A New Toolkit for Evaluating Gene Functions using Conditional Cas9 Stabilization
Summary
Here, we describe a genome-editing tool based on the temporal and conditional stabilization of clustered regularly interspaced short palindromic repeat- (CRISPR-) associated protein 9 (Cas9) under the small molecule, Shield-1. The method can be used for cultured cells and animal models.
Abstract
The clustered regularly interspaced short palindromic repeat- (CRISPR-) associated protein 9 (CRISPR/Cas9) technology has become a prevalent laboratory tool to introduce accurate and targeted modifications in the genome. Its enormous popularity and rapid spread are attributed to its easy use and accuracy compared to its predecessors. Yet, the constitutive activation of the system has limited applications. In this paper, we describe a new method that allows temporal control of CRISPR/Cas9 activity based on conditional stabilization of the Cas9 protein. Fusing an engineered mutant of the rapamycin-binding protein FKBP12 to Cas9 (DD-Cas9) enables the rapid degradation of Cas9 that in turn can be stabilized by the presence of an FKBP12 synthetic ligand (Shield-1). Unlike other inducible methods, this system can be adapted easily to generate bi-cistronic systems to co-express DD-Cas9 with another gene of interest, without conditional regulation of the second gene. This method enables the generation of traceable systems as well as the parallel, independent manipulation of alleles targeted by Cas9 nuclease. The platform of this method can be used for the systematic identification and characterization of essential genes and the interrogation of the functional interactions of genes in in vitro and in vivo settings.
Introduction
CRISPR-Cas9 which stands for "clustered regularly interspaced short palindromic repeats-associated protein 9" was first discovered as part of studies on bacterial adaptive immunity1,2. Today, CRISPR/Cas9 has become the most recognized tool for programmable gene editing and different iterations of the system have been developed to allow transcriptional and epigenetic modulations3. This technology enables the highly precise genetic manipulation of almost any sequence of DNA4.
The essential components of any CRISPR gene editing are a customizable guide RNA sequence and the Cas9 nuclease5. The RNA guide binds to the target-complementary sequence in the DNA, directing the Cas9 nuclease to perform a double-strand break at a specific point in the genome3,4. The resulting cleavage site is then repaired by non-homologous end-joining (NHEJ) or homology-directed repair (HDR), with the consequent introduction of changes in the targeted DNA sequence5.
CRISPR/Cas9 based gene editing is easy to use, and relatively inexpensive compared to previous gene-editing techniques and it has been proven to be both efficient and robust in a multiplicity of systems2,4,5. Yet, the system presents some limitations. The constitutive expression of Cas9 has often been shown to result in an increased number of off-targets and high cell toxicity4,6,7,8. Additionally, the constitutive targeting of essential and cell survival genes by Cas9 takes away from its ability to perform certain types of functional studies such as kinetic studies of cell death7.
Different inducible or conditionally controlled CRISPR-Cas9 tools have been developed to address those issues6, such as Tet-ON and Tet-Off9; site-specific recombination10; chemically-induced proximity11; intein dependent splicing3; and 4-Hydroxytamoxifen Estrogen Receptor (ER) based nuclear localization systems12. In general, most of these procedures (intein splicing and chemically induced proximity split systems) do not offer reversible control, present a very slow kinetic response to drug treatment (Tet-On/Off system), or are not amenable to high-throughput manipulation6.
To address these limitations we developed a novel toolkit that not only provides fast and robust temporal-controlled gene editing but also ensures traceability, tunability, and amenability to high throughput gene manipulation. This novel technology can be used in cell lines, organoids, and animal models. Our system is based on an engineered domain, when fused to Cas9, it induces its rapid degradation. However, it can be rapidly stabilized with a highly selective, non-toxic, cell-permeable small molecule. More specifically, we engineered the human FKBP12 mutant "destabilizing domain" (DD) to Cas9, marking Cas9 for rapid and constitutive degradation via the ubiquitin-proteasome system when expressed in mammalian cells13. The DD synthetic ligand, Shield-1 can stabilize DD conformation, thereby preventing the degradation of proteins fused to DD (such as Cas9) in a very efficient manner, and with a fast kinetic response14,15. Of note, Shield-1 binds with three orders of magnitude tighter to the mutant FKBP12 than to its wild-type counterpart14.
The DD-Cas9/Shield-1 pair can be used to study the systematic identification and characterization of essential genes in cultured cells and animal models as we previously showed by conditionally targeting the CypD gene, which plays an important role in the metabolism of mitochondria; EGFR, a key player in oncogenic transformation; and Tp53, a central gene in DNA damage response. In addition to temporally and conditionally controlled gene editing, another advantage of the method is that the stabilization of DD-Cas9 is independent of its transcription. This feature enables co-expression, under the same promoter, of traceable markers as well as recombinases, such as the estrogen receptor-dependent recombinase, CREER. In this work, we show how our method can be successfully used in vitro, to conditionally target for example, DNA replication gene, RPA3.
