Using Nanoplasmon-Enhanced Scattering and Low-Magnification Microscope Imaging to Quantify Tumor-Derived Exosomes
Summary
Clinical translation of exosome-derived biomarkers for diseased and malignant cells is hindered by the lack of rapid and accurate quantification methods. This report describes the use of low-magnification dark-field microscope images to quantify specific exosome subtypes in small volume serum or plasma samples.
Abstract
Infected or malignant cells frequently secrete more exosomes, leading to elevated levels of disease-associated exosomes in the circulation. These exosomes have the potential to serve as biomarkers for disease diagnosis and to monitor disease progression and treatment response. However, most exosome analysis procedures require exosome isolation and purification steps, which are usually time-consuming and labor-intensive, and thus of limited utility in clinical settings. This report describes a rapid procedure to analyze specific biomarkers on the outer membrane of exosomes without requiring separate isolation and purification steps. In this method, exosomes are captured on the surface of a slide by exosome-specific antibodies and then hybridized with nanoparticle-conjugated antibody probes specific to a disease. After hybridization, the abundance of the target exosome population is determined by analyzing low-magnification dark-field microscope (LMDFM) images of the bound nanoparticles. This approach can be easily adopted for research and clinical use to analyze membrane-associated exosome biomarkers linked to disease.
Introduction
Exosomes are released from most cell types and play a key role in cell-to-cell communications, including pathophysiological processes associated with various diseases, since they can be home to specific tissues or cell types, and contain a variety of nucleic acids, proteins, and lipids that reflect their cell of origin and can exert regulatory effects on their recipient cells1,2,3,4. Exosomes are often secreted at elevated levels in disease states, can interact with both adjacent and distant cells, and are found at relatively high concentration in the circulation as well as most other body fluids, including saliva, urine, pancreatic and bile juice, and bronchoalveolar lavage fluid5,6,7,8,9,10,11. This abundance and stability of exosomes in human body fluids, coupled with their information-rich nature, makes them ideal biomarkers for disease diagnosis and treatment monitoring.
This includes tumor-derived exosomes (TDEs), which contain tumor-specific or selective factors, which can serve as disease biomarkers, including tumor-associated mutant alleles. TDEs can participate in the remodeling of the tumor microenvironment to facilitate tumor development and metastasis, and regulate anti-tumor responses12. Increased TDE secretion is a common phenotype of most cancers and several features of the tumor microenvironment, including hypoxia, acidic pH, and inflammation, are known to promote exosome secretion. Surprisingly, given the number of cells that secrete exosomes, an increase in total exosome level can, itself, function as a cancer biomarker. For example, a recent study found that the total EV concentration in bile juice discriminates malignant and nonmalignant in common bile duct stenosis patients with 100% accuracy7. Similar results have been found with studies using other body fluids, including plasma. However, due to the potential for subject to subject variation, and other confounding factors, most studies investigating exosomes as disease biomarkers have focused on detection of biomarker that are selectively associated with TDEs instead of total exosome numbers.
Translating exosome biomarkers into clinical practice, however, remains challenging since most reported exosome assay approaches require time-consuming and labor-intensive isolation procedures 13. Currently popular exosome isolation methods include ultracentrifugation, density gradients, size-exclusion, co-precipitation, affinity capture, and microfluidic isolation approaches. Ultracentrifugation is the “gold standard” method, and is most commonly used for exosome isolations, but this procedure is time-consuming and results in exosome damage and exosome membrane clustering, and produces exosome samples that are contaminated with proteins, lipoproteins and other factors that can influence subsequent analyses14. Most exosome isolation methods, including ultracentrifugation, cannot separate exosomes (30–150 nm) from micro-vesicles (100–1000 nm) and apoptotic bodies (100–5000 nm), which arise through different mechanisms and have different functions, due to the size overlap among these groups, and the diversity of exosome populations15. New approaches are needed to improve the sensitivity and reproducibility of exosome assays by improving exosome recovery while reducing exosome damage and contamination, although any assays based on such methods will also need to be optimized to render them suitable for translation to applications in clinical settings.
Several recent studies have proposed to employ integrated platforms to capture and analyze exosomes directly from body fluids. These methods employ microfluidic, electrokinetic, affinity capture, and various other methods for exosome isolation, and electrochemistry, surface plasmon resonance, and other methods to detect captured exosomes. It is not clear how feasible many of these approaches will be in clinical settings, due to their complexity, expense, low-throughput or other issues.
