Visualization of Tangential Cell Migration in the Developing Chick Optic Tectum
Summary
We describe the methods for fluorescent labeling of tangentially migrating cells by electroporation, and for time-lapse imaging of the labeled cell movement in a flat-mount culture in order to visualize migrating cell behavior in the developing chick optic tectum.
Abstract
Time-lapse imaging is a powerful method to analyze migrating cell behavior. After fluorescent cell labeling, the movement of the labeled cells in culture can be recorded under video microscopy. For analyzing cell migration in the developing brain, slice culture is commonly used to observe cell migration parallel to the slice section, such as radial cell migration. However, limited information can be obtained from the slice culture method to analyze cell migration perpendicular to the slice section, such as tangential cell migration. Here, we present the protocols for time-lapse imaging to visualize tangential cell migration in the developing chick optic tectum. A combination of cell labeling by electroporation in ovo and a subsequent flat-mount culture on the cell culture insert enables detection of migrating cell movement in the horizontal plane. Moreover, our method facilitates detection of both individual cell behavior and the collective action of a group of cells in the long term. This method can potentially be applied to detect the sequential change of the fluorescent-labeled micro-structure, including the axonal elongation in the neural tissue or cell displacement in the non-neural tissue.
Introduction
The study of cell migration has been progressing with the advancing technique of live imaging. After fluorescent cell labeling, the temporal movement of labeled cells in a culture dish or in vivo can be recorded under video microscopy. In the study of neural development, the morphological changes of migrating cells or elongating axons have been analyzed using time-lapse imaging. For effective imaging, it is essential to apply a suitable method for fluorescent cell labeling and tissue preparation, based on the purpose of the experiment and analysis. For analyzing cell migration in the developing brain, slice culture has been commonly used to observe cell migration parallel to the slice section, such as radial cell migration1,2,3. The slice culture system is also used for detecting tangential cell migration4,5, but it is not suitable for directional analysis in cases where the cells disperse perpendicular to the slice section.
The optic tectum is composed of a multilayered structure, formed by radial and tangential cell migration during embryonic development. Tectal layer formation depends primarily on radial migration of postmitotic neuronal precursor cells from the ventricular zone, and their final destination in the layers correlates with their birth date in the ventricular zone6. As for tangential migration, we previously reported two streams of migrations in the middle and superficial layers in a developing chick optic tectum. In the middle layers during E6-E8, the bipolar cells with a long leading process and a thin trailing process migrate dorsally or ventrally along the axon fasciculus of tectal efferent axons that run dorso-ventrally7. After this axophilic migration, the cells differentiate into multipolar neurons located in the deep layers. In the superficial layers during E7-E14, the migrating cells disperse horizontally by reforming a branched leading process and scatter into multiple directions8. After dispersing migration, the latter cells eventually differentiate into superficial neurons of various morphologies. In both cases, a flat-mount culture is efficient to observe cell movement parallel to the pial surface.
Here, we present a protocol for time-lapse imaging to visualize tangential cell migration in the developing chick optic tectum7,8. Combination of cell labeling by electroporation in ovo, and a subsequent flat-mount culture on the cell culture insert enables detection of migrating cell movement and migration direction. The goal of this method is to facilitate detection of both individual cell behavior in the long term and the collective action of a group of cells in the horizontal plane.
Protocol
1 . Electroporation In Ovo
- Prepare the expression plasmid DNA for fluorescent labeling in high concentration. Isolate DNA from 200 mL of bacterial culture by the alkaline lysis method using anion-exchange columns according to the manufacturer's protocol (Table of Materials). Mix pCAGGS-EGFP and pCAGGS-mCherryNuc at a final concentration of 4 µg/µL each.
NOTE: Endotoxin-free plasmid DNA purification may be preferred for electroporation. - Incubate fertile chicken eggs horizontally at 38 °C in 70% relative humidity.
- After 2.5 days, eliminate 5 mL of albumen (egg white) from the pointed side of the egg using 20 mL syringe with an 18 gauge needle and seal the needle hole with tape.
- Cut the top of the egg shell with curved scissors to open a hole 2 cm in diameter and check the developmental stage of the embryo under the stereomicroscope (stage 17 of Hamburger and Hamilton9). Seal the hole again with tape.
NOTE: This step can be performed at any time during E2.5-E3.0. Steps 1.3 and 1.4 lower the level of the embryo in the egg to avoid a tight attachment of the growing embryo and the blood vessels to the top shell. - Continue incubation until E5.5.