Protocol
1.The DD-Cas9 vector
- Obtain DD-Cas9 vector from Addgene (DD-Cas9 with filler sequence and Venus (EDCPV), Plasmid 90085).
NOTE: This is a lentiviral DD-Cas9 plasmid with a U6 promoter that drives the single guide RNA (sgRNA) transcription while the EFS promoter drives the DD-Cas9 transcription. The DYKDDDDK sequence (flag-tag) is present at the C-terminal of Cas9 followed by 2A self-cleaving peptide (P2A) that separates DD-Cas9 and modified fluorescent protein Venus (mVenus).
2. Small guide RNA (sgRNA) design
- Design small guide RNA (sgRNA) that will be cloned into the DD-Cas9 vector by using one of several algorithms, targeting a specific genomic region.
NOTE: To reduce the off-target effects, select sgRNA sequences with the highest score by using the website: http://crispr.mit.edu. - Design and order two oligonucleotides per sgRNA sequence to make a complete sgRNA.
NOTE: Since the plasmid will be digested by the BsmBI enzyme, design the forward and the reverse oligonucleotide by adding BsmBI digestion overhangs to the sgRNA sequence. Use the template from Table 1.
3. Cloning of sgRNA into the lentiviral DD-Cas9 vector
- Dephosphorylate the 5′-ends of DD-Cas9 plasmid by using alkaline phosphatase (FastAP) in a restriction enzyme digestion reaction.
NOTE: Vectors can re-circularize during the ligation especially when they are linearized by a single restriction enzyme. To ensure the vector will not re-circularize, de-phosphorylate the plasmid by adding phosphatase directly into the digestion reaction.- Dephosphorylate and digest the plasmid with BsmBI enzyme by preparing a mix of 5 µg of DD-Cas9 plasmid, 6 µL of digest buffer (10x), 0.6 µL of DTT (100 mM), 3 µL of BsmBI, 3 µL of FastAP, add up to 60 µL nuclease-free water to make 60 µL the total reaction volume.
NOTE: Add the BsmBI enzyme and FastAP to the mixture in the end. - Incubate the mixture for 30 min at 37 °C heat block or thermocycler.
- Dephosphorylate and digest the plasmid with BsmBI enzyme by preparing a mix of 5 µg of DD-Cas9 plasmid, 6 µL of digest buffer (10x), 0.6 µL of DTT (100 mM), 3 µL of BsmBI, 3 µL of FastAP, add up to 60 µL nuclease-free water to make 60 µL the total reaction volume.
- Purify the digested DD-Cas9 plasmid.
- Prepare an 0.8% DNA agarose gel to remove incompletely digested plasmid and traces of enzymes. Use a wider gel-comb for better separation and run the gel at 90 V.
- Visualize bands under 360-365 nm UV light and cut the larger plasmid band out of the gel. Discard the 2 kb fragment which is corresponding to the filler.
- Purify the large cut gel-band by using a commercial gel purification kit and follow the manufacturer's instructions.
- Ligate annealed oligos with digested DD-Cas9 plasmid.
- Phosphorylate and anneal the sgRNA oligos.
- Prepare the mix of 1 µL of T4 Ligation Buffer (10x), 1 µL of Oligo 1 (100 µM), 1 µL of Oligo 2 (100 µM), 6.5 µL of nuclease-free water. Lastly, add 0.5 µL of T4 PNK. The total volume of this reaction is 10 µL.
- Add the reaction-mix to the thermocycler with the following conditions: 37 °C for 30 min, 95 °C for 5 min, and then ramp down to 25 °C with the speed of 5 °C/min.
NOTE: The PNK must be heat-inactivated before the oligonucleotides are put into the ligation otherwise the PNK will phosphorylate the vector. The phosphorylation step and annealing step are performed together in a thermocycler. The phosphorylation step where 5' phosphate is added to the sgRNA oligonucleotides is required for the ligation to occur.
- Ligate DD-Cas9 digested plasmid and sgRNA oligos.
- Dilute the annealed oligos to 1:200 in nuclease-free water and use 50 ng of plasmid DNA in the ligation reaction.
NOTE: Use the molar ratio of the 6 insert : 1 vector, to promote insert integration or use a ligation calculator to calculate the molar ratio: http://nebiocalculator.neb.com/#!/ligation. - Prepare the ligation reaction mix: 50 ng of digested and purified DD-Cas9 vector, appropriate volume of diluted annealed oligos, 1 µL of T4 Ligation Buffer (10x), 1 µL of T4 DNA Ligase, up to 10 µL nuclease-free water to obtain the total volume of the reaction 10 µL.
- If using "high concentration" ligase, incubate the reaction at room temperature for 5 min. Otherwise, incubate at room temperature for 2 h, or to achieve a higher yield of ligation, incubate at 16 °C overnight.
NOTE: Heat-inactivate if using the standard T4 DNA Ligase at 65 °C for 20 minutes. Do not heat-inactivate if you are using ligase master mixes.