We have developed a rapid and inexpensive assay that can be used for sensitive and specific quantification of total exosomes and specific exosome subtypes, including disease-associated exosomes, such as TDEs, which requires only a small amount of sample and which employs a streamlined workflow suitable for clinical environments. In this assay, a slide is coated with antibodies that bind either an exosome-specific or disease-specific marker expressed on the exosome surface in order to directly capture target exosomes present in small volume plasma or serum samples applied to wells on the slide. Captured exosomes are then hybridized with an antibody-conjugated nanoparticle that recognizes the biomarker of interest on these exosomes, which can be either a general exosome marker or a factor specific for an exosome subtype of interest. Images of these sample wells are then captured using a dark field microscope (DFM) and analyzed to measure the light scattered from nanoparticles bound to exosomes of interest captured in each sample well6,16,17. Notably, imaging an entire sample well by low-magnification DFM (LMDFM) avoids a selection bias encountered with high magnification DFM analyses when users must directly choose which fields to capture for subsequent image analysis. LMDFM image analysis is subject to light scattering artifacts from surface irregularities, including scratches and sample debris, but this background can be reduced using a simple noise-reduction algorithm we developed to run on the NIH image analysis program, ImageJ (https://imagej.nih.gov/ij/). This algorithm first applies an input contour threshold that is used to detect the boundaries of the sample well to define the region of the image for subsequent analysis. The region defined by this contour region is then split to separate signal present in the red, blue and green channels of the image and the blue channel is subtracted from the red channel to remove signal arising from surface artifacts and uneven lighting from nanorod signal.
This article describes how to use this assay to rapidly quantify either total or specific exosome levels in plasma or serum samples.
Protocol
1. Preparation of nanoparticle probes
NOTE: This assay utilizes Functionalized Gold Nanorods (AuNRs; 25 nm diameter x 71 nm length) that are covalently conjugated with neutravidin polymers (AV) and have a surface plasmon resonance peak that produces a red (641 nm peak) scattering signal upon DFM illumination.
- Wash 40 µL of AuNR-AV (2.56 x 1011 particles) three times with 200 µL PBS (pH 7.0) by centrifugation and aspiration (8,500 x g at 4 °C for 10 minutes), followed by a final centrifugation and aspiration step after which the AuNR-AV pellet is suspended in 40 µL PBS.
- Mix this AuNR-AV suspension with 10 µL biotinylated antibody (0.5 mg/mL) specific for an antigen on the surface of the exosome subtype of interest and 150 µL of PBS and then mix at 4 °C for 2 h using a mixer to allow neutravidin-biotin binding to reach completion.
- Wash the resulting antibody-conjugated AuNRs (AuNR-IgG) three times by centrifugation and aspiration (6,500 x g at 4 °C for 10 minutes), and then suspend them in 200 µL PBS and store them at 4 °C until use.
NOTE: Sterile technique and short storage times must be used to avoid contamination and degradation of the AuNR-IgG. It is best to use antibody-conjugated AuNRs within 24 hours of their conjugation.
2. Preparation of EV capture slides
- Dilute selected exosome capture antibodies to 0.025 mg/mL in PBS and add 1 µL/well of this dilution onto a multi-well protein A/G slide, and then incubate this slide at 37 °C for 1 h in a humidified chamber to allow capture antibody binding to protein A/G immobilized on the slide.
- Aspirate wells to remove unbound antibodies, and wash wells three times by the addition and aspiration of 1 µL/well of PBS, then load each well with 1 µL of blocking buffer (see Table of Materials) and incubate the slide for 2 h at 37 °C in a humidified chamber to block any remaining protein binding sites.
- Aspirate wells to remove blocking buffer, wash wells three times by the addition and aspiration of 1 µL/well of PBS, and immediately use the blocked slides for exosome capture and analysis.
3. Standard curve preparation
- To accurately quantify the absolute or relative abundance of a specific exosome subtype, the user must generate a standard curve with a pure exosome population that uniformly expresses the exosome surface biomarker of interest. This study analyzes the abundance of exosomes expressing a metastasis-associated membrane protein, Ephrin A2 receptor, which has a reported relationship with pancreatic cancer stage and prognosis6,18.
NOTE: The human pancreatic cancer cell line PANC-1 and its exosomes are known to express this protein and isolated exosomes from this cell line were used to generate a standard curve to quantify the number of exosomes that express this protein in complex exosome samples. - Culture cells for 48 hours at 37 °C in serum-free culture media to allow exosome accumulation in the media, then isolate cell culture supernatants by centrifugation of suspension cultures or direct aspiration of culture media from adherent cell cultures.