- Before electroporation, prepare 10 µL of the mentioned plasmid DNA colored with 0.5 µL of Fast Green (25 mg/mL), 10 mL of autoclaved phosphate buffered saline (PBS) containing penicillin (100 units/mL) and streptomycin (100 μg/mL), and micropipettes made from glass capillary tubes using a micropipette processor. Cut the distal tip of the micropipette to make an appropriate size of pore depending on the amount of DNA for injection.
- Cut the sealed top shell with scissors to reopen a hole 2.5 cm in diameter and set the egg stably under the stereo microscope. Add a few drops of PBS into the egg. Peel off the allantoic membrane covering the embryo without hemorrhage using two fine forceps. Remove the amnion from the head of the embryo.
- While turning the head with a microspatula, inject 0.1-1 μL of the colored DNA into the ventricular cavity of the left optic tectum using a micropipette connected to a suction tube.
NOTE: The amount of DNA injected depends on the purpose of the experiment. Concentrated DNA solution should sink and attach along the ventricular wall. - Place a pair of forcep-type electrodes (3 mm square electrodes with 7 mm distance) so that the target area of the optic tectumis placed in between (Figure 1, Step 1).
NOTE: The DNA is electroporated to the anode side. - Charge a pre-pulse of 30 V, 1 ms with a 5 ms interval and four subsequent pulses of 6 V, 5 ms with a 10 ms interval using the pulse generator.
NOTE: The electronic condition depends on the size and distance of the electrodes. It should be strong enough to achieve efficient transfection, but it must be modest enough to assure the normal development of the target tissue. - Seal the hole with tape and continue incubation. Soak the electrodes in PBS to remove albumen with an interdental brush in a plastic dish. Repeat 1.7-1.11 for each egg.
2. Flat-Mount Culture on the Cell Insert
- One day before the flat-mount culture, prepare the coated cell culture insert. Float some cell culture inserts on the autoclaved distilled water in a 10-cm cell culture dish. Load a solution of 8 µg/mL laminin and 80 µg/mL poly-L-lysine to cover the inserts and leave the floated inserts in the cell culture CO2 incubator at 37 °C overnight.
NOTE: On the following day, the coated inserts can be stored at 4 °C. - On day of the culture at E7.0, remove the laminin-poly-L-lysine solution from the insert and place the insert in a glass bottom dish (Figure 1, Step 2) filled with 1.1 mL of culture medium (60% reduced serum medium, 20% F12, 10% fetal bovine serum, 10% chick serum, 50 units/mL penicillin, 50 μg/mL streptomycin). Keep the dish in the cell culture CO2 incubator at 37 °C.
NOTE: The medium can be used throughout the culture period for three days without change. - Prepare the culture setup. Set a humid chamber unit on an inverted confocal microscope with a gas flow of 40% O2 and 5% CO2 (gas controller) at 38 °C (temperature controller).
- Cut the head of the embryo with scissors. Pinch the head out with the forceps into the ice-cold Hanks' Balanced Salt solution (HBSS) in a 6-cm cell culture dish.
- Isolate the electroporated optic tectum using two fine forceps. Transfer the tectum with a plastic dropper into another dish filled with ice-cold HBSS. Use the concaved glass dish based with black silicon as a saucer for cutting.
- Check the position of the labeling under the fluorescence stereoscopic microscope. Cut out the tectal tissue surrounding the labeling area with a microsurgical knife. Make sure the direction of the tissue in the tectum (anterior-posterior, dorsal-ventral).
- Transfer the labeled tissue with a plastic dropper to the insert so that the pia side is attached to the insert. Lay the tectal tissue in the desired direction and remove excess HBSS (Figure 1, Step 2).
- Repeat 2.4-2.7 in order to prepare other tissues on the same insert. Place the dish in the prewarmed chamber in the inverted confocal microscope (Figure 1, Step 3).
3. Time-Lapse Imaging
- Check the fluorescent labelling and focus the microscopic field in the inverted confocal microscope. Start the laser confocal units and try sampling the confocal scan to adjust the direction and position of the tissue along the X and Y-axis in the field. Use the 10X or 20X objective lens without immersion oil.
- Select the scanning size (e.g., 512×512 dpi). Decide the interval and total range of the confocal scan along the z-axis. Take a 5 or 10 µm interval for the 100 µm range. Decide the time interval of the confocal imaging and total imaging duration (e.g., 10 min intervals over 48 h).
