Purification and Analysis of Caenorhabditis elegans Extracellular Vesicles
Summary
This article presents methods for generating, purifying, and quantifying Caenorhabditis elegans extracellular vesicles.
Abstract
The secretion of small membrane-bound vesicles into the external environment is a fundamental physiological process of all cells. These extracellular vesicles (EVs) function outside the cell to regulate global physiological processes by transferring proteins, nucleic acids, metabolites, and lipids between tissues. EVs reflect the physiological state of their cells of origin. EVs are implicated to have fundamental roles in virtually every aspect of human health. Thus, EV protein and genetic cargos are being increasingly analyzed for biomarkers of health and disease. However, the EV field still lacks a tractable invertebrate model system that permits the study of EV cargo composition. C. elegans is well suited for EV research because it actively secretes EVs outside of its body into its external environment, permitting facile isolation. This article provides all the necessary information for generating, purifying, and quantifying these environmentally secreted C. elegans EVs including how to work quantitatively with very large populations of age-synchronized worms, purifying EVs, and a flow cytometry protocol that directly measures the number of intact EVs in the purified sample. Thus, the large library of genetic reagents available for C. elegans research can be tapped into for investigating the impacts of genetic pathways and physiological processes on EV cargo composition.
Introduction
The secretion of membrane-bound extracellular vesicles (EVs) facilitates the global physiological processes by actively transporting specific protein, nucleic acid, metabolite, and lipid cargos between cells1. Cells secrete EVs that span a continuum of sizes ranging from 2 μm or greater to as small as 20 nm2. Small EVs (<200 nm) are increasingly studied because of their implication in pathological processes, including metabolic disorders, cancer, cardiovascular disease, and neurodegenerative diseases3,4,5. These pathologies have also been shown to influence the protein and genetic composition of EVs cargos of small EVs. Therefore, biomarker signatures of the pathology are increasingly being uncovered through EV cargo discovery methods such as LC-MS-MS and RNAseq6,7,8,9.
C. elegans has been a useful invertebrate model for identifying evolutionarily conserved EV signaling pathways. For instance, a C. elegans flippase was first shown to induce the EV biogenesis in C. elegans embryos, and the human homolog was shown to influence EV release in human cells10,11. C. elegans EVs were reported to carry Hedgehog signals necessary for cuticle development. The delivery of Hedgehog and other morphogens was shown to play a major developmental role of EVs, and it is conserved in zebrafish, mice, and humans12,13,14,15. C. elegans is well suited for EV biomarker discovery because it secretes EVs outside of its body that function in animal-to-animal communication16,17 (Figure 1A). However, the methodology established through a previous study cannot be used because the nematodes' E. coli food source also secretes EVs18. In this method, most of the sample is comprised of E. coli contamination, limiting the power of proteomic or RNAseq approaches for the discovery of C. elegans EV cargos. The methods described here were developed to yield highly pure C. elegans EVs at abundance levels characteristic of typical cell culture experiments and thus facilitate omics approaches for EV biomarker discovery.
Large populations of worms are needed to generate a sufficient number of EVs for cargo analysis. Therefore, methods for conducting quantitative cultivation of large populations of developmentally synchronized C. elegans are also included. Typically, when a large number of worms are needed for experiments, they are cultured in liquid media. While this is effective for generating large populations of worms, the physiology of the animals is considerably different from worms cultivated under standard conditions on nematode growth medium (NGM) agar plates. Animals cultured in liquid grow more slowly, are thinner, show developmental heterogeneity, and are subjected to a high degree of batch variability. Therefore, we present a simple but effective means for the quantitative cultivation of large populations of developmentally synchronized C. elegans using 10 cm high growth plates. The media composition of high growth plates includes more peptone than regular NGM plates and are seeded with the E. coli strain NA22 which grows more robustly than OP50.
Advances in flow cytometry technology (FACS) have enabled the direct analysis of individual EVs20,21, permitting the quantification of EVs without the inherent limitations in other methods. Prior work has shown that protein is not a useful proxy for EV abundance because different purification methodologies result in significantly different EV-to-protein ratios22. Extremely pure EV fractions contain relatively little protein, making it difficult to quantify samples with BCA or Coomassie gels. Western analysis can identify relative differences of individual proteins but cannot identify how many EVs are in the sample. Robust quantification of EV number by nanoparticle tracking analysis is hampered by its narrow signal-to-noise range, inability to differentiate between EVs and solid aggregates, and the lack of transferability of methods between instruments with different specifications23. Therefore, this article also contains a generalizable flow cytometric procedure for discriminating and quantifying EVs.
Protocol
1. Preparation
- Add 2 g of gelatin to 100 mL of dH2O and heat in the microwave until it starts to boil. Then stir and let it cool for 20 min.
- Pass the solution through a 0.22 μm filter and dispense into sterile tubes. Treat pipette tips with gelatin immediately prior to the use by pipetting up and expelling the gelatin solution. Treated tips can either be used immediately or stored.
- Cut out the membrane from a sterile cap filter.
- Wet a 20 cm square of 5 μm nylon mesh fabric and place around the top of a sterile cap filter. Secure it with several thick rubber bands.
CAUTION: Carefully examine the fabric to ensure that there are no creases. These make air gaps that hinder suction.
2. Calculating Large Populations of Worms (Figure 1A)
- Vortex and pipette 10 μL of the worm suspension onto a worm cultivation plate lid or a glass slide. Repeat this process 3x. Record the number of animals in each drop.
- Adjust the worm suspension to two animals per μL.
