Stimulating and Analyzing Adult Neurogenesis in the Drosophila Central Brain
Summary
This article provides detailed protocols for inflicting Penetrating Traumatic Brain Injury (PTBI) to adult Drosophila and examining the resulting neurogenesis.
Abstract
The molecular and cellular mechanisms underlying neurogenesis in response to disease or injury are not well understood. However, understanding these mechanisms is crucial for developing neural regenerative therapies. Drosophila melanogaster is a leading model for studies of neural development but historically has not been exploited to investigate adult brain regeneration. This is primarily because the adult brain exhibits very low mitotic activity. Nonetheless, penetrating traumatic brain injury (PTBI) to the adult Drosophila central brain triggers the generation of new neurons and new glia. The powerful genetic tools available in Drosophila combined with the simple but rigorous injury protocol described here now make adult Drosophila brain a robust model for neural regeneration research. Provided here are detailed instructions for (1) penetrating injuries to the adult central brain and (2) dissection, immunohistochemistry, and imaging post-injury. These protocols yield highly reproducible results and will facilitate additional studies to dissect mechanisms underlying neural regeneration.
Introduction
Damage to the brain and nervous system is a major cause of death and disability worldwide. Approximately 1.5 million Americans suffer traumatic brain injuries (TBI) every year1, while an estimated 6 million individuals in the United States alone suffer from neurodegenerative diseases, such as Parkinson's and Alzheimer's Disease2. Both disease and injury to the brain can cause neural degeneration, leading to sensory, cognitive, and motor defects3. Developing therapeutic strategies for human brain repair has been difficult due to the complex physiology of the brain. Model organisms such as Drosophila melanogaster provide a simple system for identifying the fundamental mechanisms underlying neurodegeneration and potential therapeutic targets4.
The fruit fly Drosophila melanogaster has been a powerful model organism for more than a century, advancing the fields of genetics, developmental biology, and neuroscience5,6. The Drosophila brain comprises only ~90,000 neurons7, a million-fold fewer than the average human brain8, yet they have many similarities. Both human and fly brains utilize the neurotransmitters GABA, glutamate, acetylcholine, and the biogenic amines dopamine and serotonin9. Drosophila and human neurons also function similarly, with a shared synaptic architecture and analogous neural cell types10. The smaller brain size of Drosophila and the availability of advanced genetics techniques, in combination with the conservation of molecular, cellular, and physiological mechanisms between Drosophila and mammals, permits Drosophila researchers to ask questions that are impractical or difficult to answer in mammalian models.
Our current understanding of adult neurogenesis in Drosophila, both during homeostasis and following injury, remains limited. More is known about neurogenesis during normal development. For example, neurons and glia are created during development from precursor cells, called neuroblasts10,11. At least three different types of neuroblasts have been distinguished in the central brain. Both Type I and Type II lineage neuroblasts exit the cell cycle ~20-30 h after puparium formation12. In contrast, the mushroom body neuroblasts are the last to terminate cell division and do so via Reaper-dependent apoptosis ~85-90 h after puparium formation13. Following eclosion, the adult Drosophila brain has few dividing cells (~1 cell/brain), predominantly glia14. The adult optic lobes possess slowly cycling neuroblasts capable of neurogenesis15, while the adult central brain has no known neuroblasts. The scarcity of neural progenitors and limited cell proliferation strongly resembles the situation in the adult mammalian brain, underscoring the potential relevance of the mechanisms of adult neurogenesis in Drosophila to humans.
The discovery of low levels of adult neurogenesis in the adult Drosophila optic lobes after injury15 led to the hypothesis that the adult Drosophila central brain also might be capable of adult neurogenesis16. This protocol describes creating a rigorous, reproducible model of central brain injury in adult Drosophila that can be used to investigate neurogenesis in the adult central brain. Given the similarities between human and Drosophila brain architecture and function, these discoveries could lead to the identification of critical targets for therapeutic neurogenesis in injured and diseased human brains.
Protocol
This protocol follows the animal care guidelines of UW-Madison.
1. Generating adult Drosophila for PTBI
- For the standard cross, place 20 virgin y[1] w[1];UAS-mCD8-GFP;; OK107-GAL417 adult females and 10 y[1] w[1]17 adult male flies together in vials (see Table of Materials) containing food. To have large numbers of synchronous offspring, set up 10-20 crosses at the same time. The standard cross gives rise to F1 progeny of the genotype: y[1] w[1];UAS-mCD8-GFP/+;; OK107-GAL4/+.
- Place the vials at 25 °C for mating and egg-laying. To maximize progeny, transfer parents into new vials every 2-4 days, maintaining the earlier vials at 25 °C. Discard each vial 18 days after the parents were first placed in it to ensure that there is no F2 progeny.
NOTE: 3 sets of offspring ("broods") can be generated from each set of parents. - Check after ~10 days, when the F1 progeny will begin to eclose.
- Place the vials at 25 °C for mating and egg-laying. To maximize progeny, transfer parents into new vials every 2-4 days, maintaining the earlier vials at 25 °C. Discard each vial 18 days after the parents were first placed in it to ensure that there is no F2 progeny.
- For lineage studies, use F1 perma-twin males15 of genotype: w; FRT40A, UAS-CD2-RFP, UAS-GFP-Mir/FRT40A, UAS-CD8-GFP, UAS-CD2-Mir; tub-GAL80ts/act-GAL4 UAS-flp. For consistency, do this cross in the same direction each time.
- To generate perma-twin males, place 20 virgin w; FRT40A, UAS-CD2-RFP, UAS-GFP-Mir; tub-GAL80ts/TM6B females and 10 w; FRT40A, UAS-CD8-GFP, UAS-CD2-Mir; act-GAL4 UAS-flp/TM6B15 adult males together in vials containing food.