NOTE: To avoid laser phototoxicity, prohibit scanning in the high scanning size with short z-intervals for the long z-range, or with short time intervals during the long imaging duration. For example, when applying a high scanning size (e.g., 1024×1024 dpi), decrease the number of scans along the z-axis. - Set the parameters of the confocal running program described in 3.2 and start imaging.
- After the imaging, combine the confocal images at different z-axis to provide z-stack images with fine focal adjustment at every time point.
- Append the z-stack images at different time-point and construct a time-lapse movie in the AVI format.
Representative Results
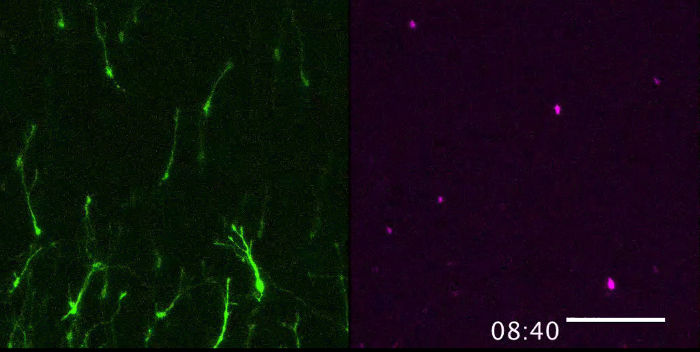
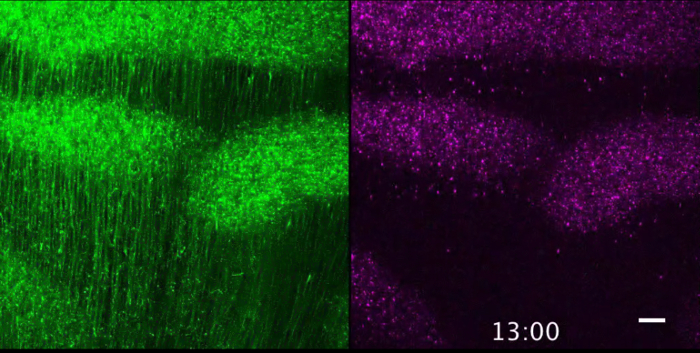
Figure 2 shows the visualized superficial tangential migration in a flat-mount culture at an elapsed time (0, 9, 18, 27 h) after onset of recording. Movie 1 is a time-lapse movie of 10 min-intervals over a period of 28 h and 50 min. The frame is selected for focusing on the migrating cells from the labeled lower-left corner of the frame to the unlabeled space (Figure 3A). The mass movement of the migrating cells (GFP; upper left panel, Movie 1) and their nuclei (mCherry-Nuc; upper right panel) can be observed with the merged movie (lower panel, Movie 1). Directionality of the cell migration can be examined by focusing on the dispersing cells from the labeled center to all directions (Movie 2, Figure 3B).
The clear images of the nuclear movement (Movie 2; mCherry-Nuc, right panel) allows us to trace the cell nuclear migration by automatic tracking using a Particle Tracker plugin10 of a Fiji image processing application of ImageJ11 (Movie 3, Figure 3B). Temporal changes of the trajectories of the tangential migration can be visualized to prove the dispersing migration in omni-directions.
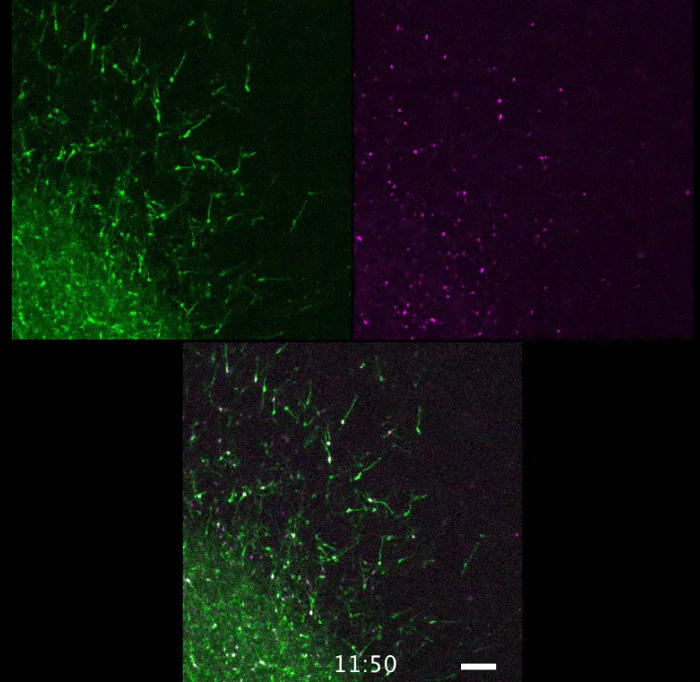
Individual cell behavior with the sequential morphological change of the leading process, trailing process and nuclei can be manifested with higher magnification images of 5 min-intervals (Movie 4, Figure 3C).
