell suspension at a final concentration of 2.4 x 107 cells/mL.
- Spin the cells down at 3,000 x g for 3 min. Aspirate the supernatant and resuspend the cell pellet in 1 mL of 17% v/v density gradient medium (see Table of Materials) in PBS. Keep it on ice until the syringe loading.
- Load a 3-mL syringe with fluorinated oil (HFE) containing a 2% w/w perfluoropolyether-polyethylene glycol (PFPE-PEG) surfactant, fit it with a 27 G needle, and place it into a syringe pump.
NOTE: Fit all syringes with a 27 G needle for the microfluidic steps in this protocol. To reduce the risk of accidental pricking, keep caps on all needles until the pump operation begins. - Load the cell suspension and molten agarose into 1-mL syringes, fit both with 27-gauge needles, and place these into syringe pumps.
- Keep the agarose syringe and pump warm with a small space heater to prevent the agarose from gelling in the syringe and inlet tubing. Set the space heater to HIGH and position it so that the heating surface is about 10 cm away from the syringe. Ensure that the temperature measured at the syringe is approximately 80 °C.
NOTE: Users are advised to maintain the heater at the recommended distance from the pumping apparatus to reduce the risk of equipment damage, including melting of the tubing. - Generate 25 µm microgel drops using the co-flow dropmaking device.
NOTE: See Figure 3A for a device schematic indicating the location of the reagent inlets and outlet.- Connect the syringe needles to the microfluidic device inlets using pieces of polyethylene (PE) tubing. Before inserting the tubes into the device, prime the pumps to remove the air from the line.
- Connect a piece of tubing to the outlet and place the free end in a 15 mL collection tube.
- Use the following (recommended) flow rates for dropmaking: 800 µL/h for the HFE 2% w/w PFPE-PEG; 200 µL/h for the cell suspension in PBS; and 200 µL/h for the 3% w/v agarose.
- After the dropmaking, place the collection tube at 4 °C for 30 min to ensure the complete gelation of the agarose.
3. Breaking and Washing the Agarose Microgels
- Remove the lower layer of oil from the collection tube using a 3 mL syringe fitted with a 20 G needle, taking care not to disturb the top layer of the agarose droplets.
- Break the emulsions with perfluorooctanol (PFO).
- Add 1 mL of 10% v/v PFO in HFE to the agarose drops. Pipet this solution up and down for 1 min to thoroughly coat the emulsions.
NOTE: For all the microgel wash steps, pipet the solutions with a 1,000 µL tip. The microgel suspension should appear homogeneous and free of clumps after pipetting it. - Spin the conical tube at 2,000 x g for 1 min to collect the agarose microgels. Remove the PFO/HFE supernatant by aspiration; the microgels are now free of their surfactant layer and will appear clear.
- Add 1 mL of 10% v/v PFO in HFE to the agarose drops. Pipet this solution up and down for 1 min to thoroughly coat the emulsions.
- Wash the microgels with a surfactant in hexane.
CAUTION: Hexane is a volatile organic solvent, and the wash in step 3.3 should be conducted in a fume hood.- Add 2 mL of 1% v/v sorbitan monooleate nonionic surfactant (see Table of Materials) in hexane to the agarose microgels. Pipet up and down 10x to mix, ensuring the complete breakup of the microgel pellet.
- Spin the tube at 1,000 x g for 1 min to collect the microgels. Aspirate the supernatant to remove the surfactant/hexane solution.
- Repeat the surfactant/hexane wash.
- Wash the microgels in an aqueous buffer to remove any residual organic solvent.
- Add 5 mL of TET buffer [0.1% v/v octylphenol ethoxylate nonionic detergent (see Table of Materials) in 1x TE] to the conical tube. Pipet up and down 10x to mix.
- Spin the conical tube at 2,000 x g for 2 min to collect the microgels. Aspirate the supernatant to remove the TET buffer.
- Repeat the TET wash 2x.
- Add 5 mL of 1x TE buffer to the conical tube. Pipet up and down 10x to mix.
- Spin the conical tube at 2,000 x g for 2 min to collect the microgels. Aspirate the supernatant to remove the TE buffer.
- Repeat the TE wash.
- Verify the cell encapsulation in the microgels under a microscope at a 400X magnification by staining a 10 µL aliquot of gels with 1x nucleic acid stain (see Table of Materials).
NOTE: Microgels will appear clear under the transparent channel while cell DNA will fluoresce under the GFP channel (497/520 nm excitation/emission wavelengths). An overlay of transparent and fluorescent channels will show the cells in the microgels as shown in Figure 4A.
4. Lysis of Cells in Agarose via Lytic Enzymes
- Prepare 1 mL of a lytic enzyme cocktail using 800 µL of TE buffer (1x), 2 µL of yeast lytic enzyme (5 U/µL stock, 10 U/mL final), 30 µL of dithiothreitol (1 M stock, 30 mM final), 60 mg of lysozyme (lyophilized powder), 15 µL of EDTA (0.5 M stock, 7.5 mM final), 2 µL of mutanolysin (100 U/µL stock, 200 U/mL final), 2 µL of lysostaphin (10 U/µL stock, 20 U/mL final), and 30 µL of NaCl (1 M stock, 30 mM final).
- Add additional TE to bring the volume to 1 mL. Mix the solution by vortexing.
- Add the entire 1 mL of the lytic enzyme solution to no more than 1 mL of the washed microgels. Mix by pipetting 10x. Incubate the mixture at 37 °C for >2 h in a shaker (the maximum is overnight incubation).
5. Detergent-based Microgel Treatment
- Following the lysis via lytic enzymes (step 4), wash the microgels.
- Spin down the microgels at 2,000 x g for 2 min and aspirate the supernatant. The droplets will appear opaque white due to cell debris dispersed in the pellet.
- Resuspend the microgels in 5 mL of 10 mM Tris-HCl buffer and pipet up and down 10x to mix.
- Spin the mixture down at 2,000 x g for 2 min, then remove the supernatant by aspiration.
- Perform a detergent treatment on the microgels.
- Resuspend the gels in a lithium dodecyl sulfate (LiDS) lysis buffer (0.5% w/v LiDS in 20 mM Tris-HCl) and 60 µL of 0.5 M EDTA to a final volume of 3 mL.
- Add 5 µL of a Proteinase K enzyme (800 U/mL stock). Pipet up and down 10x to mix, then incubate the mixture at 42 °C on a heat block for 1 h to solubilize the cell membranes and digest the proteins.