- Centrifuge the collected media at 2000 x g for 30 min to remove debris and recover the supernatant.
- Filter the clarified culture supernatant through a 0.45 µm low protein binding filter unit of appropriate capacity (e.g. a 250 mL polyethersulfone vacuum filtration unit).
- Concentrate the resulting filtrate by centrifugation at 3200 x g using a 100,000 nominal molecular weight limit filter system to a 250 µL final volume. Collect the retained volume from this filter, then wash the filter with 200 µL PBS, and combine this wash volume with the collected exosome sample volume.
- Centrifuge this sample at 21,000 x g for 45 minutes and carefully recover the supernatant, taking care not to collect any precipitated material.
- Centrifuge the recovered supernatant at 100,000 x g for 3 hours to precipitate the exosomes. Aspirate away the supernatant and collect the exosome pellet in 100 µL PBS.
- Store the resulting exosome suspensions at 4 °C if used within 24 hours or at -80 °C for long term storage.
NOTE: Do not subject exosome samples to repeat freeze-thaw cycles. - Quantify an aliquot of the exosome suspension after mixing by direct measurement of exosome numbers (e.g., by nanoparticle tracking analysis or tunable resistive pulse sensing or by measuring the protein concentration of exosome lysates by micro-bicinchoninic acid assay, or an equivalent method, as a means to approximate exosome quantity)16,19.
- Generate a set of serial dilutions of the exosome suspension to allow comparison of nanoparticle signal to input exosome number or protein content.
- Transfer 1 µL of each exosome standard to each of its replicate wells on the assay plate.
NOTE: Standard curves can be used to calculate the slope of the correlation line between nanoparticle signal and exosome concentration to (1) evaluate assay performance and (2) determine the relative concentration of target exosomes in experimental samples.
4. Processing human plasma or serum samples
- Collect plasma or serum samples by standard methods and store at -80 °C until needed for exosome analysis. Rapidly thaw samples in a room temperature water bath. Repeatedly mix the thawed samples by inversion to promote homogenous suspension.
NOTE: Results from serum and plasma samples may not be equivalent, since there is a significant release of exosomes during the clotting reaction. - Centrifuge plasma or serum samples at 500 x g for 15 min to precipitate protein aggregates and other debris. Transfer an aliquot of the plasma or serum sample to a fresh tube and add PBS to generate a 1:1 dilution. Mix the diluted sample by gentle vortexing or inversion, as appropriate. Transfer 1 µL of each plasma or serum suspension to each of its replicate wells on the assay plate.
5. Exosome capture and detection
- Load wells of a blocked EV capture slide with 1 µL/well of exosome sample, using 8 replicates per sample, and incubate the slide overnight at 4 °C in a humidified chamber. Aspirate all sample wells and then add 1 µL/well of PBS to wash wells and remove unbound exosomes and other contaminants from the loaded exosome sample.
- Load sample wells with 1 µL/well of a previously prepared AuNR-IgG suspension (see section 1 above) and incubate the slide for 2 h at 37 °C in a humidified chamber. Aspirate the nanoparticle solution and wash the slide in PBS supplemented with 0.01% Tween-20 (PBST) for 10 min using a mixer, then aspirate and wash all sample wells with deionized water for 10 min using a rotating mixer, and air-dry for subsequent LMDFM imagery.
NOTE: Inter-assay coefficients of variation (CVs) are assessed from eight replicates of the same sample. Samples that exhibit CVs >20% are considered non-informative and should be repeated, if there is sufficient sample.
6. DFM Image Capture
- Capture images for exosome quantification under consistent lighting using a digital camera attached to a microscope equipped with a dark-field condenser (1.2 < NA < 1.4) and a 4x objective employing a 1/220 s exposure time.
- Open the image capture software.
NOTE: We use NIS-Elements microscope imaging software (see Table of Materials) for the protocol described below, but it is possible to use another software that can match its image capture parameters. NIS-Elements Viewer imaging software is a free standalone program to view image files and data sets that contains analysis, visualization and archiving tools. The parameters below are also for a microscope with autofocus and an automated stage that permits multiple images to be automatically captured and stitched into a single image. - Place the slide upside down on the microscope stage, adjust the slide position and apply a small drop of immersion oil on the back of the slide, where the condenser lens contacts the slide.
- Click the live button in the software interface, and adjust the exposure time against a high concentration standard well to ensure the image is not saturated.
- Open the Scan Large Image window from the Acquire tab and set the software interface parameters as follows: Macro Image Optical conf = current; Objective: 2:10x, Scanning Optical conf = current, Objective: 2:10x; Stitching Overlap = 20%; Stitching via = Optimal Path.