Another type of tangential migration in the middle layers7 can be visualized using a similar protocol (Movie 5, Figure 3D). Bidirectional linear migration along the axon fasciculus running dorsal to ventral (top to down) is evident7.

Figure 1. Protocol flow chart. Step 1: Electroporation in ovo. Step 2: Laying tectal tissue so that the pia side is attached to the insert. Step 3: Inverted fluorescent microscope with laser confocal unit. Please click here to view a larger version of this figure.
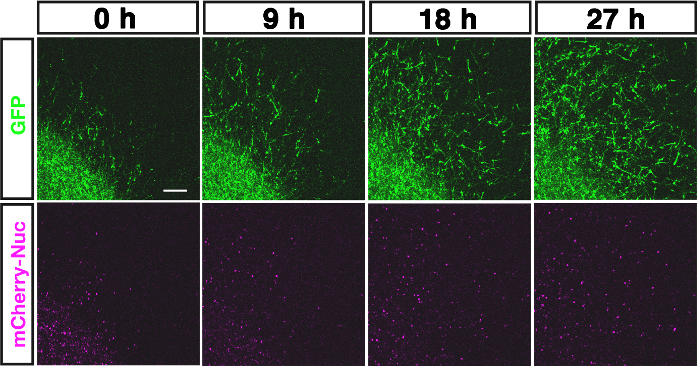
Figure 2. Visualization of superficial tangential migration in flat-mount culture. The tangentially migrating cells (GFP; upper panel) and the nucleus (mCherry-Nuc; lower panel) in the superficial layers of the optic tectum are shown at 0, 9, 18, 27 h after onset of the culture from E7.0. The cells at the lower-left corner of the frame are labeled at 0 h (see Figure 3A). Scale bar: 100 µm. Please click here to view a larger version of this figure.
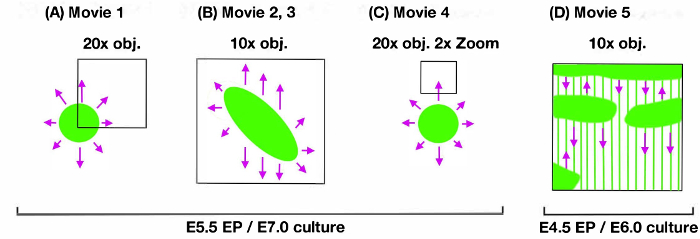
Figure 3. Schematic figure illustrating the mount conditions for time-lapse imaging. The shape of the fluorescent labeling (green), initial direction of cell migration (magenta), and video frame (black square) were illustrated with magnification of the objective lens (obj) and digital zoom (zoom) in the upper field, with day of electroporation (EP) and onset of culture (culture) in the bottom field. (A) Movie 1, (B) Movie 2 and 3, (C) Movie 4, (D) Movie 5. Please click here to view a larger version of this figure.
Movie 1. Visualization of superficial tangential migration in a flat-mount culture. Movement of the tangentially migrating cells (GFP; upper left panel) and their nuclei (mCherry-Nuc; upper right panel) in the superficial layers of the optic tectum after onset of the culture from E7.0. The merged image is shown in the lower panel. The cells at the lower-left corner of the frame are labeled at 0 h (see Figure 3A), and the time-lapse images were captured over 28 h and 50 min. Scale bar: 100 µm. Please click here to view this video. Right-click to download.
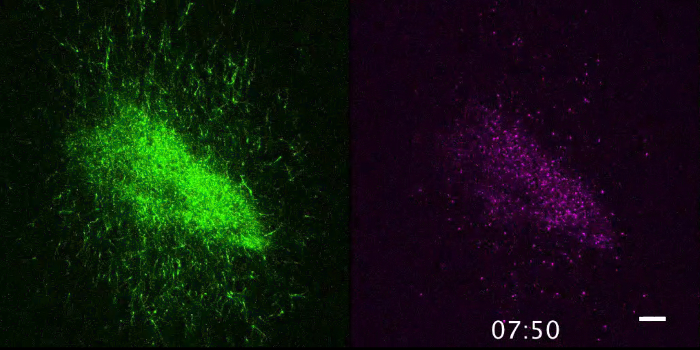
Movie 2. Dispersing movement of the tangentially migrating cells (GFP; left panel) and their nuclei (mCherry-Nuc; right panel) from the center of both panels are shown over 48 h (see Figure 3B). Scale bar: 100 µm. Please click here to view this video. Right-click to download.