- Following the detergent treatment, wash the microgels.
- Spin the conical tube with the microgels down at 2,000 x g for 2 min. Remove the supernatant by aspiration.
- Wash the microgels with 10 mL of 2% v/v polysorbate 20 in water. Pipet up and down 10x to mix.
- Spin the conical tube down at 2,000 x g for 2 min, then remove the supernatant by aspiration.
- Wash the microgels with 10 mL of 100% ethanol to inactivate any residual enzyme. Pipet up and down 10x to mix.
- Spin the conical tube down at 2,000 x g for 2 min, then remove the supernatant by aspiration.
- Wash the microgels with 10 mL of 0.02% v/v polysorbate 20 in water. Pipet up and down 10x to mix.
- Spin the conical tube down at 2,000 x g for 2 min, then remove the supernatant by aspiration.
- Repeat the polysorbate 20 wash 3x. Pass the solution through a 100-µm cell strainer prior to the last wash to remove any large clumps.
- Resuspend the microgels in 5 mL of 10 mM Tris-HCl buffer to prevent DNA degradation. The microgels may be stored at 4 °C for up to 1 week prior to the tagmentation (step 7).
6. Generating Barcode Droplets by Digital PCR
- Prepare a 500-pM stock of BAR primer (Table 2) in a 1x TE buffer in a low-bind tube. Prior to each use, dilute the primer to a working stock of 1 pM and heat it to 70 °C for 1 min on a heat block.
- Prepare a 150-µL PCR reaction mix using 75 µL of high-fidelity hot start master mix (see Table of Materials) (2x), 42 µL of PCR-grade water, 3 µL of DNA_BAR primer (10 µM stock, 0.2 µM final), 3 µL of P7_BAR primer (10 µM stock, 0.2 µM final), 6 µL of barcode dilution (1 pM stock, 40 fM final), 6 µL of polysorbate 20 (50% v/v stock, 2% final), and 15 µL of PEG 6k (50% w/v stock, 5% final). Mix by pipetting up and down 10x.
- Prepare an HFE-backed syringe by drawing 200 µL of HFE oil into a syringe and fit it with a needle. Attach a section of PE tubing to the needle and prime the line by hand. Insert the end of the PE tubing into the target solution and carefully draw all 150 µL of the PCR mix into the PE tubing and syringe. Load the syringe into a syringe pump.
- Load a 1 mL syringe with fluorinated oil (HFE) containing a 2% w/w perfluoropolyether-polyethylene glycol (PFPE-PEG) surfactant, fit it with a needle, and place it into a syringe pump.
- Generate 25 µm barcode drops using the co-flow dropmaking device.
NOTE: See Figure 3A for a device schematic indicating the location of the reagent inlets and outlet.- Plug the Cells inlet of a co-flow dropmaker device with a small piece of lead solder.
- Connect the syringes loaded with HFE and PCR mix to the microfluidic device inlets using pieces of PE tubing, using the Molten Agarose inlet for the barcode PCR mix. Before inserting the tubes into the device, prime the pumps to remove the air from the line.
- Connect a piece of tubing to the outlet and place the free end in a 0.2 mL PCR tube. Use the following (recommended) flow rates for dropmaking: 600 µL/h for the HFE 2% w/w PFPE-PEG and 200 µL/h for the PCR mix. Collect the drops into the PCR tubes with approximately 50 µL of drops in each tube.
- After the dropmaking, carefully remove the lower layer of HFE oil from the emulsions using gel-loading pipet tips and replace it with FC-40 fluorinated oil containing a 5% w/w PFPE-PEG surfactant. Thermal cycle with the following protocol: 98 °C for 3 min, 40x of (98 °C for 10 s, 62 °C for 20 s, 72 °C for 20 s), 72 °C for 5 min, and then hold at 12 °C.
NOTE: The thermal-cycled droplets may be stored at 4 °C for up to 1 day. - Verify the barcode amplification and encapsulation rate.
- Prepare a 1x nucleic acid stain (see Table of Materials) in HFE with a 2% w/w PFPE-PEG surfactant; the stain is marginally miscible in HFE and will bind to the DNA in the droplets.
- Add 1 µL of thermal-cycled barcode emulsion to 10 µL of staining oil. Incubate them for 5 min at room temperature.
- Image droplets by fluorescent microscopy (GFP channel, 497/520 nm excitation/emission wavelengths) at a 200X magnification and count the barcode encapsulation rate. Note that the signal will be discrete: the droplets containing amplified barcodes will fluoresce brightly, whereas the empty drops will appear dark (Figure 4B).
7. Tagmentation of Genomic DNA in Droplets
NOTE: See Figure 2B.
- Prepare 500 µL of tagmentation solution using reagents from a next-generation sequencing library preparation kit (see Table of Materials). Use 7 µL of tagmentation enzyme, 250 µL of tagmentation buffer, and 243 µL of PCR-grade water. Mix them by vortexing and spin the mixture down to collect. Load this solution into an HFE oil-backed 1 mL syringe and fit it with a needle.
- Prepare the microgels for the reinjection.
- Spin down the microgel tube for 2 min at 2,000 x g and aspirate the supernatant. Transfer 200 µL of gels to the top of an HFE-backed syringe with a gel-loading pipet tip and seal the nozzle with a small piece of tape.
- Using a 3D-printed centrifuge adapter (see Supplemental Files 1 and 2), spin the microgel syringe for 3 min at 3,000 x g.
- Remove the liquid supernatant from the syringe using a gel-loading pipet tip. Push the microgel layer to the base of the syringe nozzle. Fit the syringe with a needle.
- Load a 3 mL syringe with fluorinated oil (HFE) containing a 2% w/w perfluoropolyether-polyethylene glycol (PFPE-PEG) surfactant, fit it with a needle, and place it into a syringe pump.
- Re-encapsulate the microgels into droplets containing tagmentation reagents.
NOTE: See Figure 3B for a device schematic indicating the location of the reagent inlets and outlet.- Connect the syringes containing HFE, tagmentation mix, and microgels to the microfluidic device inlets using pieces of PE tubing. Before inserting the tubes into the device, prime the pumps to remove the air from the line.
- Connect a piece of tubing to the outlet and place the free end in an empty 1-mL syringe with the plunger drawn to the 1 mL line.
- Use the following (recommended) flow rates for dropmaking: 2,000 µL/h for the HFE 2% w/w PFPE-PEG, 200 µL/h for the microgels, and 500 µL/h for the tagmentation mix.