- Choose Create Large Image, Close active shutter during stage movement, Wait before each Capture: 20 ms, Focus manually at start, and Use step-by-step focus every 20 field. These setting will be saved with the scanned images.
- Move the microscope stage to define the left top right bottom limits of the target scan field. Adjust the focus to achieve a clear image on the monitor and adjust the condenser settings and environmental lighting as necessary to minimize any lighting irregularities in the focused image.
- Name the image output file in the software. Click the Scan button and allow the microscope to scan and create and save a stitched image of the entire slide.
- Open the saved image with the image capture software you are using and save it at 1/8 scale for subsequent analysis on the DSM plugin in ImageJ.
7. DFM image analysis
- Download the ImageJ program (https://imagej.nih.gov/ij/). Install the DSM algorithm plugin into ImageJ using the instructions listed at https://imagej.net/Plugins#Installing_plugins_manually.
- Open the ImageJ software, then set the following input parameters within the DSM algorithm: Contour threshold (Ct) = 253.020, Type = Red, Center scale (S) = 0.8, Low (Lt)/High (Ht) quantification limit = 0/62.
- Open the saved image from section 6.8 with ImageJ. Choose the DSM Scan button from the Plugins tab, then define the number of columns and rows according to the opened image. The program can recognize the detecting areas and analysis the scatter intensity of nanoparticle in according areas automatically. Set the following input parameters within the DSM Scan window: Resize Percentage = 25, Spot Diameter (in pixels) = 190 – 200, Diameter Range = 32, Increment diameter (in pixels) = 8, DSM Configuration – Low limit = 0, High limit = 62, Adjacent Distance = 100, Subtract Bias = 0.
NOTE: The results of scatter intensity of nanoparticle reflect the quantity of bound exosomes on the slide.
Representative Results
Multi-well protein A/G coated slides (Figure 1A) were functionalized with anti-EphA2 antibody and then used to specifically capture EphA2-positive exosomes from serum samples of patients with and without pancreatic cancer (1 µL/well) and incubated with gold nanorods conjugated with an anti-CD9 antibody (Figure 1B). DFM images of light scattered from these nanoparticles were analyzed using the DSM plugin in ImageJ software to quantify the bound EphA-2 positive exosomes in each well. The DSM algorithm automatically defines the boundary of a sample well, filters the noise from artifacts, calculates the scattered signal from each well, and outputs this information (Figure 2). The DSM algorithm strongly attenuates light scattering artifacts from scratches or debris present in the sample and improves the sensitivity and reproducibility of nanoparticle detection and can automatically process a batch of slide images for high-throughput use. This algorithm uses ImageJ commands and parameters input by the user to subtract image background, calculate the scattering signal from each well, and output a data and image file (Figure 3). Regions of interest are defined by the high intensity well boundaries in the capture image using the contour threshold function of the ImageJ macro program. Image analyses use a predefined contour threshold and image parameters to calculate the intensity of nanoparticle scattering for each well.
As reported in our previous work (supplementary information of Liang et al.6), exosomes isolated from PANC-1 cell cultures characterized by transmission electron microscopy and Western blot exhibited the size range, morphology, and protein marker expression consistent with a high purity exosome sample. The PANC-1 exosomes, prepared with the same procedure as our previous work, were used here to validate our nPES assay for exosome quantification. This assay used an anti-EphA2 antibody to capture a large population of exosomes from the total exosome population and an antibody against the general exosome protein CD9 to detect captured exosomes. Results obtained using serially diluted PANC-1 exosome samples, with protein concentrations ranging from 0.24 to 1.2 µg/µL, showed good reproducibility in replicate wells (Figure 4A) and a strong linear correlation between the scatter response and exosome protein concentration (Figure 4B).
To demonstrate the potential application of this method, serum samples from patients with and without pancreatic cancer were analyzed to detect the abundance of serum exosomes expressing the cancer-associated biomarker EphA2, using an anti-EphA2 antibody to directly capture target exosomes from serum and nanoparticles conjugated with an anti-CD9 antibody to detect bound exosomes. This analysis revealed that serum samples from the pancreatic cancer patients had significantly higher levels of EphA2+ exosomes (Figure 5) than their controls.