- Dilute the annealed oligos to 1:200 in nuclease-free water and use 50 ng of plasmid DNA in the ligation reaction.
- Phosphorylate and anneal the sgRNA oligos.
4. Bacterial transformation
- Pre-warm 10 cm LB-ampicillin agar plates at room temperature.
- Thaw 50 µL of "One shot Stbl3" competent bacterial cells on ice.
- Add 2-3 µL of ligation mixture to 50 µL of competent bacterial cells and gently mix by flicking the bottom of the tube with a finger 4 times. Incubate on ice for 30 min.
- Heat-shock the cells at exactly 42 °C for 40 seconds, using a timer. Cool down the bacterial cells on ice for 5 min.
- Add 500 µL of SOC media without antibiotic to the bacterial cells and grow them in the shaking incubator at 250 rpm for 45 min, at 37 °C.
- Spread 100 µL of transformed bacteria on pre-warm LB-ampicillin plates and incubate overnight at 37 °C.
5. Mini/maxi-prep of ligated plasmid
- Prepare a maxi/mini-prep starter culture.
- The next day pick a single bacterial clone and inoculate a starter culture with 8 mL of LB media containing ampicillin.
- Grow the bacteria in the shaking incubator at 250 rpm, 37 °C for 10-12 h.
- Use 3-5 mL of bacteria for mini-prep or use it as a starter culture for maxi-prep. Use the rest of the bacteria as a bacterial glycerol-stock by adding 100% glycerol in the ratio of bacterial culture:glycerol is 1:1.
NOTE: Here, we will describe the mini-prep procedure.
- Mini-prep procedure
- Harvest 3-5 mL of bacterial cells and centrifuge at 12,000 x g for 1 min.
- Resuspend the pellet of bacterial cells with 200 µL of P1 buffer, containing RNase A and stored at 4 °C.
- Add 200 µL of buffer P2 and mix gently by inverting the tube 10 times until the liquid clarifies.
- Add 300 µL of buffer P3 and mix by inverting the tube 10 times. The liquid will become cloudy. Proceed immediately with centrifuging the samples at 12,000 x g for 10 min.
- Transfer clear supernatant to the spin column and centrifuge at 12,000 x g for 1 min.
Discard the flow-through.
NOTE: Make sure the supernatant is clarified. Particles can clog the spin column and reduce the column efficacy. - Add 400 µL of PD buffer and centrifuge at 12,000 x g for 1 min. Discard the flow-through.
- Add 600 µL of PW buffer to wash the spin column and centrifuge at 12,000 x g for 1 min. Discard the flow-through.
- Centrifuge again the spin column at top speed for 3 min to remove the buffer residues. Discard the flow-through and let it sit for 2 min.
- Place the spin column to a fresh tube and add 50 µL of elution buffer. Incubate for 5 min and centrifuge at top speed for 2 min.
- Use the nanodrop device to measure the concentration of purified DD-Cas9 plasmid with the sgRNA insert (ng/µL).
- Validate the sgRNA cloning by DNA sequencing of the DD-Cas9 plasmid. Use the U6 primer sequence in Table 2.
6. Lentiviral preparation
- Prepare HEK293T packaging cells for transfection.
- Use healthy HEK293T up to passage 10 and seed them evenly at density 1-2 x 106 cells per 10 cm tissue culture plate. Use 10 mL of Glucose Dulbecco's Modified Eagle Medium (DMEM) with 10% filtered Fetal Bovine Serum (FBS). Incubate cells in the tissue culture incubator, at 5% CO2 and 37 °C for 20 h.
- Next day, confirm that cells reached 70% confluency by brightfield microscopy and that they are evenly dispersed across the plate. Change media to cells 1 h before the transfection with the final volume of 10 mL.
NOTE: The media should be absent of antibiotics and antimycotics. - Prepare the mixture of 2 tubes with 500 µL of warm media (e.g., OptiMEM).
- Add 25 µL of transfection reagent to tube 1 containing warm 500 µL of media, and incubate at room temperature for 5 min.
- Meanwhile prepare a mix of plasmids to tube 2 containing 3.5 µg of DD-Cas9 containing cloned sgRNA, 6 µg of the packaging plasmid (psPAX2), and 3 µg of the envelope plasmid (pMD2.G) into warm 500 µL of media.
- Mix tube 1 with tube 2 to form a transfection mixture and incubate at room temperature for 20 min.
- Dropwise the transfection mixture to HEK293T cells and incubate plate in the tissue culture incubator, at 5% CO2 and 37 °C overnight.
- After 18 h, carefully change the media to 10 mL of fresh DMEM with 10% FBS to remove the transfection reagent. Incubate for the next 48 h.
- After 48 h, collect the supernatant with a 10 mL syringe and pass it through a 0.45 µm filter.
- Aliquot and store the virus supernatant at -80 °C.
7. Determining virus titer and transduction efficacy with flow cytometry
- Seed the HEK293T cells with 5 x 105 cells/well into a 6-well plate. Seed cells into two 6-well plates. Use one plate for counting the next day.