- Verify the microgel encapsulation rate under a light microscope at a 400X magnification. Approximately 80 – 90% of the drops should contain a microgel, as shown in Figure 4C.
- Fit the syringe containing the tagmentation emulsions with a needle and incubate it upright in a heat block or oven for 1 h at 55 °C to fragment the genomic DNA.
8. Single-cell Barcoding by Microfluidic Double Merger
NOTE: See Figure 2C.
- Prepare the barcode droplets for the merger by replacing the FC-40 oil fraction with HFE 2% w/w PFPE-PEG. Carefully transfer these drops into a 1 mL syringe, fit it with a needle, and place it into a syringe pump.
- Load the incubated and tagmented microgel droplet syringe into a syringe pump.
- Prepare 500 µL of PCR mix. Add the reagents in the following order to prevent precipitates from forming: 140 µL of PCR-grade water, 10 µL of P5_DNA primer (10 µM stock, 0.2 µM final), 10 µL of P7_BAR primer (10 µM stock, 0.2 µM final), 50 µL of PEG 6k (50% w/v stock, 5% final), 50 µL of polysorbate 20 (50% v/v stock, 5% final), 250 µL of Taq Master Mix (2x) (see Table of Materials), 10 µL of isothermal polymerase (see Table of Materials), and 10 µL of neutralization buffer (see Table of Materials). Mix them by pipetting up and down 10x and spin the mixture down to collect it. Load this solution into an HFE-backed 1-mL syringe, fit it with a needle, and place it into a syringe pump.
- Load three 3-mL syringes with fluorinated oil (HFE) containing a 2% w/w perfluoropolyether-polyethylene glycol (PFPE-PEG) surfactant, fit each with a needle and place them into syringe pumps.
- Load three 1 mL syringes with 2 M NaCl, fit each with a needle and set them aside.
- Merge the barcode droplets, tagmented genome droplets, and PCR mix using the double merger device.
NOTE: See Figure 5 for a device schematic indicating the location of the reagent and electrode inlets and outlet.- Connect the 3 NaCl syringes to the 2 electrode inlets and single moat inletusing pieces of PE tubing. For the electrodes, apply pressure to the syringes manually until the electrodes are completely filled with salt solution. After filling the electrodes, apply manual pressure to the moat syringe until the moat is filled. Plug the free end of the moat with a small piece of lead solder.
- Connect the 3 HFE syringes mounted on pumps to the 2 spacer oil inlets and dropmaking oil inletusing pieces of PE tubing. Before inserting the tubes into the device, prime the pumps to remove the air from the line.
- Connect the PCR mix syringe, microgel drops syringe, and barcode drops syringe to their respective inlets using PE tubing. Shoot all droplet reinjection tubing with an antistatic gun before connecting them to the syringe needles; the antistatic treatment reduces the risk of droplet coalescence induced by static charges on the PE tubing.
- Connect a piece of PE tubing to the outlet and place the free end in a 0.2 mL PCR tube.
- Connect the needle of the electrode syringe to a cold cathode fluorescent inverter using an alligator clip. Set the inverter's DC power supply to 2 V.
- Run the double merger device with the recommended flow rates: 300 µL/h for the tagmented microgel drops, 100 µL/h for the barcode drops, 1500 µL/h for the HFE 2% w/w PFPE-PEG (dropmaking oil), 600 µL/h for the amplification mix, 200 µL/h for the HFE 2% w/w PFPE-PEG (barcode spacer oil), and 700 µL/h for the HFE 2% w/w PFPE-PEG (microgel spacer oil). Collect the drops into PCR tubes with approximately 50 µL of emulsion in each tube.
- Prior to the thermal cycling, carefully remove the lower layer of HFE oil from the emulsions using gel-loading pipet tips and replace them with FC-40 fluorinated oil containing a 5% w/w PFPE-PEG surfactant. Thermal cycle with the following protocol: 65 °C for 5 min, 95 °C for 2 min, 30x (95 °C for 15 s, 60 °C for 1 min, 72 °C for 1 min), 72 °C for 5 min, then hold at 12 °C.
- Recover the barcoded DNA from the thermal-cycled droplets.
- Pool the droplets into a microcentrifuge tube and break the emulsions using 20 µL of PFO. Vortex them for 10 s to mix.
- Spin the tube at 10,000 x g for 1 min to fractionate the mixture into aqueous (top) and oil (bottom) phases. Carefully remove the upper aqueous layer from the tube using a pipet and transfer it to a new microcentrifuge tube. Discard the oil phase.
- Purify the barcoded PCR product in a spin column according to the manufacturer's protocol and elute it in 20 µL of 1x TE buffer.
- Proceed to the performed sequencing and analysis as per steps 9 and 10.
9. Library Preparation and Sequencing
- Prepare the single-cell library for the sequencing by following the manufacturer's protocols for fragment size-selection and quantification.
- Sequence the paired-end library with default chemistry for Read 1 and Read 2. Use the custom Index 1 primer I7_READ (Table 2) for a 15-bp index read, corresponding to the single-cell barcode.
10. Single-cell Data Analysis
NOTE: Custom Python scripts for quality control and preliminary analysis of SiC-seq data can be downloaded from https://www.github.com/AbateLab/SiC-seq.
- Run the script "barcodeCleanup.py" to perform quality control on the barcoded reads and export the single-cell data to an SQLite database. For a control experiment, use this script with the "-align" flag set to align the reads to known reference genomes.
- Analyze the purity of the barcode groups (for a control experiment) using the script "purity.py" and confirm the high purity values consistent with Figure 6B.
Representative Results
The SiC-seq experimental workflow contains 3 PDMS microfluidic devices fabricated using a soft lithography procedure (Figure 1). A co-flow dropmaker (Figure 3A) generates 25 µm of digital barcode droplets for labeling genomic DNA with a unique single-cell identifier. The barcode oligonucleotides consist of a 15 bp degenerate sequence flanked by PCR handles for amplification (Table 2, BAR primer). The barcodes are diluted to a femtomolar concentration to achieve the single-molecule encapsulation, and all droplets receive either 0 or 1 barcode fragment(s). The droplets containing a barcode are amplified, yielding many copies of double-stranded barcode amplicons. A nucleic acid stain is used to verify the successful amplification and quantify the encapsulation rate of the barcode fragments (Figure 4B). The microgels are generated by co-flowing a bacterial cell suspension and a molten agarose gel at equal flow rates (Figure 2A). The agarose is prepared at twice the desired final concentration, as the co-flow dropmaking process effectively dilutes the aqueous solutions by a factor of 2. As the agarose cools, it solidifies into a 25 µm diameter microgel occupying the spherical volume of the droplet.