- Vortex the suspension and pipette nine drops (10 μL) on a slide or a plate lid. Manually count the number of animals. Record the number of animals in each drop.
- Calculate the mean and standard error of the mean (S.E.M.) as a percentage of each worm population.
- Calculate the total number of animals in each population by dividing the mean value by 10 μL and then multiplying by the total volume of the worm suspension in μL.
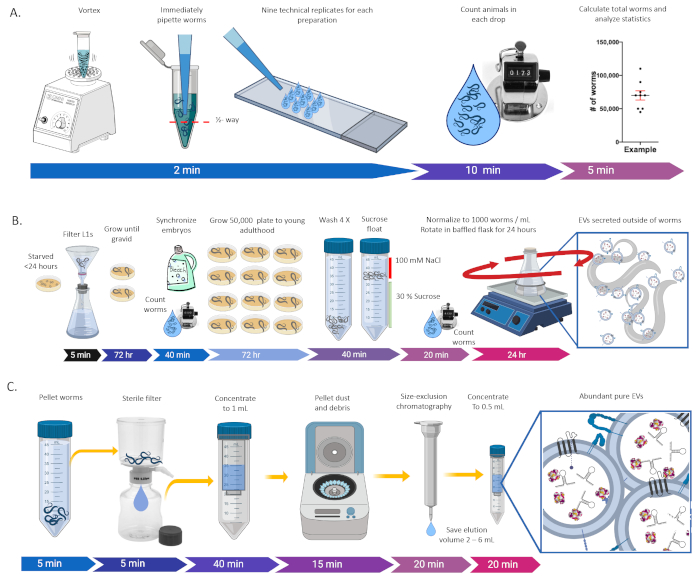
Figure 1: Procedure overviews for generating, quantifying, and purifying C. elegans EVs. (A) Schematic for counting large populations of animals. (B) Schematic for generating worms for harvesting EVs. (C) Schematic for purifying EVs from C. elegans supernatent. Please click here to view a larger version of this figure.
3. Cultivating C. elegans for EV Purification
- Generate a large population of developmentally synchronized C. elegans. To do so, follow the steps below.
- Add a single animal on a 6 cm normal growth media plate. Incubate for two generations and then wash the animals onto two high growth medium plates.
- Incubate until the worms have exhausted the food.
- Filter L1 worms with 5 μm nylon mesh prepared in step 1.4. Place the screw cap on the sterile bottle, start the vacuum, and pour worms over the filter. Pass the same volume of the medium over the worm aggregates on the filter.
- Quantify the worms and calculate the total number (see step 2). Recently starved (<24 h) worms grown on a high growth plate result in about 80,000 L1 larvae.
- Centrifuge L1 larvae at 2,000 x g for 3 min. Resuspend to ~100,000 animals per mL.
- Place ~50,000 animals per high growth plate. Cultivate the animals at 20 °C until they are gravid adults (about 72 h).
- Bleach the gravid adults by standard means and allow embryos to hatch in 10 mL of S Basal solution with 2.5 μg/mL cholesterol.
- Quantify L1 larvae (see step 2) and then add 50,000 worms to each high growth plate.
- Cultivate plates at 20 °C until animals are young adults.
- Prepare C. elegans for secretome generation.
- Wash worms from plates by adding 15 mL of sterile M9 with 50 μg/mL carbenicillin to each high growth plate. Let the saturated plates sit for 5 min. Dislodge worms by circling the buffer around the plate. Avoid sloshing. Pour the buffer from each plate into a separate 50 mL conical tube. Repeat the wash step 2x more for a total of 45 mL of buffer per plate.
- Let the worms settle for 15 min. Decant the supernatant and add 50 mL of fresh M9 buffer to the tube. Repeat for a total of 4x.
- Float the worms on the top of a 30% sucrose step-gradient and recover animals in the S Basal solution24.
- Briefly resuspend the worms in 15 mL of ice-cold 100 mM NaCl, add 15 mL of ice-cold 60% sucrose, and invert to mix. With a glass pipette gently layer 5 mL of ice-cold 100 mM NaCl on the top of the sucrose mixture.
- Centrifuge at 1,500 x g for 5 min. Worms will be concentrated at the interface between the NaCl and the sucrose.
- Gently pipette out the animals from the interface into a new 50 mL conical tube. Add S Basal solution to 50 mL. Allow worms to settle 5 min.
- Decant the supernatant and add fresh 50 mL of S Basal solution. Repeat this at least 3x.
- Quantify the number of worms as described in step 2.
- Resuspend the worms to a density of one animal per μL with sterile S Basal solution containing 2.5 μg/mL of cholesterol and 50 μM of carbenicillin.
- To prepare the sample for secretome generation, add up to 400 mL of the suspension to a sterile 2 L bottom-baffled flask.
- Place flasks on the circular rotator in a 20 °C incubator at 100 rpm for 24 h.
4. EV Purification
- Harvesting and fractionation
- Remove the 2L flask of worms from the incubator and pellet the animals in 50 mL conical vials (500 x g for 2 min).
- Pour the supernatant through a 0.22 μm vacuum filter to remove any particulate debris.
NOTE: At this point, the filtered supernatant containing the secretome and the vesicles can be stored at -80 °C. - Count the number of animals as described in step 2.
- Determine the viability of the worms by placing a drop of suspended worms onto a bacterial lawn. Wait for 15 min and then score animals as moving, paralyzed, or dead.
- Concentrate the 0.22 μm filtered supernatant to 700 μL using regenerated nitrocellulose 10 kDa filter units. Add 150 μL S Basal solution and then pipette over the filter and vortex. Repeat 2x.