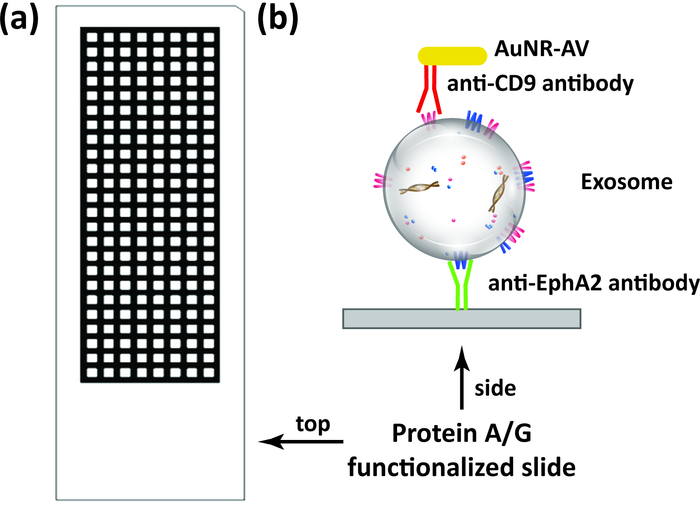
Figure 1: Exosome quantification scheme. (A) Schematic of multi-well protein A/G slides (192 wells) used in this assay. (B) Target exosomes are directly captured from samples, including serum and plasma, by surface immobilization on the capture antibody (e.g. anti-EphA2 antibody) bound to the slide, and then incubated with gold nanorods conjugated with a detection antibody (e.g. anti-CD9 antibody) before analysis by DFM image analysis. Please click here to view a larger version of this figure.
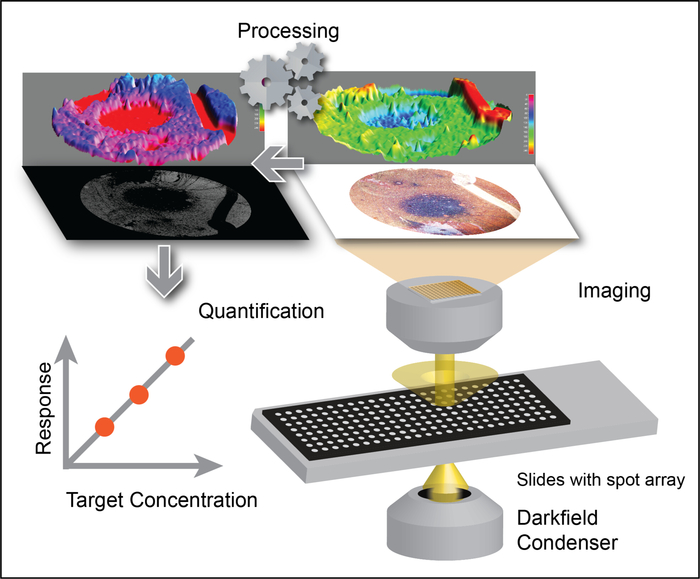
Figure 2: Exosome quantification by applying the DSM algorithm to LMDFM images. Low-magnification images are processed with the DSM algorithm to eliminate background signal and signal artifacts that can arise from scratches, mixing voids, debris, and uneven sample illumination to allow robust detection of gold nanorod signal, which correlates with exosome concentration. This figure has been adapted with permission from Sun, D. et al. Noise Reduction Method for Quantifying Nanoparticle Light Scattering in Low Magnification Dark-Field Microscope Far-Field Images. Analytical Chemistry. 88 (24), 12001-12005 (2016). Copyright (2016) American Chemical Society. Please click here to view a larger version of this figure.