A series of wash and lysis steps purifies the high-molecular-weight genomic DNA in the microgels (Figure 2B). After breaking the emulsions, aqueous washes are carried out in large volumes to dilute out trace organic solvents which can inhibit the downstream enzymatic treatments. The washed microgels are observed under a microscope to verify the cell encapsulation rate (Figure 4A). A cocktail of enzymes with broad lytic activity is added to the microgel suspension to digest the cell walls of the bacteria and eukaryotic microbes19. A second treatment with Proteinase K and detergent degrades the proteins and solubilizes cell debris.
Tagmentation of the purified DNA is carried out in droplets to avoid potential cross-contamination resulting from the diffusion of small tagmented DNA fragments between the microgels18. A droplet encapsulation device (Figure 3B) compartmentalizes each microgel with a buffer and tagmentation enzyme, which simultaneously fragments double-stranded DNA while also "tagging" it with a preloaded oligonucleotide20. The microgels are loaded into the droplets as close-packed particles, achieving encapsulation rates approaching 1 microgel for every drop with few doublets21 (Figure 4C).
In the final step of the microfluidic workflow (Figure 2C), a device performs a double merger operation combining 1 barcode drop, 1 microgel-containing drop, and the amplification mix in a controlled two-step process. First, a droplet containing PCR reagent is paired and merged with a barcode drop in the region shown in yellow (Figure 5). Saltwater electrodes in the microfluidic channel produce a high electric field gradient which triggers the droplet merger. In a similar manner, the first merged droplet is paired with a microgel droplet and merged a second time in the region shown in red. The droplets are collected and thermal cycled off-chip in a single-overlap extension (SOE) PCR. The overlapping complementary ends of the barcode and the tagmented genomic DNA allow fusion and exponential amplification of only properly barcoded constructs.
The sequencing data is first filtered by a read quality and then parsed by grouping the reads according to their 15-bp single-cell barcode sequence. For a barcode group to be considered valid, it should contain a minimum number of reads; this thresholding limits the analysis to cells with a useful amount of sequencing data and removes the PCR-mutated barcode "orphans" from the dataset. In this sample run, the minimum is set to 7.5 kbps per group (50 reads of 150 bp each). A histogram of the barcode counts versus the group size shows that a significant portion of the valid barcode groups is just above the threshold size (Figure 6A).
In a control experiment where the microbial community composition is known, the purity and relative abundance metrics are used to evaluate the quality of a SiC-seq run. Here, a synthetic 10-cell community consisting of 3 gram-negative bacteria, 5 gram-positive bacteria, and 2 yeasts is analyzed. The purity of a given barcode group is defined as the number of reads mapping to the most common genome in the group divided by the total number of reads in the group. The vast majority of the barcode groups have purities greater than 0.95 (Figure 6B). Relative abundance of the cell types is calculated by counting the raw reads and by counting the barcode groups, where the groups are assigned a cell type corresponding to the consensus of its member reads (Figure 6C). The abundance of reads and barcode groups track in roughly equal proportions, indicating that the cell populations are being sampled such that certain species are not contained in disproportionately small or large barcode groups. Plotting the aggregate coverage of all barcode groups from a single species indicates a high coverage across the entire genome, with few or no dropout regions (Figure 7). The uniformity of coverage can be verified with a frequency distribution of normalized coverage values, with most values centered around the average (Figure 7, inset).
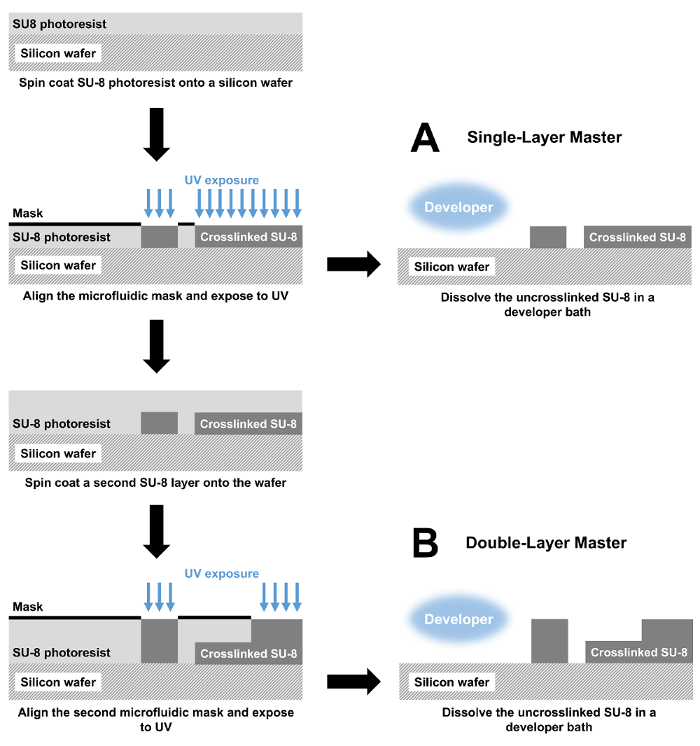
Figure 1: Fabrication of microfluidic devices by photolithography. (A) Master molds with a single feature height are fabricated by spin coating a layer of SU-8 photoresist onto a silicon wafer. The photoresist is then patterned with a photolithographic mask and UV light, crosslinking the exposed SU-8. Finally, uncrosslinked SU-8 is dissolved in a developer bath. The resulting mold is used to cast PDMS which is bonded to a glass slide to produce the complete microfluidic device. (B) For a double-layer device, the fabrication similarly begins with spin coating and exposure steps. These steps are then repeated to create a two-layer device. Please click here to view a larger version of this figure.