- Centrifuge the concentrated supernatant at 18,000 x g for 20 min at 4 °C and transfer the supernatant to a new tube. This step removes any potential debris or larger particles that might have accumulated during handling.
- Add the protease inhibitor cocktail + EDTA per the manufacturer's instructions.
NOTE: At this point the concentrated supernatant containing the secretome and the vesicles can be stored at -80 °C.
- Size exclusion chromatography
- Pour 80–200 μm agarose resin (pore size fractionation range of 70,000 to 40,000,000 kDa for globular proteins) into an empty gravity flow column cartridge. Flow S Basal solution until the resin bed is packed at a final volume of 10 mL.
NOTE: If too much resin is poured into the column cartridge, then disperse the top of the column and remove the excess with a pipette. - Pass 40 mL of sterile filtered S Basal solution through the column and let it flow under gravity.
CAUTION: Check that the resin is not disturbed. - Let the buffer flow until the top of the resin is not submerged and then cap the bottom of the column cartridge. Place a collection tube underneath the column.
- Remove the cap placed on the cartridge. Add the concentrated secretome obtained in step 2.1 dropwise to the top of the column bed. Pipette 1 mL of S Basal solution dropwise. Place a fresh collection tube under the column.
- Slowly fill the upper column reservoir with 5 mL S Basal solution to not disturb the resin. Collect the first 2 mL of eluate then quickly change tubes and collect the next 4 mL. This is the fraction that is enriched for EVs.
- Concentrate the eluate to 300 μL with a regenerated nitrocellulose 10 kDa molecular weight cut off (MWCO) filter and the transfer retentate to a low-binding microcentrifuge tube.
- Wash filter membrane 2x with 100 μL of S Basal solution by vortexing for 20 s and pipetting the buffer across the filter. Add to the initial sample for a final volume of 500 µL.
- Add protease inhibitor cocktail per the manufacturer's instructions.
- Perform the downstream experiments immediately (RNAseq, LC-MS-MS, GC-MS-MS, Western, FACS, etc.) or store at -80 °C.
- Pour 80–200 μm agarose resin (pore size fractionation range of 70,000 to 40,000,000 kDa for globular proteins) into an empty gravity flow column cartridge. Flow S Basal solution until the resin bed is packed at a final volume of 10 mL.
5. Flow Cytometry Quantification of EV Abundance
- Dye preparation
- Prepare a 10 mM stock of DI-8-ANEPPS using fresh DMSO. Distribute into 10 μL aliquots and store at -20 °C.
- Prepare a 1 mM working solution by adding 90 μL of sterile filtered PBS to a tube with 10 μL of dye stock.
- Experimental sample preparation
- Add 840 μL of S Basal solution into a microcentrifuge tube. Then add 60 μL of the purified EVs generated in section 4.2 and vortex.
- Aliquot 300 μL of the sample into two new microcentrifuge tubes. There is now an experimental set of three tubes containing 300 μL of diluted EVs each. Label the tubes #1–3. To samples #2 and #3 add 7 μL of the DI-8-ANEPPS stock prepared in 5.1.2. Then add 7 μL of 1% Triton-X 100 to tube #3. Mix each tube by vortexing.
- Sonicate sample #3 by placing the sonicator tip in the middle of the sample and pulsing 10 times at 20% power 30% duty cycle.
CAUTION: Ensure that the tip of the sonicator probe is in middle of the sample so that foam generation is kept to a minimum. Foam may compromise the sample by causing the EVs to aggregate. - Add 300 μL of S Basal solution to two microcentrifuge tubes. Add 7 μL of the DI-8-ANNEPS working reagent prepared in step 5.1 to the second tube and label "Dye Only". Label the other tube "Buffer Only".
NOTE: Prepare the Dye Only and Buffer Only control samples and the experimental samples with the same S Basal source. - Incubate the samples away from the direct light and at room temperature for 1 h.
- Conduct FACS experiments.
- Set the FACS excitation filter to 488 nm (blue) and the emission filter to 605 nm (orange). Set the flow rate to 1.5 μL per min.
- Run nanobead FACS calibration mix (optional but recommended).
- Run each sample on a FACS machine for 3 min (180 s). Note the exact running times in seconds.
- Analyze the FACS data.
- Open the files with FACS analysis software.
- Switch the Y-axis to 488-Org (Area) by clicking on the Axis, selecting from the drop-down menu.
- Set a rectangular gate starting at the top of the plot, spanning from SALS level 102 to 104 (<300 nm sized particles). Extend the gate downwards until it contains 2.5% of the total events. Name the gate "DI-8-ANEPPS+" events.
- Quantification of sample EV abundance
- Copy this gate and paste it onto the other two samples in your experimental set as well as the Buffer Only and Dye Only controls. Export the analysis to a spreadsheet.
- If the FACS samples were not run for the recommended 3 min (180 s), normalize all event counts in the sample to events per 180 s. For example, if a sample was run for 250 s, the DI-8-ANEPPS+ event number is scaled by 250 s/180 s.
- Remove the dye background events by subtracting the number of DI-8-ANEPPS+ events in the Dye Only control from those in sample #2.
- Remove isotype background events by subtracting the number of DI-8-ANEPPS+ events in sample #1.
- Remove detergent-insensitive events by subtracting the number of DI-8-ANEPPS+ events in sample #3. This value is the number of bona fide EVs.
- Calculate the number of EVs in 1 μL of sample by dividing the value calculated in step 5.5.5 by the volume analyzed in μL (4.5 μL analyzed as described in step 5.3) and multiplying by the dilution factor (10x as described in step 2.5). Multiply this value by the number of μL remaining in the EV preparation. This is the number of EVs available for downstream analysis.