- Incubate the plates in the incubator at 37 °C, 95% humidity, 5% CO2 overnight to reach 50-60% confluency the next day.
- The next day, use one of the 6-well plates to count the cells in 6 wells.
- Thaw the virus aliquot that will be used to perform the serial dilution and add 8 µg/mL of polybrene reagent.
- Prepare DMEM with 10% FBS for serial dilution of the virus and add 8 µg/mL of polybrene reagent.
- Prepare 2 mL of ten-fold serial dilutions of the lentivirus from 1 x 10-1 to 1 x 10-4 in polybrene-containing media.
- Remove media from 6-well plate and add 1 mL of viral dilutions to wells. Leave one well with media alone as negative control and one well with 100% virus.
- Incubate cells with the virus in the incubator for 24 h.
- The next day, remove the media with virus from 6-well plates and change it for 2 mL of fresh DMEM with 10% FBS. Incubate cells for 48-72 h. Observe GFP by using a fluorescent microscope every day.
- After 48-72 h, detach the cells and resuspend them in MACS buffer.
- Use a flow cytometer to determine the percentage of GFP expression.
- Calculate virus titer by using the following formula: TU/mL = (Number of cells transduced x Percent fluorescent x Dilution Factor)/(Transduction Volume in mL)
8. Lentiviral transduction of target cells
- Plate the cell line of interest to a 10 cm plate and incubate overnight to reach confluency 50-60% the next day.
NOTE: We used the A549 cell line expressing DD-Cas9 and two independent RPA3-gene sgRNAs. We also used the A549 cell line expressing vector with Renilla control. We seeded those cells at the density of 2 x 103cells/cm2 and incubated them at 37 °C, 95% humidity, 5% CO2 overnight to reach 50-60% confluency the next day. We used RPMI media with 10% filtered FBS. We proceeded with the next steps as follow: - The next day, infect the cells with 500-2000 µL of virus particles in 10 mL total volume of culture medium. Incubate viral media for 24 h.
- The next day, change the viral media to culture media and determine the percentage of GFP positive cells by using flow cytometry.
- Select GFP positive cells by FACS cell sorting or by using bleomycin selection.
- Expand positively selected cells and freeze a stock.
9. Conditional induction of Cas9 mediated gene editing
- Plate positively selected cells 24 h after infection as well as untransduced cells to 12-well plate separately. Wait until the cells attach and change the media to cell culture media containing 200 nM of Shield-1. Replace the media in the plates with transduced and untransduced-cells, leaving 2 wells per plate with regular media as a negative control.
- Incubate and extract proteins from each well at different time points: time 0, 2 h, 6 h, 12 h, 24 h, 48 h, and 72 h after adding the Shield-1 to the wells.
- Then remove the media with Shield-1 from the rest of the wells and change it for regular cell media.
- Collect the proteins from those wells 2 h, 6 h, and 12 h after changing the media.
- Visualize by western blot analysis the reversibility and rapidity of destabilized DD-Cas9 protein regulation after the addition and the withdrawal of its ligand, Shield-1. Use antibody directed towards DYKDDDDK Tag to visualize proteins and a control antibody targeting for example beta-tubulin.
- Determine the optimal dose of Shield-1 by dose-response curve observed by Western Blot analysis.
10. Validation of gene editing
NOTE: The GFP expression assays, such as flow cytometry analysis and bleomycin selection marker only confirm successful CRISPR reagent delivery but they do not determine if the desired sequence was successfully targeted. The most common assays to confirm successful gene targeting by the CRISPR experiment are Sanger DNA sequencing, Next-generation sequencing, the Surveyor Nuclease Assay, the Tracking of Indels by Decomposition (TIDE) Assay, or western blot analysis16,17,18.
- Plate positively selected cells and change media to 200 nM of Shield-1 containing media. Incubate cells for 5 days, changing media with Shield-1 every 3 days.
- Use the above-mentioned validation techniques 5 days after DD-Cas9 induction by Shield-1.
Representative Results
To enable the conditional expression of Cas9, we developed a dual lentiviral vector construct consisting of a U6-driven promoter to constitutively express sgRNA, and an EF-1α core promoter to drive the expression of the DD-Cas9 fusion protein (Figure 1A)19. As a paradigm to illustrate the robustness and efficiency of the system, we transduced the lung carcinomatous A549 cell line with the lentiviral construct. The levels of Cas9 in the presence or absence of the ligand Shield-1 were measured by reverse transcription-polymerase chain reaction (RT-PCR) and Western blot analysis using an anti-Flag specific antibody. We used uninfected cells and mock-infected cells (the vehicle) as controls. The cells were treated with a concentration of Shield-1 ranging from 10 to 1000 nM for 7 d. As shown, the treatment with Shield-1 was able to regulate the expression of DD-Cas9 in a strong dose-dependent manner (Figure 1B and Figure 2A). RT-PCR analysis with specific primers for DD-Cas9 and control primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) confirmed that levels of mRNA expression of DD-Cas9 were similar amongst the transduced cells and the vehicle notwithstanding the presence or absence of Shield-1 (Figure 2B). This confirms that the induction of Cas9 protein is a post-transcriptional event.