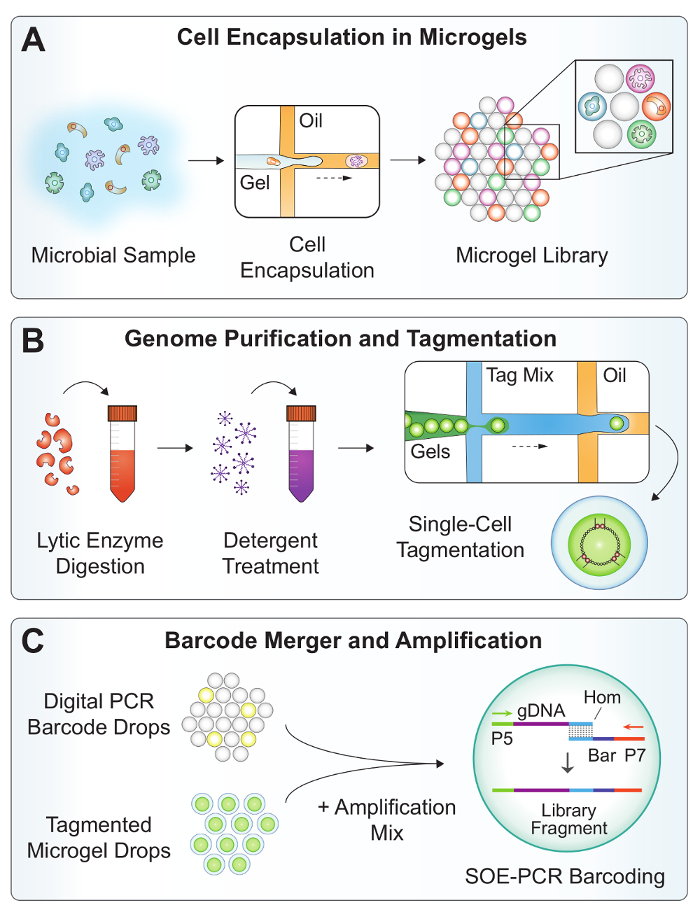
Figure 2: Overview of the SiC-seq workflow. (A) A microbial suspension is co-flowed with molten agarose in a dropmaker device to encapsulate single cells in microgels. (B) The microgels are subjected to a series of washes to purify the bacterial genomic DNA. Lytic enzymes digest the cell walls of gram-positive bacteria and yeasts, and detergent solubilizes the cellular debris. The microgels are re-encapsulated into droplets for the tagmentation to reduce cross-contamination. (C) The microfluidic merger combines a digital PCR barcode, a tagmented microgel genome, and an amplification mix at a rate >1 kHz. Off-chip SOE-PCR splices a unique single-cell barcode onto the tagmented genome and selectively amplifies fully barcoded constructs. Please click here to view a larger version of this figure.
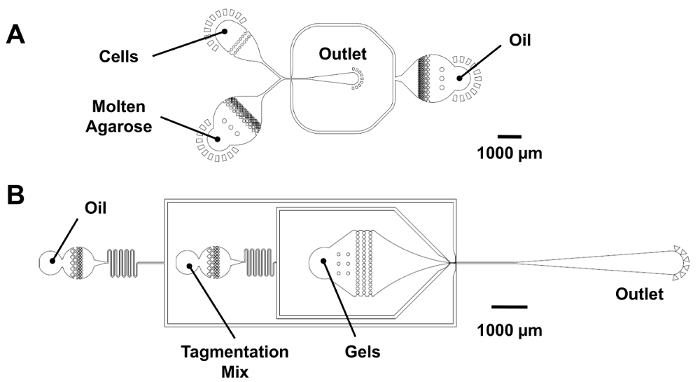
Figure 3: Microfluidic devices for dropmaking and microgel re-encapsulation. (A) This panel shows a co-flow dropmaker (25 µm of feature height). Cells and molten agarose are introduced into the device at equal flow rates to produce 25 µm droplets at a 25 µm x 25 µm junction. For the digital barcode dropmaking, the cell inlet is plugged, and a PCR mix is introduced into the agarose inlet. (B) This panel shows a microgel re-encapsulation device (25 µm of feature height). The microgels flow into a funnel-shaped inlet to maintain their close-packed ordering and receive a volume of tagmentation mix prior to the re-encapsulation at a 25 µm x 30 µm junction. Please click here to view a larger version of this figure.
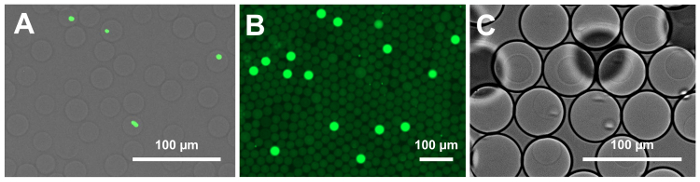
Figure 4: Micrographs of droplets and microgels. (A) This panel shows washed 25 µm microgels prior to enzymatic lysis. The bacteria are fluorescently stained for the quantification of the encapsulation rate. Poisson loading statistics dictate that the cells should be encapsulated at a rate of 1 in 10 drops or less to minimize the frequency of multiple-encapsulation events. (B) This panel shows a fluorescence microscopy image of 25 µm digital barcode droplets treated with a nucleic acid stain. The droplets containing amplified barcode fragments produce a strong fluorescence signal. (C) This panel shows microgels re-encapsulated in 50 µm drops. The close-packing of the microgels allows for encapsulation rates approaching 1 gel per drop with few doublets. Please click here to view a larger version of this figure.
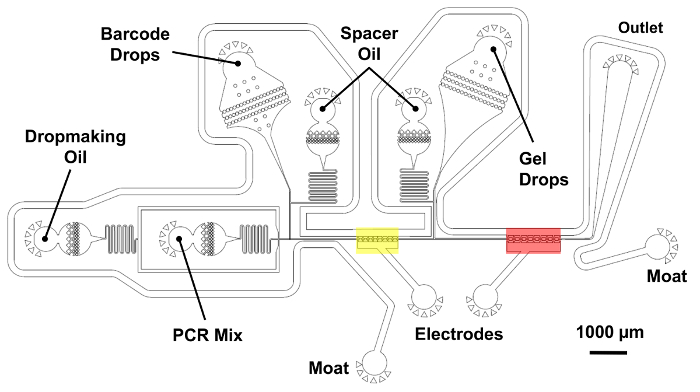
Figure 5: Microfluidic double merger device for single-cell genome barcoding. A two-step merger operation pairs barcode droplets with tagmented genomes at a high-throughput. A droplet of PCR mix is first generated and merged with a barcode droplet in the region shown in yellow using saltwater electrodes. Next, a droplet containing a microgel is introduced and merged a second time in the region shown in red. Oil inlets allow for precise control of the spacing between the reinjected droplets. The barcode reinjection chamber and its spacer oil are placed on the shorter 25 µm layer, shaded in blue. All other device features belong to the thicker layer with 45 µm of total height. Please click here to view a larger version of this figure.