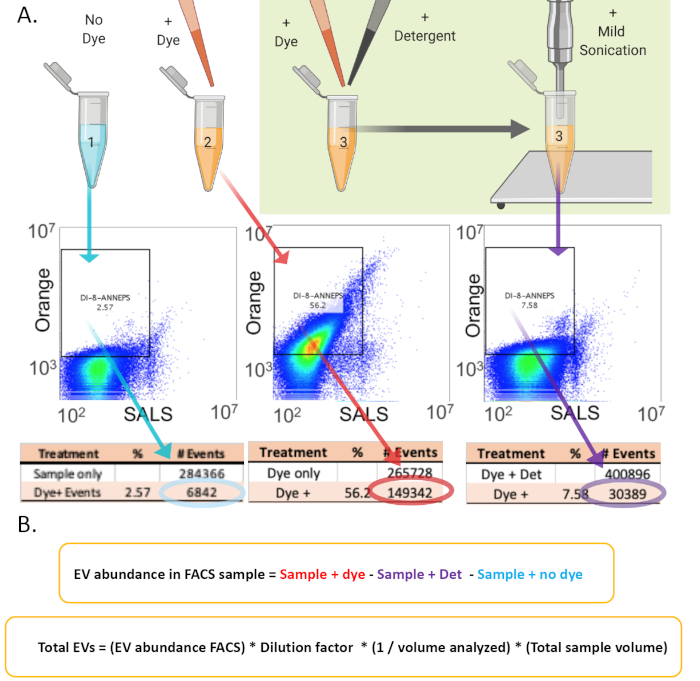
Figure 2: Schematic of flow cytometry sample preparation to quantify EV abundance. (A) The EV preparation is divided into three identical samples. Sample # 1 is the No Dye negative control. DI-8-ANEPPS is then added to samples #2 and #3. Triton-X 100 is then added to sample #3. The green arrow indicates that only sample #3 is subsequently sonicated. The data from the flow help determine the absolute number of detergent sensitive EVs in the FACS samples and then calculate the abundance of EVs in the total preparation. The gate for determining dye-positive events is set with sample #1, which contains no dye. The red writing on the spreadsheet data is to illustrate that dye-positive events from the detergent-treated sample are subtracted from the sample with dye. (B) Equations for quantifying EVs in a FACS analyzed sample and calculating the total number of EVs in the sample. Please click here to view a larger version of this figure.
Representative Results
A schematic for the processes necessary for generating and purifying EVs is shown in Figure 1. The typical times required to complete each step are shown underneath. Figure 2 shows a schematic for preparing samples for FACS analysis using DI-8-ANEPPS (Figure 2A) as well as the calculations necessary to estimate total number of EVs in a sample (Figure 2B).
Representative results from 10 biological replicates are shown in Figure 3A. The variability between replicates is not significant and the typical S.E.M. of population size is a little over 10% (Figure 3B). Filtering the worms and cultivating on the fresh high growth plates will generate a large population of gravid adults suitable for bleach synchronization. A single starved plate processed in this way can produce an experimental population of 1 x 106 or more synchronized progeny because each gravid adult will contain about 10 eggs. It is possible to maintain large populations of developmentally synchronized populations by filtering eggs and larvae through a 40 μm filter to obtain EVs from older animals. Retained adults are transferred to fresh high growth plates at a density of 20,000 animals per plate. This process was repeated every 2 days. Figure 3C shows three biological replicates of worm populations being moved across three plate transfers. The transferring of animals between plates results in about a 10% loss of animals (Figure 3C) while about 20% of the animals are lost in the sucrose flotation step (Figure 3D).
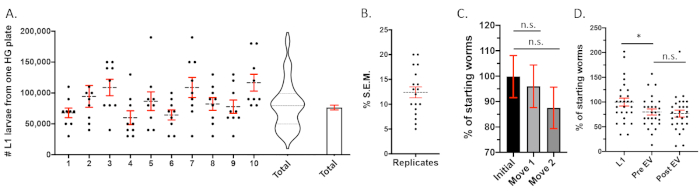
Figure 3: Quantification of large populations of C. elegans for EV analysis. (A) A comparison of the number of L1 larval stage worms isolated from a single recently starved (<24 h) high growth plate. The values of each of 10 biological replicates is plotted. The summation of all the data points in the 10 replicates is also presented as a violin plot and bar with S.E.M. This shows that a single plate of recently starved worms will yield ~80,000 L1 larval stage worms. (B) The S.E.M. of the 15 different plate population measurements in panel A plotted as individual points. Generally, the S.E.M. is slightly greater than 10%. (C) Three biological replicates of worm populations were estimated after each of two transfers between plates every 4 h. This transfer of animals between plates did not result in a significant decrease in population size. This indicates that the methodology presented here for moving worms does not result in a significant loss of animals. (D) Population measurements taken along the course of three EV experimental replicates: The numbers of animals that were estimated for the original L1 larvae that started the experiment, the number of young adult animals recovered from the sucrose float and ready for the 24 h incubation period, the number of animals counted from EV preparation upon recovery of the secretome. There is a small but not significant decrease of population size between the initial population estimation at the L1 larval stage and later when the animals are young adults. The mean L1 population measurement was designated as 100% and all other measurements were scaled by their relation to this value. Error bars S.E.M. * = p value < 0.05, N.S. = not significant in a parametric paired t-test. Please click here to view a larger version of this figure.