This system enables fast and reversible stabilization of the DD-Cas9 protein. Figure 3 shows sufficient induction of Cas9 expression in the transduced A549 cell line 2 h after treatment with 200 nM of Shield-1 compared to the uninfected A549 cells. However, withdrawal of Shield-1 results in a rapid decrease of the DD-Cas9 protein, which becomes negligible within 6 to 12 h (Figure 3).
The RPA3 protein is a component of the human replication protein A (RPA) heterotrimer. It is a single-stranded DNA binding complex that plays an important role in DNA replication, recombination, and repair. To validate the use of the system to study essential genes, we targeted the RPA3 gene. To this end, we used two independent locus-specific single guide RNAs (guides 25 and 44) as well as Renilla as a control (guide 208). The A549 cells transduced with the DD-Cas9 lentiviral construct were treated with 200 nM of Shield-1 for 3 d. A decrease in cell number in Shield-1-treated transduced A549 cells containing the RPA3 guide RNA was apparent after 48 h of treatment, and no effect was observed on the cell number in the Renilla sample (Figure 4A). To validate the depletion of the RPA3 protein, we performed an immunoblotting analysis using an antibody against RPA3 3 d after Sheild-1 induction (Figure 4B). Furthermore, we confirmed gene editing inversion or indel mutations using Surveyor nuclease assays and DNA sequencing (Figure 4C).
The lentiviral vector construct we designed bears a unique feature: the regulation of DD-Cas9 protein stability is independent of its mRNA expression. This enables the generation of bicistronic systems to express another gene of interest under the same EF-1a promoter without being modulated by the destabilized DD-Cas9 (Figure 5A,5B). We also added a 2A self-cleaving peptide (P2A) between DD-Cas9 and mVenus, a modified fluorescent protein that can be used to trace infected cells (Figure 5A). As mVenus is placed after P2A, as shown by the results of the Western Blot analysis in Figure 5B, the expression of the mVenus protein was observed in the vehicle and A549 transduced cells independent of DD-Cas9 expression and Shield-1 treatment.
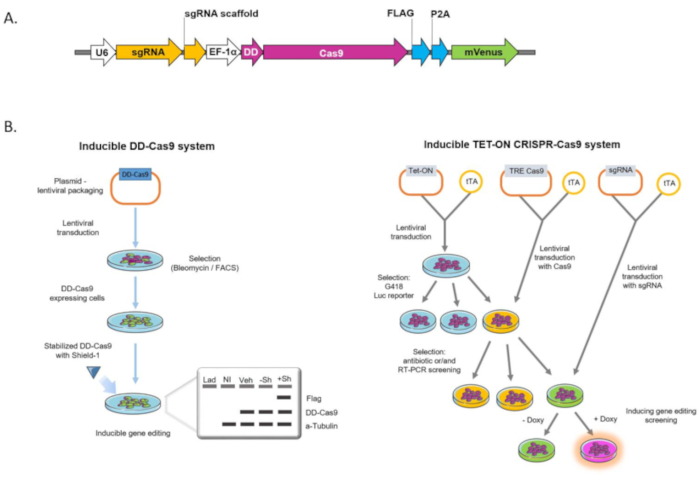
Figure 1: Schematic of the lentiviral construct and different gene-editing tools. A) The DD-Cas9 lentiviral backbone contains a U6 promoter, sgRNA, EF-1a promoter, DD, spCas9, nucleoplasmin NLS, and Flag-tag. B) Comparison between DD-Cas9 system (left panel) and different Tet-On system (right panel) used as gene-editing tools. Lad-ladder, NI-non-infected cells, Veh-vehicle, -Sh cells treated without Shield-1, + Sh cells treated with 200 nM Shield, – Doxy cells without doxycycline treatment, + Doxy cells treated with doxycycline. Please click here to view a larger version of this figure.
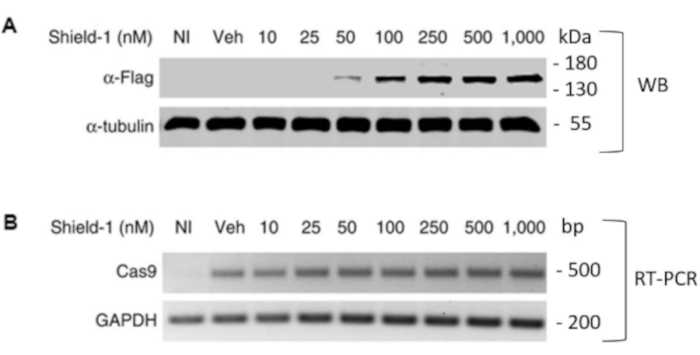
Figure 2: Representative results of dose-dependent DD-Cas9 stabilization by Shield-1. A) Western Blot analysis using an anti-Flag-tag antibody of stabilized DD-Cas9 expression in cells treated with increasing concentrations of Shield-1. As a control, the uninfected cells (NI) and mock-treated cells (Veh) were used. The fusion protein DD-Cas9 was undetectable in both controls. B) The RT-PCR results of mRNA expression levels of DD-Cas9 in transduced cells in the absence or dose-dependent treatments of Shield-1 were similar amongst transduced cells and vehicle, using GAPDH primers as an internal control. This figure has been modified from Serif et al19. Please click here to view a larger version of this figure.