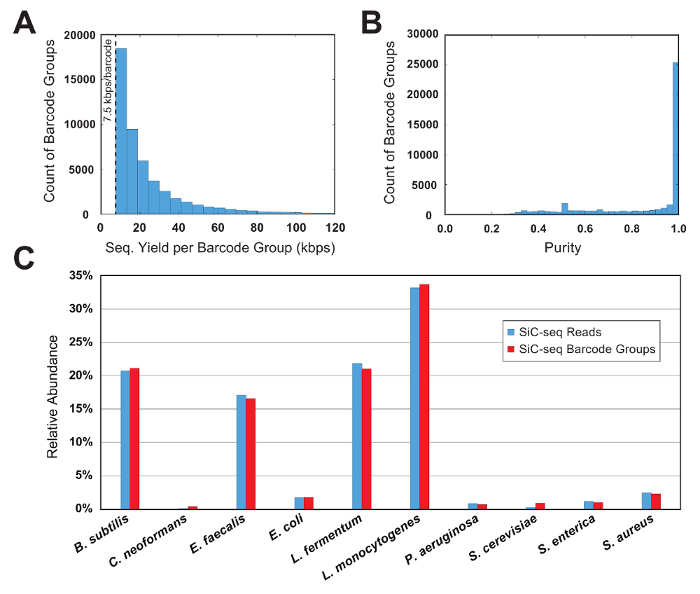
Figure 6: Barcode group metrics for a 10-cell synthetic microbial community. (A) This panel shows the distribution of the barcode group sizes. The number of groups of a given size decreases exponentially as the group size increases. A minimum threshold of 7.5 kbps per group limits the analysis to groups with a sufficient amount of information and removes the PCR-mutated sequence "orphans." (B) This panel shows the distribution of barcode group purities. The vast majority (>90%) of groups are of very high purity (>95%). (C) This panel shows the relative abundance of 10 species calculated at the read and barcode group level. The 2 counting methods produce similar results, indicating that the barcode group sizes are consistent across species. Please click here to view a larger version of this figure.
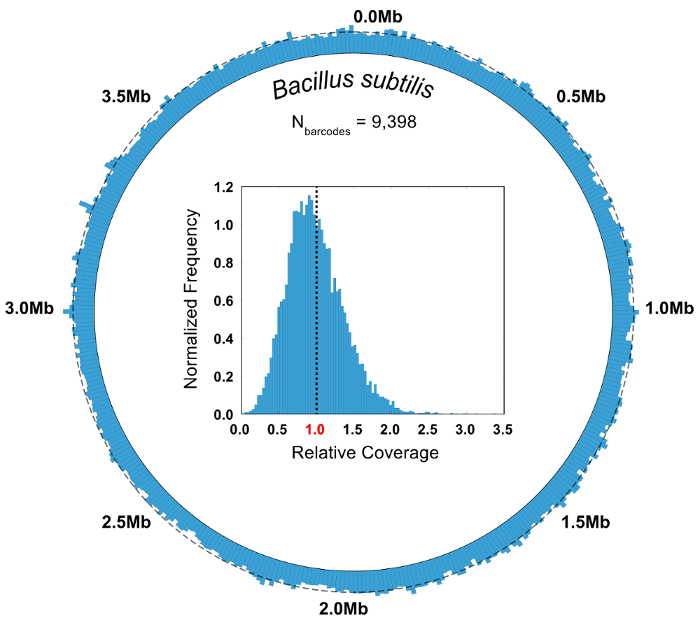
Figure 7: Aggregate genomic coverage of Bacillus subtilis barcode groups. The reads from all the barcode groups mapping to the bacterium B. subtilis (N = 9,398) are pooled and analyzed in aggregate. A circular coverage map illustrates the coverage uniformity of the SiC-seq reads, with no observable dropout regions. A dashed line around the circumference indicates the average coverage (5.55x). The inset histogram of the relative coverage frequencies shows that a bulk of the bases are covered at a depth near the genome-wide average, represented by the dashed line. Please click here to view a larger version of this figure.
| Device | 1st Layer Height (µm) | 1st Layer Spin Speed (rpm) | 2nd Layer Height (µm) | 2nd Layer Spin Speed (rpm) |
| Co-flow dropmaker | 25 | 4000 | N/A | N/A |
| Gel re-encapsulator | 25 | 4000 | N/A | N/A |
| Double merger | 25 | 4000 | 20 | 5000 |
Table 1: Microfluidic device fabrication parameters. This table shows a listing of the microfluidic devices used in the SiC-seq workflow with their required speeds for photoresist spin coating (based on the manufacturer's specifications for SU-8 3025).
| Label | Sequence (5' > 3') | ||||
| BAR | GCAGCTGGCGTAATAGCGAGTACAATCTGCTCTGATGCCGCATAGNNNNNNNNNNNNNNNTAAGCCAGCCCCGACACT | ||||
| DNA_BAR | CTGTCTCTTATACACATCTCCGAGCCCACGAGACGTGTCGGGGCTGGCTTA | ||||
| P7_BAR | CAAGCAGAAGACGGCATACGAGATCAGCTGGCGTAATAGCG | ||||
| P5_DNA | AATGATACGGCGACCACCGAGATCTACACTCGTCGGCAGCGTC | ||||
| I7_READ | GCCCACGAGACGTGTCGGGGCTGGCTTA | ||||
Table 2: Primer sequences.
Supplemental File 1: Please click here to download this file.
Supplemental File 2: Please click here to download this file.
Discussion
The SiC-seq microfluidic workflow produces single-cell genome sequencing data from thousands of bacterial cells. Digital barcodes spliced onto the genomes of microgel-encapsulated cells allow for the in silico deconvolution of NGS data into groups of barcoded reads originating from the same cell. A control experiment with a microbial community of known composition is necessary for evaluating the purity of the barcode groups. A large fraction of low-purity groups indicates that the cell encapsulation rate is too high or that there is significant droplet cross-contamination occurring during the microfluidic processing steps. According to Poisson statistics, the barcodes and cells should be encapsulated at a target ratio of 1 particle for every 10 drops to limit the rate of multiple encapsulation events to less than 5% of all non-empty droplets. An encapsulation rate higher than this increases the rates of doublets exponentially, so the verification of the encapsulation ratio during the dropmaking process is of critical importance. Users should be particularly cautious of the encapsulation of multiple cells in a single microgel because reads from different cells sharing the same barcode sequence cannot be bioinformatically separated. In the case that 1 cell receives 2 different barcodes, the barcode group purity is unaffected although the abundance metrics are skewed when counting by barcode sequence.