Protein-to-animal ratio is a useful metric for characterizing an EV preparation. This metric can provide a quick means for determining the consistency between EV preparations. On average, 1,000 wild type young adult animals will secrete 1 μg of proteins larger than 10 kDa into their environment (Figure 4A). When this fraction is further separated by the size exclusion chromatography, the total protein elution profile shows a small protein peak between 2–6 mL and a large peak after 8 mL. The EVs are contained in the first 5 mL of the column eluate (Figure 4B). Nanoparticle tracking analysis the first 5 mL of the column eluate contains a monodisperse population of small, ~150 nm EVs (Figure 4C). Transmission electron microscopy of size exclusion fractions reveals abundant EVs in the 2–6 mL fraction of eluate but not in the later eluate volumes. EVs are known for their cup-like shape and are therefore easy to discriminate from solid particles that appear as punctate bright dots when prepared under the negative staining conditions (Figure 4D).
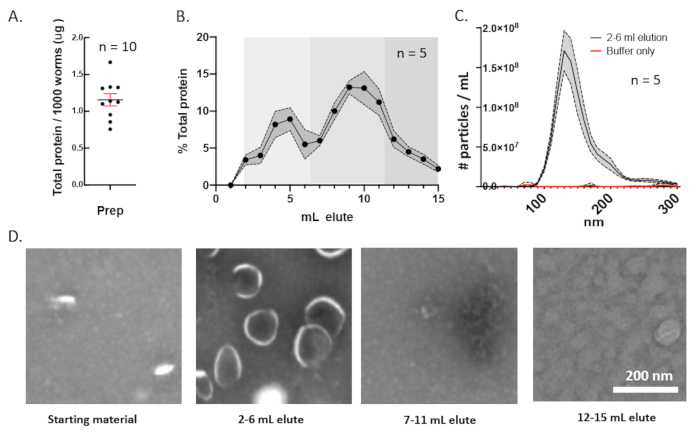
Figure 4: Purification of C. elegans EVs from total secreted biomass. (A) The ratio of the total proteins larger than 10 kDa to the number of worms incubated in the preparation for 10 biological replicates was plotted. Most preparations contained 1 μg of protein per 1,000 animals. (B) Protein elution profile from a 10 mL resin column loaded with 1 mL of the concentrated secretome. The fractions that are consolidated in further analyses are shaded into groups. (C) Nanoparticle tracking analysis of the consolidated resin eluted fractions 2–6 mL. The 95% confidence interval is shaded gray. (D) TEM analysis of the consolidated resin eluted fractions. The starting material had both vesicle-like particles and non-vesicle particles. The consolidated 2–6 mL elution fraction was enriched for vesicles with few non-vesicle particles. The 7–11 mL consolidated fractions contained few vesicle-like particles but many non-vesicle particles. The 12–15 mL elution fractions contained only non-vesicle particles. All micrographs are shown at the same magnification. Scale bar = 200 nm. Please click here to view a larger version of this figure.
To directly compare the flow cytometry characteristics between C. elegans and human EVs, we purified EVs from the conditioned media of cell cultures of human neurons with this method. Flow cytometry separates particles based on light scattering. The small-angle light scattering (SALS) roughly correlates with the size, and the long-angle light scattering (LALS) correlates with internal membrane structure. The majority of the total sample events from both cell culture and C. elegans EV preparations are tightly focused in a cluster centered around 103 SALS and 104 LALS (Figure 5A). A histogram of C. elegans EV sample events sorted by SALS shows that all preparations peak at ~103 (Figure 5B).
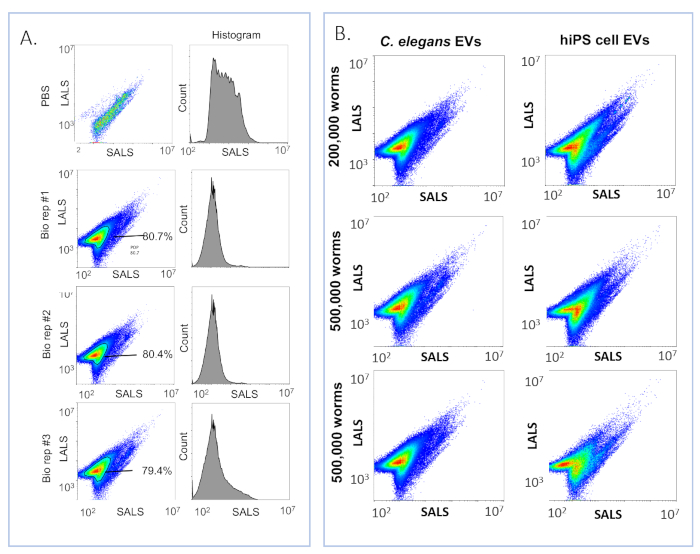
Figure 5: C. elegans EVs are clearly defined by their light scattering properties. (A) Histogram of SALS events in three biological replicates of C. elegans EVs. ~80% of the total sample events are contained within a tight, clearly defined population. Similar to nanoparticle tracking analysis, it shows that the size distribution of C. elegans EVs is monodisperse. The histogram of the S Basal buffer used to dilute the samples is shown for comparison. (B) Comparison of refractive properties of C. elegans and human cell culture EVs purified with the methods described in this text. Both EV types consistently present comparable short and long-angle light scattering distributions (SALS and LALS). Please click here to view a larger version of this figure.