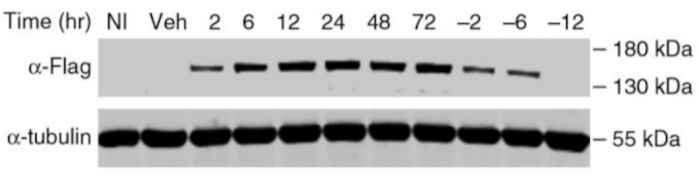
Figure 3: The Western Blot analysis illustrates the reversibility and rapidity of destabilized DD-Cas9 protein regulation after the withdrawal of its ligand Shield-1. Transduced A549 cell line with DD-Cas9 and uninfected A549 as control were treated 24 h after infection with 200 nM of Shield-1 ligand for the indicated time points. The protein level of DD-Cas9 in mock control cells was undetectable. This figure has been modified from Serif et al19. Please click here to view a larger version of this figure.
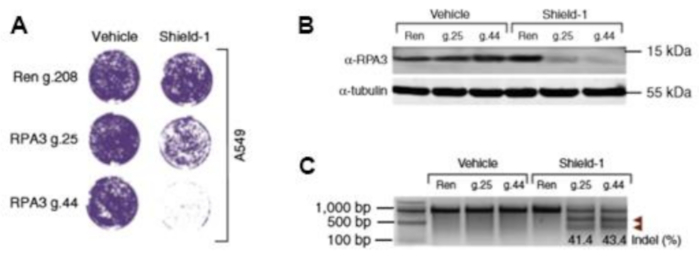
Figure 4: The DD-Cas9 system can induce robust gene editing in "in-vitro" and "in-vivo" settings. A) The cell line A549 transduced with a vector expressing sgRNA for RPA3 gene and DD-Cas9 (RPA3). As a control A549 transduced with a vector expressing sgRNA for Renilla (Ren) was used. Cells were treated with Shield-1 (200 nM) for 3 days which resulted in a rapid decrease in cell viability in cells expressing RPA3 sgRNA, with no effect on cell number in the Renilla control sample. The efficiency of RPA3 gene editing in the A549 cell line was validated by B) Western Blot analysis using the antibody against Flag-tag in RPA3 A549 and Renilla A549 in the presence and absence of 3 days Shield-1 (200 nM) treatment and C) by SURVEYOR nuclease assay. The arrows in panel C) show the fragments of the SURVEYOR nuclease assay. This figure has been modified from Serif et al19. Please click here to view a larger version of this figure.
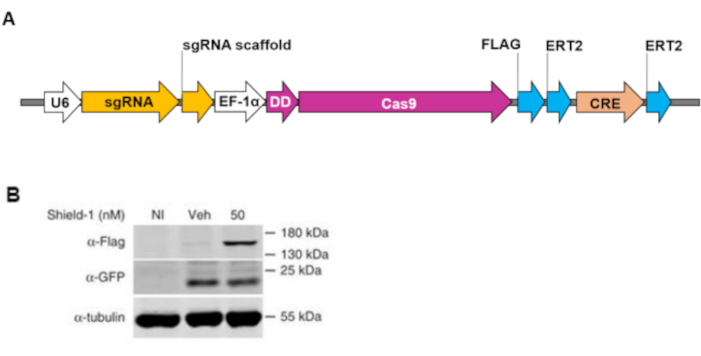
Figure 5: Scheme of a bicistronic DD-Cas9 lentiviral construct to drive the expression of mVenus independently of DD-Cas9. A) The construct consists of U6 promoter, sgRNA, EF-1a promoter, DD-Cas9, P2A, and mVenus. B) Transduced A549 cells with a lentiviral plasmid containing DD-Cas9, P2A, and mVenus were treated with 50 mM ligand Shield-1 for 3 days. On the third day, the western blot analysis was performed with cell lysate, using the antibody against GFP and Flag-tag separately. Figure 5B has been modified from Serif et al19. Please click here to view a larger version of this figure.
| Forward (Oligo 1) | 5’-CACCGNNNNNNNNNNNNNNNNNNNN-3’ |
| Reverse (Oligo 2) | 5’-AAACNNNNNNNNNNNNNNNNNNNNC-3’ |
Table 1:The design of the forward and reverse sgRNA oligonucleotide. The forward (Oligo 1) and the reverse (Oligo 2) oligonucleotide is designed by adding BsmBI enzyme digestion overhangs the your sgRNA sequence. “N” denotes the different nucleotides present in your sgRNA sequence, the rest are overhangs for BsmBI digestion.
| U6 primer sequence | 5’-GACTATCATATGCTTACCGT-3’ |
Table 2: The U6 promoter sequence for sgRNA cloning validation. To validate how successful is the sgRNA cloning, use the U6 promoter sequence for DNA sequencing of the DD-Cas9 plasmid.