Droplet cross-contamination may also arise due to suboptimal merger conditions. During a successful operation, the microfluidic merger device (Figure 5) can controllably pair 1 barcode droplet with 1 microgel and a volume of PCR reagent. Non-ideal flow rates will result in a droplet pairing at incorrect ratios: 1 barcode could be paired with 2 microgels, for example. All flow rates listed in the protocol are intended to be estimates and may need to be adjusted depending on slight variations in the device geometry and droplet sizes. Users with access to cameras with high-speed recording capabilities (>10,000 frames/s) should verify the correct droplet merger at the beginning and over the course of the microfluidic operation. Users without access to a high-speed camera can collect a small volume of the merged output and manually measure the droplet sizes under a microscope. The droplet size should be uniform: an excess of unmerged barcodes or microgel drops indicates that the reinjection rates should be reduced accordingly.
Several general precautions should be taken when handling microgels and microdroplets to preserve their integrity. Microgels, though mechanically robust, must be sufficiently cooled prior to the breaking and washing steps to ensure complete gelation. Non-spherical microgels are an indication that the agarose was not given adequate time to solidify. When washing microgels, spin the suspensions down at the required speeds to avoid a loss of product. Agarose hydrogel has a refractive index closely matching that of water and may be difficult to see in a tube22, so users should carefully identify the gel-liquid boundary prior to aspiration. Water-in-oil droplets are susceptible to coalescence by the build-up of static forces23 on laboratory gloves and tubing. For this reason, we recommend loading the droplet reinjection syringes with bare hands and treating all the reinjection lines with an anti-static gun prior to the pump priming. Large coalesced droplets can be removed by slowly rotating the emulsions in a syringe and manually aspirating the larger drops, which accumulate near the top due to their larger buoyant force.
SiC-seq is the first technology to demonstrate single-cell genome sequencing of >50,000 bacterial cells. This platform offers significant advantages in throughput over existing approaches and enables a deeper sampling of heterogeneous microbial communities. To date, microfluidic technologies for single-cell genome sequencing have employed microchambers9 and microwells24 for cell isolation and amplification, but with throughputs in the range of only tens to hundreds of cells. The flow sorting of single cells into wellplates5,6 requires no specialized microfluidic instrumentation but possesses a similarly low throughput. Given that soil and water samples from the environment commonly have alpha diversities of >1,000 at the species level25,26, SiC-seq is highly advantageous by virtue of its ability to sample a far greater number of organisms. The SiC-seq workflow is adaptable to cell inputs from laboratory culture, the natural environment, or a living host. A cell sample need only be in an aqueous suspension and free of large particles (>10 µm) to be suitable for microfluidic encapsulation. For example, the method has been previously applied to a sample of seawater using a series of wash and filtering steps to pre-process the cells prior to encapsulation17.
The SiC-seq protocol generates a relatively sparse amount of sequencing data from each single cell and may not be suitable for all applications. Some bioinformatics algorithms such as de novo genome assembly or single nucleotide variant (SNV) calling require higher coverage depths to work effectively. Instead, barcode groups can be clustered in silico by taxonomic binning methods27 so that algorithms can be applied on larger sets of reads. The relatively low overall barcoding efficiency of the SiC-seq workflow may also present challenges in cases where the availability of the input sample is low. SiC-seq relies on a Poisson-distributed barcode encapsulation step, therefore approximately 10% of the cells receive a molecular barcode and are amplified during the final library preparation step. While this is comparable to other microdroplet-based barcoding schemes10, users working with precious cell samples may have difficulty achieving the adequate library yield for sequencing and may need to increase the number of PCR cycles in the final amplification step. Another potential solution for users with microfluidic expertise is to sort positive barcode droplets after the digital PCR step, thus bringing the overall barcoding efficiency to >85%28.
A potential future direction for SiC-seq technology is adapting the workflow for use with mammalian cells, paving the way for new clinical single-cell studies. As an example, an analysis of the copy number variation among single cancer cells may further our understanding of the role of heterogeneity in cancer pathology2. Alternatively, integrating SiC-seq with existing methods to probe and enrich DNA sequences of interest29 would enable the targeted single-cell sequencing of subpopulations or rare strains of cells. With environmental samples, genes from within a known metabolic pathway could be targeted and analyzed contextually alongside neighboring genes to identify novel genomic islands. From within a human host environment, low-titer pathogenic bacteria samples could be isolated and sequenced at the single-cell level to examine more closely their genotypic origins of virulence.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Science Foundation through a CAREER Award (grant number DBI-1253293); the National Institutes of Health (NIH) (grant numbers HG007233-01, R01-EB019453-01, 1R21HG007233, DP2-AR068129-01, R01-HG008978); and the Defense Advanced Research Projects Agency Living Foundries Program (contract numbers HR0011-12-C-0065, N66001-12-C-4211, HR0011-12-C-0066).