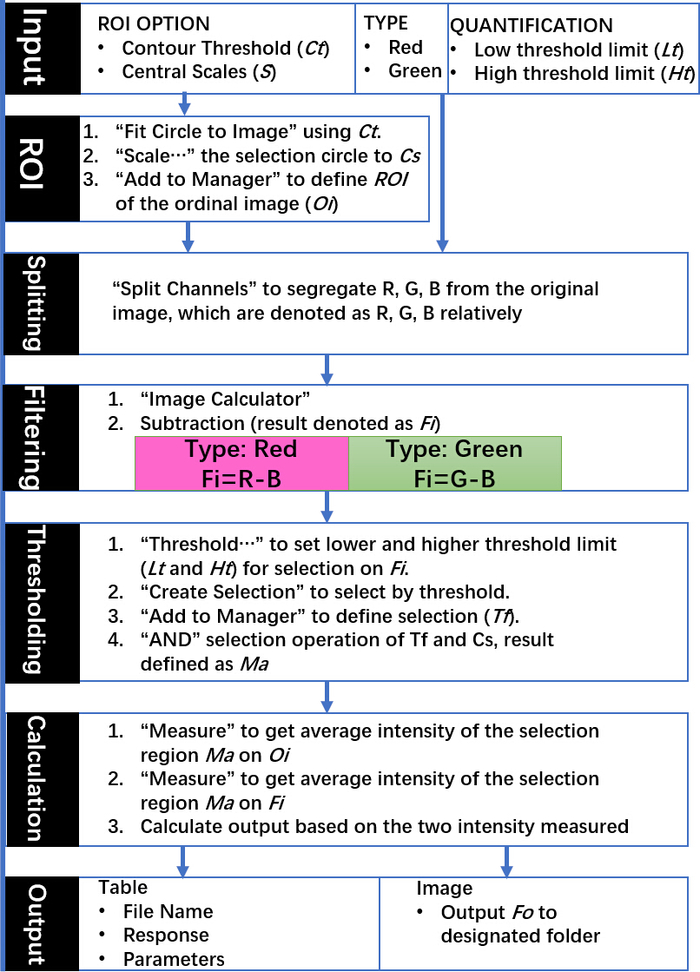
Figure 3: Schematic of the DSM algorithm commands and outputs. Indicated steps use native commands from ImageJ and all input parameters are selected according to the requirements of experiments via graphical user interface. This figure has been adapted with permission from Sun, D. et al. Noise Reduction Method for Quantifying Nanoparticle Light Scattering in Low Magnification Dark-Field Microscope Far-Field Images. Analytical Chemistry. 88 (24), 12001-12005 (2016). Copyright (2016) American Chemical Society. Please click here to view a larger version of this figure.
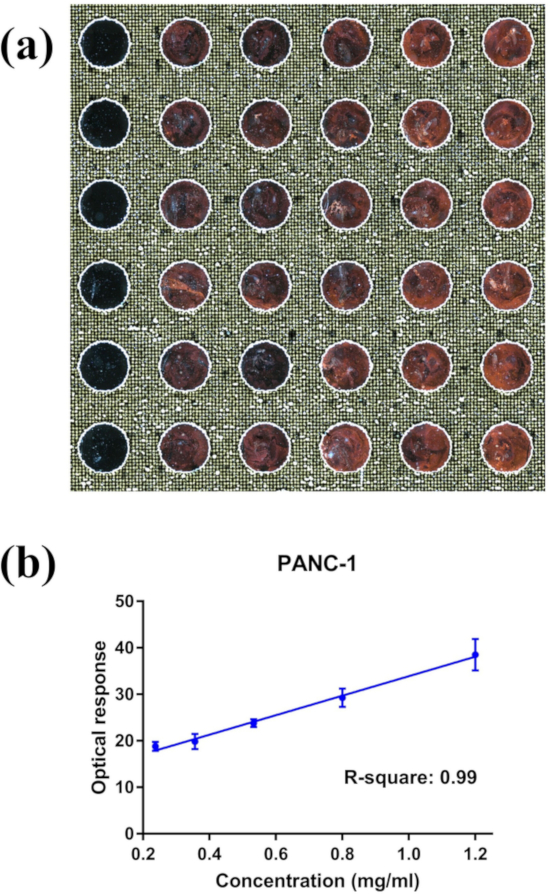
Figure 4: Representative nPES LMDFM images and DSM output data. (A) LMDFM image of an nPES assay analyzing a concentration gradient of PANC-1 exosomes and (B) the linear correlation of the optical signal and exosome concentration from this slide (left to right: 0.24, 0.356, 0.53, 0.80, 1.20 µg/µL respectively for each column). Data are presented as Mean±SE, n=6, with a Pearson correlation coefficient R2 = 0.99 and coefficient of variation for the replicates of each concentration < 0.2. Please click here to view a larger version of this figure.
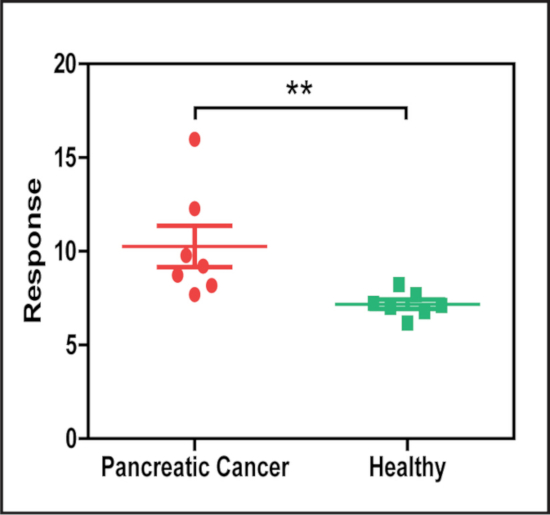
Figure 5: LMDFM signal of EphA2+ exosomes differs in serum from patients with and without pancreatic cancer. Serum samples analyzed by nPES using an anti-EphA2 capture antibody (cancer-associated) and an anti-CD9 detection antibody (general exosome marker) exhibited a significant difference in the concentration of EphA2+ exosomes in serum samples of patients with and without pancreatic cancer (N = 7/group). Results are presented as Mean±SE. **p = 0.002 by Mann-Whitney U-test (two-sided). This figure has been modified from [Sun, D. et al. Noise Reduction Method for Quantifying Nanoparticle Light Scattering in Low Magnification Dark-Field Microscope Far-Field Images. Analytical Chemistry. 88 (24), 12001-12005 (2016)]. Please click here to view a larger version of this figure.