The DI-8-ANEPPS robustly labels C. elegans EVs, consistently highlighting a majority of the total events in both the C. elegans and cell culture derived samples. The calibration beads and controls are presented in Figure 6A. The individual FACS scatter plots are shown from a C. elegans sample prepared from 200,000 animals (#1) and two prepared from 500,000 animals (Figure 6B). EVs from either 100 mL (#1) or 200 mL (#2,3) of cell culture were also analyzed (Figure 6C). Both C. elegans and cell culture derived samples showed abundant fluorescent events that disappeared when treated with 0.05% Triton-X 100 and light sonication. This demonstrates that the labeled events are detergent labile phospholipid structures (vesicles) rather than detergent-insensitive solid lipoprotein aggregates. The EV abundance is calculated as the difference between the number of events in the DI-8-ANEPPS+ gate between the fractions without detergent (-) and with detergent (+) minus the events in the Dye Only control. For clarity, the data presented individually in Figure 6B and Figure 6C is summarized as bar graphs in Figure 6D and Figure 6E. The abundance of the purified EVs was not significantly different between the C. elegans and the cell culture derived preparations (Figure 6F).
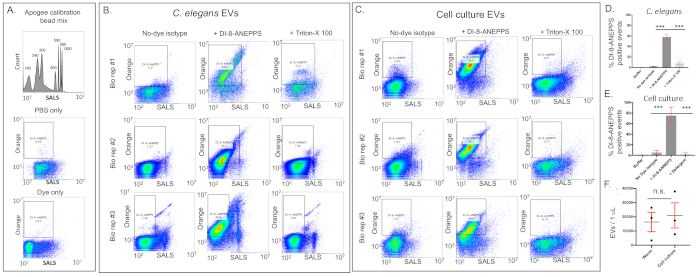
Figure 6: Examples of flow cytometry data used to calculate EV abundance. (A) To ensure that the number of events can be directly compared between samples, the Buffer Only and the Buffer and Dye Only controls need to be run at the same flow rate and collection time as the EV sample fractions. There are very few events in the DI-8-ANEPPS gate. (B) Representative results of the DI-8-ANEPPS and detergent treatment protocol from C. elegans. Three biological replicates are shown. Biological replicate #1 was prepared from 200,000 animals while #2 and #3 were prepared from 500,000 animals. The disappearance of EVs with the detergent treatment is clear in all cases. (C) Representative results of the DI-8-ANEPPS and detergent treatment protocol using human cell culture conditioned media. Biological replicates #1 and #2 were prepared from 200 mL of conditioned media while biological replicate #3 was prepared from 100 mL. (D) Bar graph of the data collected from the three biological replicates of C. elegans EVs. Error bars = S.E.M. Paired two-tailed t-tests between treatments. *** = p value < 0.001. (E) Bar graph of the data collected from the three biological replicates of cell culture derived EVs. Paired two-tailed t-tests between treatments. *** = p value < 0.001 (F) The number of dye-labeled detergent-sensitive events per μL was calculated for all the biological replicates of C. elegans and cell culture. The values between the two EV sample sources is not significantly different. Paired two-tailed t-tests between treatments p value = 0.685. Please click here to view a larger version of this figure.
Table 1 contains the raw experimental values for the full 4.5 μL of each sample type analyzed. The total number of events collected from the EV fractions purified from 500,000 animals was 10–20x over the background number of particles collected by buffer alone while the fraction purified from 200,000 animals contained about 2x as many events as buffer alone. The number of events within the DI-8-ANEPPS+ gate is also shown for each sample. These metrics can be imported into a spreadsheet to calculate the number of dye-positive, detergent-sensitive EVs as described above. For example, the number of EVs in 1 mL of the first C. elegans biological replicate shown would be calculated like this:
(18,974 – 3,853 – 1,487) x (10/4.5) x 1,000 = 30,297,778.
Table 1: Tabulated flow cytometry data. The data shown in Figure 6 is presented as a spreadsheet. For each sample the total events and DI-8-ANEPPS+ gated events are shown. Each biological replicate is arranged in the order of samples #1–3 in the protocol. These metrics reveal that the total number of events collected from the C. elegans samples prepared from 500,000 animals or 200 mL of cell culture conditioned media was 10–20x over the background number of particles collected by buffer alone while the biological replicate purified from 200,000 animals contained about 2x as many total events as buffer alone. The number of events within the DI-8-ANEPPS+ gate is also shown for each sample. These metrics can be exported to a spreadsheet to calculate the total number of EVs. Please click here to view this table (Right click to download).
Discussion
A fundamental challenge of the EV field is separating the wide variety of EV subtypes2. The methods described here use filtration, differential centrifugation, and size-exclusion chromatography to generate a pure population of small EVs because ~100 nm EVs were previously shown to be secreted into the external environment and function in physiologically relevant communication pathways16. EVs purified through size-exclusion chromatography may still be a mixture of exosomes and small microvesicles because these EV subclasses are similarly sized. Vertebrate EV subclasses are commonly separated by immunoprecipitation because this method isolates EVs through binding to selective membrane protein markers. The methods described facilitate the identification of C. elegans EV membrane marker proteins. Therefore, in the future it may be possible to further separate small EV subclasses through analogous immunoprecipitation methods. In theory, immunoprecipitation could isolate EVs from well-fed worms because E. coli EVs should not interact with the antibody. Researchers interested in identifying the protein and genetic cargos of larger EV subclasses (>200 nm) can do so by skipping the 0.22 μm filtration step and then analyzing the pellet rather than the supernatant. Uncovering the protein and genetic cargo abundances of large EVs will establish a more comprehensive understanding of the physiological processes that function through the C. elegans secretome. In addition to being secreted into the environment, C. elegans EVs are transferred internally between tissues. Therefore, these methods only isolate a subset of total C. elegans EVs. However, cargo discovery of internal EVs is not possible because there is no method for separating EVs from lysed worms. Cargo analysis of this externally secreted EV subset provides a means to identify potential general EV protein markers capable of labeling internal EVs as well.