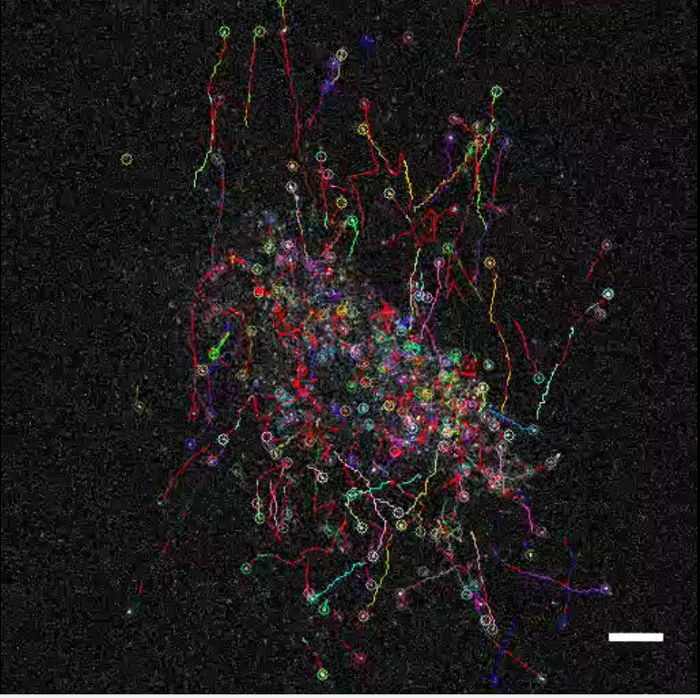
Movie 3. Trajectories of tangential migration. Displacement of the cell nucleus (the right panel of Movie 2) was tracked to visualize the trajectories of the tangential migration. Scale bar; 100 µm. Please click here to view this video. Right-click to download.
Movie 4. Individual cell behavior in higher magnification. Movement of the individual cells (GFP; left panel) and their nuclei (mCherry-Nuc; right panel) are shown in higher magnification over 24 h (see Figure 3C). Branching process of the leading process can be recognized. Scale bar: 100 µm. Please click here to view this video. Right-click to download.
Movie 5. Middle layer migration. Movement of the tangentially migrating cells (GFP; left panel) and their nucleus (mCherry-Nuc; right panel) in the tectal middle layers are shown over 24 h after onset of the culture from E6.0 (see Figure 3D). Linear migration along dorso-ventral axis (top to down) is remarkable. Scale bar: 100 µm. Please click here to view this video. Right-click to download.
Discussion
The protocol described above is optimized for detecting cell migration in superficial layers6,8. It is applicable for detecting middle layer migration streams (Movie 5)6,7, just by shifting the timing of the electroporation (E5.5 to E4.5) and the onset of culture and imaging (E7.0 to E6.0).
The presented procedure is composed of cell labeling by electroporation in ovo, flat-mount culture and time-lapse confocal imaging (Figure 1). First of all, it is a prerequisite that the culture conditions should be optimized to keep the tissue healthy and growing normally as in vivo. It is also crucial to ensure that the orientation of the tissue is suitable for detecting cell migration. For such purposes, we apply a flat-mount culture on cell insert, which facilitates observation of horizontal cell dispersion and supplies rich medium with high oxygen. After ensuring the culture condition and orientation, it is critical for visualization to adjust the conflicting conditions of better fluorescent labelling with the least electronic and photo damages. For better labeling, we can choose an expression vector with an efficient promoter and electroporate concentrated DNA. For achieving least damage, it is important to moderate the electric condition of the electroporation, lower the DNA concentration, and minimize the total time of laser irradiation.
An advantage of this protocol using the flat-mount culture on the cell insert is that we can observe tangential cell migration in the long term. Generally, it is difficult to determine how long the tissue in culture maintains the physiological conditions in comparison to that in ovo. At the very least, superficial tangential migration continues over 72 h after onset of culture at E7.0, which shows normal cellular dispersion similar to that in ovo8. In addition, fresh tissue is prepared at the start of the culture and imaging to observe migration at later stages. It is also advantageous that cell displacement can be followed over a long period because the cells remain moving horizontally in the superficial flat sheets of the tectum, which is close to the objective lens. Using the confocal system also facilitates tracking cell movement along the z-axis. On the other hand, a disadvantage of this method is that the flat-mount culture may not always be relevant to recapitulate other types of migration such as radial migration. When the method is applied to observe radial migration in the tectal slice on the insert, the thickness of the tectal tissue does not increase as rapidly as that in ovo. The slice culture method in the collagen gel may be better to recapitulate such layer development.