Discussion
The CRISPR/Cas9 technology has revolutionized the capability of functionally interrogate genomes2. However, the inactivation of genes often results in cell lethality, functional deficits, and developmental defects, limiting the utility of such approaches for studying gene functions7. Additionally, constitutive expression of Cas9 may result in toxicity and the generation of off-target effects6. Different approaches have been developed to temporally control CRISPR-Cas9 based genome editing technologies6. These systems are based either on the transcriptional control of Cas9 and/or sgRNA, or Cas9 post-transcriptional and post-translational activation21,22,23. As an alternative to these systems, we developed a novel toolkit based on the conditional destabilization of Cas9.
The critical step in this protocol is the design of at least two or three sgRNA for a specific gene to avoid off-target effects and facilitate efficient gene editing. A rate-limiting step is an efficient transformation of the cell line with the DD-Cas9 vector. The quality of lentivirus particles (high titers) relies on the HEK-293T cells. It is crucial to use a low passage number (up to passage 10) of the HEK-293T cells that were properly maintained (split when they reached 70% confluency, usually twice a week in a 1:6 ratio). As an alternative, the HEK-293 Lenti-X cell line, which was clonally selected to yield 30× higher viral titers than regular HEK-293T cells can be used24. Another crucial step is the optimization of the ratio of the DD-Cas9 plasmid, psPAX2 packaging plasmid, and pMD2.G envelope plasmid. In our experience, both the volume in which the transfection mix with lipofectamine is prepared, as well as the plasmid ratio, have a substantial effect on the transfection efficiency. We recommend following the steps in our protocol for the best results. The next crucial step for efficient gene editing is the optimization of Shield-1 concentrations. We recommend a final concentration of 200 nM but to achieve the best results, the concentration should be optimized to a specific transduced cell line. The DD/Shield-1 system has been successfully used in different cell cultures, germ cells, the protozoan Entamoeba histolytica, the flatworm Caenorhabditis elegans, transgenic xenografts, transgenic mice, and medaka25. One of the biggest limitations is the high cost of the Shield-1 molecule, especially when being used in in vivo settings. Additionally, it has previously been shown that the proteins that are targeted to certain cell compartments, such as the mitochondrial matrix or lumen of the endoplasmic reticulum can be stabilized or accumulated in the absence of Shield-1. This is because different proteins have different local protein quality control machinery26. While DD fusions in the cytoplasm or nucleus can be very efficiently degraded in mammalian cells and stabilized by Shield-1, to overcome the limitations mentioned above, we recommend using an alternative destabilizing domain derived from the bacterial dihydrofolate reductase27. This system uses trimethoprim as a binding molecule to stabilize the destabilization domain and is also a less expensive alternative to Shield-127.
Besides achieving the transcription of DD-Cas9 as its independent expression, the advantages of this method, compared to other inducible or conditionally controlled CRISPR-Cas9 gene editing tools, are temporally and conditionally controlled gene editing, less off-target effect, and lower cell toxicity4,6,7,8. The efficiency and simplicity of the Shield-1/DD-Cas9 method enable the generation of a variety of tools that can be easily adapted and utilized in a multiplicity of applications. The system can be easily used in in vivo settings as well, and it has been shown that Shield-1 can efficiently penetrate through the blood-brain barier19,25,28,29. Although this paper described the use of CRISPR-Cas9 technology for the characterization of essential cell genes, the same approach could be easily implemented for the identification of genes required for the survival or progression of tumors.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank previous members of our laboratory and scientist Serif Senturk for previous work. We thank Danilo Segovia for critically reading this manuscript. This study was possible and supported by Swim Across America and the National Cancer Institute Cancer Target Discovery and Development Center program.
Materials
| 100mM DTT | Thermosfisher | ||
| 10X FastDigest buffer | Thermosfisher | B64 | |
| 10X T4 Ligation Buffer | NEB | M0202S | |
| colorimetric BCA kit | Pierce | 23225 | |
| DMEM, high glucose, glutaMax | Thermo Fisher | 10566024 | |
| FastAP | Thermosfisher | EF0654 | |
| FastDigest BsmBI | Thermosfisher | FD0454 | |
| Flag [M2] mouse mAb | Sigma | F1804-50UG | |
| Genomic DNA extraction kit | Macherey Nagel | 740952.1 | |
| lipofectamine 2000 | Invitrogen | 11668019 | |
| Phusion High-Fidelity DNA Polymerase | NEB | M0530S | |
| oligonucleotides | Sigma Aldrich | ||
| pMD2.G | Addgene | 12259 | |
| polybrene | Sigma Aldrich | TR-1003-G | |
| psPAX2 | Addgene | 12260 | |
| QIAquick PCR & Gel Cleanup Kit | Qiagen | 28506 | |
| secondary antibodies | LICOR | ||
| Shield-1 | Cheminpharma | ||
| Stbl3 competent bacterial cells | Thermofisher | C737303 | |
| SURVEYOR Mutation Detection Kit | Transgenomic/IDT | ||
| T4 PNK | NEB | M0201S | |
| Taq DNA Polymerase | NEB | M0273S | |
| α-tubulin [DM1A] mouse mAb | Millipore | CP06-100UG |
References
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816-821 (2012).