The scale of the EV preparation will depend on the demands of the experiment. A total of 500,000 young adults provides enough EVs for conducting flow cytometry, LC-MS-MS, and RNAseq analysis in parallel. This number can be scaled up or down depending on the experimental needs. The largest practical factor for scaling up is the number of 10 cm high growth plates required. High growth plates prepared with 500 μL of 20x concentrated overnight NA22 culture will support a population of ~50,000 adults from the L1 larval stage until the first day of adulthood. Therefore, to conduct an experiment with 500,000 adults, 13 high growth plates are needed: one plate to generate the population of L1s, two plates to generate the adults for bleaching, and 10 plates for the growth of the experimental animals. These metrics are contingent on the seeding and bacterial growth conditions of the high growth plates. Therefore, it is recommended to make all plates in a standardized way and then calibrate with known quantities of animals.
Worms are counted at two stages during the cultivation process: 1) before seeding worms on plates and 2) before generating the conditioned media. Animal cultivation density has been shown to impact behavior, development, and stress responses15,16,17,18 and may, therefore, also influence EV cargos. Therefore, for consistent cultivation density it is necessary to accurately estimate worm populations. Quantifying the number of worms in nine drops gives statistical confidence of population estimations. It is essential to treat each drop individually, vortexing between each drop and inserting the pipette the same distance into the worm suspension. Taking these precautions will ensure S.E.M. values of ~5–10% of the total population. If the S.E.M. for a worm population is higher than 20%, then something went awry.
After the 24 h bacteria-free incubation period, young, wild type C. elegans crawl immediately upon addition to a bacterial lawn. This indicates that the incubation does not severely impact their health. However, this step does influence the physiology of the animals and therefore may influence the composition of the EV cargos. Therefore, when working with very sick genotypes or older animals, it is essential to check viability after the incubation step. If all animals do not survive the incubation, then increase the experimental population size and decrease the time of incubation. When working with large volumes of conditioned media, it is more practical to use a stirred-cell concentrator with a filter made of regenerated nitrocellulose. EVs do not bind to this filter chemistry as strongly as other filter types14.
The ratio of the total protein recovered from the conditioned media to the number of animals incubated to make the preparation is a useful metric. If this ratio is much higher than expected, then it is possible that the population estimations were off or that animals died and deteriorated in the conditioned media. If this ratio is much lower than expected, then some protein may have been lost on the filters during the concentration process. While the FACS methods will directly quantify the EVs in the preparation, this metric is useful because it can highlight sample anomalies early in the process. Another useful method for verifying the quality of the EV preparation is transmission electron microscopy, as shown in Figure 4D. Although the protocol for transmission electron microscopy is not formally included in this methods article due to length, it can be conducted by standard methods. It is recommended to conduct this type of analysis, at least initially, as a complimentary method to FACS for assessing the quality of EV preparations. For best results use freshly discharged formvar-carbon grids and stain the samples with 2% phosphotungstic acid instead of uranyl acetate.
DI-8-ANEPPS was chosen because it has been shown to quantitatively label EV membranes, outperforming other general EV dyes with human biopsy samples and liposomes25. The measurements are quick, taking only 3 min of collection time for each sample. The quantification of detergent-sensitive EVs has direct functional significance because it capitalizes on a fundamental physical distinction between EVs and solid lipoprotein aggregates. Importantly, this method is not influenced by the amount of total protein or number of non-EV aggregates and therefore can quantify EVs obtained through different purification methods in an unbiased manner. The FACS-verified EV metric we describe here would be especially helpful for studies that demonstrate physiological impacts of purified EVs. It could also benefit the EV field in general to establish a FACS methodology as a universal metric so that absolute EV abundances can be directly compared across studies. Vital dyes can also label C. elegans EVs, but they are not as bright as EVs labeled with Di-8-ANEPPS.
This purification approach capitalizes on the active secretion of EVs outside the bodies of intact, living worms. This permits C. elegans EVs to be isolated at comparable purity and abundance as from cell culture, specifications that are sufficient for identifying hundreds of protein and RNA cargos via LC-MS-MS and RNAseq analysis26. Thus, the large library of reagents available for C. elegans research can be leveraged for investigating the influence of the genetic and physiological perturbation on EV cargo composition.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We gratefully acknowledge Nick Terzopoulos for worm plates and reagents; the Caenorhabditis Genetics Center (CGC) at the NIH Office of Research Infrastructure Programs (P40 OD010440) for the N2 nematode line; Lucia Vojtech, PhD, for assistance with the nanoparticle tracking analysis; Jessica Young, PhD, and Marie Claire, MD, PhD, for hiPSC-derived neuronal conditioned cell media; Wai Pang for assistance with TEM imaging. This work was supported by NIH grant P30AG013280 to MK and NIH grant AG054098 to JCR.