Materials
| 3" silicon wafers, P type, virgin test grade | University Wafers | 447 | |
| SU-8 3025 photoresist | Microchem | 17030192 | |
| Spin coater | Specialty Coating Systems | G3P-8 | |
| Photomasks | CadArt Servcies | (custom) | See Supplemental Files for mask designs |
| PGMEA developer | Sigma-Aldrich | 484431 | |
| Isopropanol | Sigma-Aldrich | 109827 | |
| Sylgard 184 silicone elastomer kit | Krayden | 4019862 | |
| Degassing chamber | Bel-Art | 42025 | |
| 0.75 mm biopsy punch | World Precision Instruments | 504529 | |
| Glass microscope slides (75 mm x 50 mm) | Corning | 294775X50 | |
| Aquapel (hydrophobic glass treatment) | Pittsburgh Glass Works | 47100 | |
| PE-2 polyethylene tubing | Scientific Commodities | B31695-PE/2 | |
| 1 mL syringes | BD | 309628 | |
| 27 gauge needles | BD | 305109 | |
| Syringe pump | New Era Pump Systems | NE-501 | |
| Novec HFE-7500 fluorinated oil (HFE) | 3M | 98-0212-2928-5 | |
| FC-40 fluorinated oil | Sigma-Aldrich | F9755 | |
| PEG-PFPE surfactant | Ran Biotechnologies | 008-FluoroSurfactant | |
| Space heater | Lasko | CD09250 | |
| Agarose, low gelling temperature | Sigma-Aldrich | a9414 | |
| TE (10X) | Rockland | mb-007 | |
| PBS 1X, pH 7.4 | E&K Scientific Products | EK-65083 | |
| OptiPrep (density gradient medium) | Sigma-Aldrich | d1556 | |
| 1H,1H,2H-Perfluoro-1-Octanol (PFO) | Sigma-Aldrich | 370533 | |
| Span 80 (sorbitane monooleate) | Sigma-Aldrich | s6760 | |
| Hexane | Sigma-Aldrich | 139386 | |
| Tween 20 (polysorbate 20) | Sigma-Aldrich | p2287 | |
| Lysozyme Type IV | MP Biomedicals | 195303 | |
| Mutanolysin | Sigma-Aldrich | M9901 | |
| Zymolyase (yeast lytic enzyme) | Zymo Research | e1004 | |
| Lysostaphin | Sigma-Aldrich | L7386 | |
| Sodium chloride | Sigma-Aldrich | S9888 | |
| EDTA | Sigma-Aldrich | E6758 | |
| Tris-HCl, pH 7.5, 1M | Invitrogen | 15567-027 | |
| Dithiothreitol (DTT) | Teknova | d9750 | |
| Lithium dodecyl sulfate | Sigma-Aldrich | L9781 | |
| Proteinase K | New England Biosciences | P8107S | |
| Ethanol, 200 Proof (100%) | Koptec | V1001 | |
| SYBR Green I (nucleic acid stain) | Invitrogen | S7563 | |
| PEG 6k | Sigma-Aldrich | 81260 | |
| Triton X-100 (octylphenol ethoxylate) | Sigma-Aldrich | t8787 | |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1030 | |
| Phusion Hot Start Flex Master Mix (High-Fidelity Hot Start Master Mix) | New England Biosciences | m05365 | |
| Platinum Multiplex PCR Master Mix (Taq Master Mix) | Applied Biosystems | 4464263 | |
| Warmstart 2.0 Bst Polymerase (isothermal polymerase) | New England Biosciences | m0538m | |
| NT buffer from Nextera XT kit (neutralization buffer) | Illumina | FC-131-1024 | |
| Cold cathode fluorescent inverter | (custom) | (custom) | |
| DC power supply | Mastech | HY1503D | |
| Zerostat 3 anti-static gun | Milty | 5036694022153 | |
| 3D-printed centrifuge syringe holder | (custom) | (custom) | See Supplemental Files for 3D print file |
| Zymo DNA Clean & Concentrator-5 | Zymo Research | D4003 |
References
- Navin, N., et al. Tumour evolution inferred by single-cell sequencing. Nature. 472 (7341), 90-94 (2011).
- Ni, X., et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 110 (52), 21083-21088 (2013).
- Schmidt, H., Hensel, M. Pathogenicity islands in bacterial pathogenesis. Clinical Microbiology Reviews. 17, 14-56 (2004).
- Martínez, J. L., Baquero, F. Interactions among strategies associated with bacterial infection: pathogenicity epidemicity, and antibiotic resistance. Clinical Microbiology Reviews. 15 (4), 647-679 (2002).
- Rinke, C., et al. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nature Protocols. 9 (5), 1038-1048 (2014).
- Rinke, C., et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 499 (7459), 431-437 (2013).
- Zhang, H., Liu, K. K. Optical tweezers for single cells. Journal of the Royal Society Interface. 5 (24), 671-690 (2008).
- Xu, L., Brito, I. L., Alm, E. J., Blainey, P. C. Virtual microfluidics for digital quantification and single-cell sequencing. Nature Methods. 13 (9), 759-762 (2016).
- Gawad, C., Koh, W., Quake, S. R. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proceedings of the National Academy of Sciences of the United States of America. 111 (50), 17947-17952 (2014).
- Macosko, E. Z., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161 (5), 1202-1214 (2015).
- Klein, A. M., et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 161 (5), 1187-1201 (2015).
- Rotem, A., et al. High-throughput single-cell labeling (Hi-SCL) for RNA-Seq using drop-based microfluidics. PLoS One. 10 (5), 1-14 (2015).
- Amini, S., et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nature Reviews Genetics. 46 (12), 1343-1349 (2014).
- Zheng, G. X. Y., et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nature Biotechnology. 34 (3), 303-311 (2016).
- Lan, F., Haliburton, J. R., Yuan, A., Abate, A. R. Droplet barcoding for massively parallel single-molecule deep sequencing. Nature Communications. 7, 11784 (2016).
- Rotem, A., et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature Biotechnology. 33 (11), 1165-1172 (2015).
- Lan, F., Demaree, B., Ahmed, N., Abate, A. R. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nature Biotechnology. 35 (7), 640-646 (2017).
- Novak, R., et al. Single-cell multiplex gene detection and sequencing with microfluidically generated agarose emulsions. Angewandte Chemie Internation Edition. 50 (2), 390-395 (2011).
- Gill, C., Van De Wijgert, J. H. H. M., Blow, F., Darby, A. C. Evaluation of lysis methods for the extraction of bacterial DNA for analysis of the vaginal microbiota. PLoS One. 11 (9), 1-16 (2016).
- Picelli, S., et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Research. 24 (12), 2033-2040 (2014).
- Abate, A. R., Chen, C. H., Agresti, J. J., Weitz, D. A. Beating Poisson encapsulation statistics using close-packed ordering. Lab on a Chip. 9 (18), 2628 (2009).
- Jain, A., Yang, A. H. J., Erickson, D. Gel-based optical waveguides with live cell encapsulation and integrated microfluidics. Optic Letters. 37 (9), 1472 (2012).
- Karbaschi, M., Shahi, P., Abate, A. R. Rapid chemical-free breaking of microfluidic emulsions with a hand-held antistatic gun. Biomicrofluidics. 11 (4), 1-6 (2017).
- Gole, J., et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nature Biotechnology. 31 (12), 1126-1132 (2013).
- Chao, Y., et al. Metagenomic analysis reveals significant changes of microbial compositions and protective functions during drinking water treatment. Scientific Reports. 3 (1), 3550 (2013).
- Fierer, N., et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences of the United States of America. 109 (52), 21390-21395 (2012).
- Mande, S. S., Mohammed, M. H., Ghosh, T. S. Classification of metagenomic sequences: methods and challenges. Briefings in Bioinformatics. 13 (6), 669-681 (2012).
- Eastburn, D. J., et al. Microfluidic droplet enrichment for targeted sequencing. Nucleic Acids Research. 43 (13), e86 (2015).
- Clark, I. C., Abate, A. R. Finding a helix in a haystack: nucleic acid cytometry with droplet microfluidics. Lab on a Chip. 17 (12), 2032-2045 (2017).