Discussion
Exosomes arise from regulated invaginations of the outer endosome membrane that produce multivesicular bodies, a specialized subset of endosomes that contain a large number of intraluminal vesicles that undergo fusion with the plasma membrane to release mature exosomes into the extracellular space. Due to this biogenesis pathway, exosomes can carry membrane-bound factors associated with membrane fractions that comprise or fuse with the endosome membrane, as well as multiple different types of cytosolic components, and thus contain cargoes of proteins, DNA and various RNA subtypes (mRNAs, microRNAs, long non-coding RNAs) that can reflect the phenotype of their cell of origin20. Since exosomes are secreted by most if not all cell types, can exhibit increased secretion from diseased or malignant cells, and accumulate in most body fluids, exosomes are the subject of comprehensive and systematic investigation as a promising minimally invasive means to detect specific disease conditions and monitor their responses to treatment21.
Exosome isolation, which is required for most current exosome analyses, is a lengthy and labor-intensive procedure, restricting the clinical translation of exosome-associated biomarkers with potential medical relevance. Many common isolation methods (ultracentrifugation, size-exclusion, precipitation, etc.) often do not sufficiently distinguish exosomes (30–100 nm) from micro-vesicles (100–1000 nm) and apoptotic bodies (100–5000 nm) due to overlaps in their size ranges or physical properties or may damage exosome integrity15. New approaches are under development that may permit more rapid exosome analyses, but it is not clear how feasible it may be to implement many of these platforms in clinical settings due.
In this report, we present a novel approach that allows nanoparticle-based high-throughput exosome quantification using low magnification dark field microscope images. This method does not require exosome purification, expensive dedicated equipment, or novel technical expertise and should thus be amenable to rapid translation in most research and clinical settings. Our assay can be applied to precisely quantify the concentration of a target exosome population bearing a specific biomarker when assay results are compared to a standard curve, since our results exhibit there is a strong linear correlation (R2 = 0.99) between optical response and exosome concentration. To demonstrate the real world potential of this approach, we have provided data where we employed this method to quantify the concentration of an exosome biomarker associated with pancreatic cancer in serum samples obtained from patients with and without pancreatic cancer.
In LMDFM, the entire sample well is imaged to avoid the selection bias found in high-magnification DFM analyses, where users must directly choose the sample fields to capture for subsequent image analysis, but is subject to light scattering artifacts from surface irregularities, including scratches and sample debris. This background can be reduced to detect target exosome-derived signal using our DSM noise-reduction algorithm that runs on the NIH image analysis program, ImageJ, but care must still be taken to avoid introducing such artifacts which may reduce the dynamic range of the assay.
Materials used in this assay:
Multi-well SuperProtein A/G slides holding 1 µL/well were purchased from Arrayit Corporation (AGMSM192BC). Nanoparticles were obtained from Nanopartz (C12-25-650-TN-DIH-50-1, 6.4 x 1012/mL). DFM images were captured with a Nikon DiR2 digital camera attached to a Nikon Ti-Eclipse microscope, with consistent lighting and a 1/220 s exposure time. The PANC-1 cell line used in this study was purchased from the American Type Culture Collection.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The work was primarily supported by research funding provided by NIH (U01CA214254, R01HD090927, R01AI122932, R01AI113725 and R21Al126361-01), Arizona Biomedical Research Commission (ABRC) young investigator award.