Materials
| 0.22 filters units | Genesee | 25-233 | For clairifying the conditioned media from debris |
| 1% solution of Triton X-100 | ThermoFisher | HFH10 | Add this to 0.05 % to lyse EVs |
| 10 cm high growth plates | N/A | N/A | For cultivating large populations of worms |
| 2-liter bottom baffled flasks | ThermoFisher | 4110-2000PK | For conducting size exclusion separation of EVs |
| 40 μm mesh | Amazon | CMY-0040-C/5PK-05 | Use this to separate adult worms from larval stages |
| 5 μm mesh | Amazon | CMN-0005-C | Use this to separate L1s from other worm stages |
| 6 cm normal growth medium worm cultivation plate | N/A | N/A | For cultivating small populations of worms |
| Amicon Ultra-15 Centrifugal Filter 10 kD mwco | MilliporeSigma | C7715 | For concentrating the size exclusion elute |
| Amicon Ultra-4 Centrifugal Filter 10 kD mwco | MilliporeSigma | C78144 | For concentrating the conditioned media |
| Apogee A50 flow cytometer | Apogee | This flow cytometer is specialized for resolving nanometer size particles | |
| ApogeeMix | Apogee | 1493 | Assess light scatter and fluorescence performance of FACS |
| DI-8-ANEPPS | ThermoFisher | D3167 | Add this at 20 uM to label EVs |
| Disposable chromatography Column | BioRad | 7321010 | For conducting size exclusion separation of EVs |
| Geletin | MilliporeSigma | G9391-100G | To treat pipette tips so that worms do not stick |
| HALT protease inhibitor | ThermoFisher | 87785 | Add to EV sample to prevent protein degradation |
| Low-protein binding collection tubes | ThermoFisher | 90410 | Use these for EV samples |
| NaCl | Sigma | S7653-1KG | For sucrose floatation of worms |
| S Basal buffer | N/A | N/A | Recipe in WormBook |
| Sepharose CL-2B resin | MilliporeSigma | CL2B300-100ML | For conducting size exclusion separation of EVs |
| Small orbital shaker | ABC Scientific | 83211301 | Use this for worm S Basal incubation |
| Sucrose | Sigma | S8501-5KG | For sucrose floatation of worms |
References
- Maas, S. L. N., Breakefield, X. O., Weaver, A. M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends in Cell Biology. 27, 172-188 (2017).
- Raposo, G., Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of Cell Biology. 200, 373-383 (2013).
- Lakhter, A. J., Sims, E. K. Minireview: Emerging Roles for Extracellular Vesicles in Diabetes and Related Metabolic Disorders. Molecular Endocrinology. 29, 1535-1548 (2015).
- Anderson, H. C., Mulhall, D., Garimella, R. Role of extracellular membrane vesicles in the pathogenesis of various diseases, including cancer, renal diseases, atherosclerosis, and arthritis. Laboratory Investigation; a Journal of Technical Methods and Pathology. 90, 1549-1557 (2010).
- Lai, C. P. -. K., Breakefield, X. O. Role of exosomes/microvesicles in the nervous system and use in emerging therapies. Frontiers in Physiology. 3, 228 (2012).
- Duijvesz, D., et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 8, 82589 (2013).
- Zhou, H., et al. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney International. 70, 1847-1857 (2006).
- Cheng, L., Sharples, R. A., Scicluna, B. J., Hill, A. F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. Journal of Extracellular Vesicles. 3, (2014).
- Michael, A., et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Diseases. 16, 34-38 (2010).
- Wehman, A. M., Poggioli, C., Schweinsberg, P., Grant, B. D., Nance, J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Current Biology. 21, 1951-1959 (2011).
- Naik, J., et al. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS One. 14, 0213069 (2019).
- Liégeois, S., Benedetto, A., Garnier, J. -. M., Schwab, Y., Labouesse, M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. Journal of Cell Biology. 173, 949-961 (2006).
- Simon, E., Aguirre-Tamaral, A., Aguilar, G., Guerrero, I. Perspectives on Intra- and Intercellular Trafficking of Hedgehog for Tissue Patterning. Journal of Developmental Biology. 4, (2016).
- Qi, J., et al. Exosomes Derived from Human Bone Marrow Mesenchymal Stem Cells Promote Tumor Growth Through Hedgehog Signaling Pathway. Cellular Physiology and Biochemistry. 42, 2242-2254 (2017).
- Sigg, M. A., et al. Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways. Developmental Cell. 43, 744-762 (2017).
- Wang, J., et al. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Current Biology. 24, 519-525 (2014).
- Beer, K. B., Wehman, A. M. Mechanisms and functions of extracellular vesicle release in vivo-What we can learn from flies and worms. Cell Adhesion and Migration. 11, 135-150 (2017).
- Deatherage, B. L., Cookson, B. T. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infection and Immunity. 80, 1948-1957 (2012).
- Stiernagle, T. Maintenance of C. elegans. C. elegans. 2, 51-67 (1999).
- Kibria, G., et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Scientific Reports. 6, 36502 (2016).
- Rose, J. A., et al. Flow cytometric quantification of peripheral blood cell β-adrenergic receptor density and urinary endothelial cell-derived microparticles in pulmonary arterial hypertension. PLoS One. 11, 0156940 (2016).
- Lobb, R. J., et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. Journal of Extracellular Vesicles. 4, 27031 (2015).
- Gardiner, C., Ferreira, Y. J., Dragovic, R. A., Redman, C. W. G., Sargent, I. L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of Extracellular Vesicles. 2, (2013).
- Fernandez, A. G., et al. High-throughput fluorescence-based isolation of live C. elegans larvae. Nature Protocols. 7, 1502-1510 (2012).
- de Rond, L., et al. Comparison of Generic Fluorescent Markers for Detection of Extracellular Vesicles by Flow Cytometry. Clinical Chemistry. 64 (4), 680-689 (2018).
- Russell, J. C., et al. Isolation and characterization of extracellular vesicles from Caenorhabditis elegans for multi-omic analysis [Internet]. bioRxiv. , (2018).