- Al-Attar, S., Westra, E. R., van der Oost, J., Brouns, S. J. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biological Chemistry. 392, 277-289 (2011).
- Hsu, P. D., Lander, E. S., Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157, 1262-1278 (2014).
- Mali, P., et al. RNA-guided human genome engineering via Cas9. Science. 339, 823-826 (2013).
- Barrangou, R., et al. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Research. 43, 3407-3419 (2015).
- Zhang, J., Chen, L., Zhang, J., Wang, Y. Drug Inducible CRISPR/Cas Systems. Computational and Structural Biotechnology Journal. , (2019).
- Wade, M. High-Throughput Silencing Using the CRISPR-Cas9 System: A Review of the Benefits and Challenges. Journal of Biomolecular Screening. 20, 1027-1039 (2015).
- Egeler, E. L., Urner, L. M., Rakhit, R., Liu, C. W., Wandless, T. J. Ligand-switchable substrates for a ubiquitin-proteasome system. Journal of Biological Chemistry. 286, 31328-31336 (2011).
- Cao, J., et al. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Research. 44, 149 (2016).
- Oakes, B. L., et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nature Biotechnology. 34, 646-651 (2016).
- Kamiyama, D., et al. Versatile protein tagging in cells with split fluorescent protein. Nature Communications. 7, 11046 (2016).
- Roper, J., et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nature Biotechnology. 35, 569 (2017).
- Chu, B. W., et al. Recent progress with FKBP-derived destabilizing domains. Bioorganic & Medicinal Chemistry Letters. 18 (22), 5941-5944 (2008).
- Banaszynski, L. A., Sellmyer, M. A., Contag, C. H., Wandless, T. J., Thorne, S. H. Chemical control of protein stability and function in living mice. Nature Medicine. 14, 1123-1127 (2008).
- Banaszynski, L. A., Chen, L. C., Maynard-Smith, L. A., Ooi, A. G., Wandless, T. J. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 126, 995-1004 (2006).
- Sentmanat, M. F., Peters, S. T., Florian, C. P., Connelly, J. P., Pruett-Miller, S. M. A Survey of Validation Strategies for CRISPR-Cas9 Editing. Scientific Reports. 8, 888 (2018).
- Hua, Y., Wang, C., Huang, J., Wang, K. A simple and efficient method for CRISPR/Cas9-induced mutant screening. Journal of Genetics and Genomics. 44 (4), 207-213 (2017).
- Guschin, D. Y., et al. A rapid and general assay for monitoring endogenous gene modification. Methods in Molecular Biology. 649, 247-256 (2010).
- Senturk, S., et al. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nature Communications. 8, 14370 (2017).
- McJunkin, K., et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proceedings of the National Academy of Sciences. 108, 7113 (2011).
- Davis, K. M., Pattanayak, V., Thompson, D. B., Zuris, J. A., Liu, D. R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nature Chemical Biology. 11, 316-318 (2015).
- Dow, L., et al. Inducible in vivo genome editing with CRISPR-Cas9. Nature Biotechnology. 33, 390-394 (2015).
- Wright, A. V., et al. Rational design of a split-Cas9 enzyme complex. Proceedings of the National Academy of Sciences of the United States of America. 112, 2984-2989 (2015).
- Albrecht, C., et al. Comparison of Lentiviral Packaging Mixes and Producer Cell Lines for RNAi Applications. Molecular Biotechnology. 57, 499-505 (2015).
- Froschauer, A., Kube, L., Kegler, A., Rieger, C., Gutzeit, H. O. Tunable Protein Stabilization In Vivo Mediated by Shield-1 in Transgenic Medaka. PLOS One. , (2015).
- Sellmyer, M. A., et al. Intracellular context affects levels of a chemically dependent destabilizing domain. PLOS One. 7 (9), 43297 (2012).
- Iwamoto, M., et al. A general chemical method to regulate protein stability in the mammalian central nervous system. Chemistry & Biology. 17 (9), 981-988 (2010).
- Tai, K., et al. Destabilizing domains mediate reversible transgene expression in the brain. PLOS One. 7 (9), 46269 (2012).
- Auffenberg, E., et al. Remote and reversible inhibition of neurons and circuits by small molecule induced potassium channel stabilization. Scientific Reports. 6, 19293 (2016).