Materials
| Eppendorf Repeater stream | Fisher Scientific | 05-401-040 | |
| Eppendorf Research plus | Eppendorf | 3120000011 | 0.1 – 2.5 µL, dark gray |
| Functionalized Gold Nanorods | Nanopartz | C12-25-650-TN-DIH-50-1 | In vitro neutravidin polymer functionalization |
| HulaMixer Sample Mixer | Thermo Fisher Scientific | 15920D | |
| Incu-shaker 10L | Benchmark Scientific | H1010 | |
| Inverted Research Microscope | Nikon | Ti-DH | With Dark field condenser, DS-Ri2 camera, and Ti-SH-U universal holder, and motorized stage |
| NIS-Elements | Nikon | Microscope imaging software | |
| Phosphate Buffered Saline (1X) | GE Healthcare Life Sciences | SH30256.02 | HyClone |
| Protein A/G Treated Glass Substrate Slides | Arrayit Corp. | AGMSM192BC | Premium microarray substrate |
| Q500 Sonicator | Qsonica, LLC | Q500-110 | With standard probe (#4220) |
| Superblock blocking buffer | Thermo Scientific | ||
| TWEEN 20 | Sigma Life Sciences | 9005-64-5 |
References
- Andaloussi S, E. L., Mager, I., Breakefield, X. O., Wood, M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature Reviews Drug Discovery. 12 (5), 347-357 (2013).
- Choi, D. S., Kim, D. K., Kim, Y. K., Gho, Y. S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 13 (10-11), 1554-1571 (2013).
- Schorey, J. S., Cheng, Y., Singh, P. P., Smith, V. L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Reports. 16 (1), 24-43 (2015).
- Hoshino, A., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 527 (7578), 329-335 (2015).
- Zaborowski, M. P., Balaj, L., Breakefield, X. O., Lai, C. P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience. 65 (8), 783-797 (2015).
- Liang, K., et al. Nanoplasmonic Quantification of Tumor-derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nature Biomedical Engineering. 1, (2017).
- Severino, V., et al. Extracellular Vesicles in Bile as Markers of Malignant Biliary Stenoses. Gastroenterology. 153 (2), 495-504 (2017).
- Osteikoetxea, X., et al. Detection and proteomic characterization of extracellular vesicles in human pancreatic juice. Biochemical and Biophysical Research Communications. 499 (1), 37-43 (2018).
- Bulacio, R. P., Nosetto, E. C., Brandoni, A., Torres, A. M. Novel finding of caveolin-2 in apical membranes of proximal tubule and first detection of caveolin-2 in urine: A promising biomarker of renal disease. Journal of Cellular Biochemistry. , (2018).
- Nair, S., Tang, K. D., Kenny, L., Punyadeera, C. Salivary exosomes as potential biomarkers in cancer. Oral Oncology. 84, 31-40 (2018).
- Kim, J. E., et al. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: A pilot study. Thoracic Cancer. 9 (8), 911-915 (2018).
- Boussadia, Z., et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. Journal of Experimental & Clinical Cancer Research. 37 (1), 245 (2018).
- An, M., Wu, J., Zhu, J., Lubman, D. M. Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum. Journal of Proteome Research. , (2018).
- Brenner, A. W., Su, G. H., Momen-Heravi, F. Isolation of Extracellular Vesicles for Cancer Diagnosis and Functional Studies. Methods in Molecular Biology. 1882, 229-237 (2019).
- Li, P., Kaslan, M., Lee, S. H., Yao, J., Gao, Z. Progress in Exosome Isolation Techniques. Theranostics. 7 (3), 789-804 (2017).
- Sun, D., et al. Noise Reduction Method for Quantifying Nanoparticle Light Scattering in Low Magnification Dark-Field Microscope Far-Field Images. Analytical Chemistry. 88 (24), 12001-12005 (2016).
- Sun, D., Hu, T. Y. A low cost mobile phone dark-field microscope for nanoparticle-based quantitative studies. Biosensors and Bioelectronics. 99, 513-518 (2018).
- Koshikawa, N., Minegishi, T., Kiyokawa, H., Seiki, M. Specific detection of soluble EphA2 fragments in blood as a new biomarker for pancreatic cancer. Cell Death & Disease. 8 (10), e3134 (2017).
- Clayton, A., Turkes, A., Navabi, H., Mason, M. D., Tabi, Z. Induction of heat shock proteins in B-cell exosomes. Journal of Cell Science. 118 (Pt 16), 3631-3638 (2005).
- Henne, W. M., Buchkovich, N. J., Emr, S. D. The ESCRT pathway. Developmental Cell. 21 (1), 77-91 (2011).
- Panagiotara, A., Markou, A., Lianidou, E. S., Patrinos, G. P., Katsila, T. Exosomes: a cancer theranostics road map. Public Health Genomics. 20 (2), 116-125 (2017).