Since our method enables the observation of horizontal movement parallel to the culture insert, it can potentially be applied for detecting sequential change of the fluorescent-labeled micro-structure, including the axonal elongation in the neural tissue or cell displacement in the non-neural tissue. For example, after labeling commissural axons in the developing neural tube by electroporation, movements of pre- and post-crossing axons over the floor plate can be visualized in the open-book culture incising the roof plate. Provided that the appropriate culture condition on the cell insert is available for reproducing in vivo conditions, our method provides an effective technique to visualize horizontal movements of various cell types and structures.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 15K06740 to Y.W.
Materials
| Materials | |||
| NucleoBond Xtra Midi Plus EF | MACHEREY-NAGEL | 740422.5 | endotoxin-free plasmid DNA purification kit |
| 20 ml syringe | TERUMO | SS-20ESZ | |
| 18 gauge needle | TERUMO | NN-1838R | |
| Fast Green | Wako | 061-00031 | |
| 100x penicillin and streptomycin | Gibco | 15140-122 | |
| glass capillary tube | Narishige | G-1 | |
| cell culture insert | Millipore | Millicell CM-ORG | |
| Laminin | SIGMA | L2020 | coating of culture insert |
| poly-L-Lysine | Peptide Institute | 3075 | coating of culture insert |
| glass bottom dish | Matsunami | D11130H | |
| Opti-MEM | Gibco | 31985-070 | culture medium |
| F12 | Gibco | 11765-054 | culture medium |
| fetal bovine serum | Gibco | 12483 | culture medium |
| chick serum | Gibco | 16110082 | culture medium |
| 10xHBSS | Gibco | 14065-056 | |
| microsurgical knife | Surgical specialties cooperation | 72-1501 | |
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| curved scissors | AS ONE | No.11 | |
| micropipette processor | SUTTER INSTRUMENT | P97/IVF | |
| forceps-type electrode | BEX | LF646P3x3 | |
| pulse generator | BEX | CUY21EX | electroporator |
| fluorescence stereoscopic microscope | Leica | MZ16F | |
| inverted fluorescence microscope | Olympus | IX81 | |
| gas controller | Tokken | MIGM/OL-2 | |
| temperature controller | Tokai Hit | MI-IBC | |
| laser confocal unit | Olympus | FV300 |
References
- Marín, O., Rubenstein, J. L. R. Cell migration in the forebrain. Annual Reviews of Neuroscience. 26, 441-483 (2003).
- Nadarajah, B., Alifragis, P., Wong, R. O. L., Parnavelas, J. G. Neuronal migration in the developing cerebral cortex: Observations based on real-time imaging. Cerebral Cortex. 13, 607-611 (2003).
- Tabata, H., Nakajima, K. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. Journal of Neuroscience. 23, 9996-10001 (2003).
- Martini, F. J., et al. Biased selection of leading process branches mediates chemotaxis during tangential neuronal migration. Development. 136, 41-50 (2009).
- Polleux, F., Whitford, K. L., Dijkhuizen, P. A., Vitalis, T., Ghosh, A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 129, 3147-3160 (2002).
- Watanabe, Y., Yaginuma, H. Tangential cell migration during layer formation of chick optic tectum. Development, Growth and Differentiation. 57, 539-543 (2015).
- Watanabe, Y., Sakuma, C., Yaginuma, H. NRP1-mediated Sema3A signals coordinate laminar formation in the developing chick optic tectum. Development. 141, 3572-3582 (2014).
- Watanabe, Y., Sakuma, C., Yaginuma, H. Dispersing movement of neuronal tangential migration in superficial layers of the developing chick optic tectum. Developmental Biology. 437, 131-139 (2018).
- Hamburger, V., Hamilton, H. L. A series of normal stages in the development of the chick embryo. Journal of Morphology. 88, 49-92 (1951).
- Cordelières, F. P., et al. Automated cell tracking and analysis in phase-contrast videos (iTrack4U): development of Java software based on combined mean-shift processes. PLOS One. 8, e81266 (2013).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9, 676-682 (2012).