- Place the vials at 17 °C for mating and egg-laying. Transfer parents into vials of new food every 7 days, maintaining all vials at 17 °C. Discard each vial 35 days after the parents were first placed in it to ensure that there are no F2 progeny.
- Check after ~21 days, when the F1 progeny will begin to eclose.
2. Penetrating traumatic brain injury (PTBI; Figure 1)
- Sort newly eclosed F1 flies. Select young males within 6 h post-eclosion. Place these males in clean vials containing food, with 40 or fewer flies per vial.
NOTE: This is most easily accomplished in mid-morning by anesthetizing flies on the CO2 pad and identifying adult males that still have meconium (visible as a dark greenish spot through the abdominal wall) in their guts. - Pre-feed with 5-ethynyl-2'-deoxyuridine (EdU) (see Table of Materials) for 6 h before the injury if planning to label dividing cells with EdU. See step 3 for details.
- Sanitize Minutien pins (see Table of Materials) for at least 5 min by placing ~100 pins in a 1.5 mL microcentrifuge tube filled with 70% ethanol.
- Sanitize the CO2 pad and a paintbrush by spraying 70% ethanol and wipe dry with a clean lint-free tissue. Once the tools are clean and dry, transfer 40 or fewer sorted F1 males back onto the clean pad.
- Separate the F1 males into 2 groups on the fly pad. One group will serve as a control, uninjured flies. The second experimental group will be subjected to PTBI.
- Using forceps, pull 4-5 new Minutien pins out of the microcentrifuge tube and place them near the edge of the CO2 pad. Under the dissecting scope, choose a straight Minutien pin with a sharp point.
NOTE: Using a single Minutien pin for an experiment will reduce variability. Reuse sharp pins. Place damaged or blunt pins in a separate 1.5 mL microcentrifuge tube containing 70% ethanol for safe disposal. - If working with flies from the standard cross, switch on the stereo microscope light-emitting diode (LED) lamp equipped with the appropriate excitation and emission filters for green fluorescence protein (GFP). This permits excitation at 440-460 nM and allows visualization of 500-560 nM.
NOTE: With this lamp and filter set, the cell bodies of the mushroom body will fluoresce green through the head cuticle (Figure 1C). For perma-twin flies or flies of other genotypes, a standard white light illuminator for the stereo microscope can be used to visualize landmarks on the head cuticle for targeting the injury to the mushroom body (Figure 1C). - Use the forceps to pick up and hold the selected Minutien pin in one hand (for right-handed people, this is usually the right hand) and the paintbrush in the other (usually left) hand. Choose a fly from the experimental group and position the fly such that you have a dorsal view of the head capsule with the fly's head to the right. Place the brush on the anterior of the dorsal thorax and push down gently to stabilize the fly.
- Aim the Minutien pin's tip at the mushroom body's cell bodies on the right side of the head and penetrate the head capsule. If using landmarks, target the dorsal head cuticle between the ocelli and the dorsal rim of the eye (Figure 1).
- After completing the injury, use the paintbrush to push the head gently off the Minutien pin.
- If using the brain for RNA-Seq or qRT-PCR, make a second injury on the left side of the head.
- Repeat steps 2.8-2.10 to injure all of the flies in the experimental group.
- Once all flies have been injured, place the control and injured flies into labeled, separate vials containing food. Lay the vials horizontally (i.e., on their sides) while the flies recover from the anesthesia and during subsequent aging to prevent flies from getting caught in the food.
- Place standard cross flies at 25 °C, and perma-twin flies at 30 °C to age.
- For flies aged longer than 24 h, place them on clean food every 1-2 days.
3. EdU labeling
- Prepare a stock of 10 mM 5-ethynyl-2'-deoxyuridine (EdU) in dimethyl sulfoxide (DMSO). This can be stored at -20 °C for up to 12 months.
- Prepare 200 µL of 50 µM EdU in 10% sucrose. Pre-feed flies with EdU for 6 h before PTBI.
- Place 200 µL of 50 µM EdU in 10% sucrose on a 23 mm round grade 3 filter paper (see Table of Materials) in an otherwise empty vial.
- Place the flies in the vial. Then seal the vial with a cotton plug.
- Lay the vial horizontally in a humidified incubator at 25 °C for 6 h.
- Carry out PTBI as described in steps 2.3-2.10.
- Place the flies back in the EdU-containing vial. Seal the vial with a cotton plug.
- Lay the vial horizontally in a humidified incubator at 25 °C for up to 24 h. Follow one of the steps described below (step 3.8.1-3.8.3).
- Dissect and fix brains as described in step 4 using fixative, wash buffer, and blocking buffer without azide and with the EdU detection reaction carried out before antibody staining.
- Transfer the flies to a clean vial containing a new filter paper and 200 µL of 50 µM EdU in 10% sucrose. Lay the vial horizontally in a 25 °C incubator. Repeat every 24 h during the duration of the labeling. Then, dissect and fix brains as described in step 4 using buffers without azide with the EdU detection reaction carried out before antibody staining.
- To pulse-chase label with EdU, feed EdU for the pulse period, transfer the flies every 24 h to a clean vial containing a new filter paper and 200 µL of 50 µM in 10% sucrose. After the pulse period (e.g., 4 days), transfer the flies to a vial containing standard Drosophila food. Place the vial on its side in a 25 °C incubator for an additional 3 days. Then, dissect and fix brains as described in step 4 using buffers without azide with the EdU detection reaction carried out before antibody staining.
4. Dissection, immunohistochemistry, and mounting
- Prepare 1.5 mL microcentrifuge tubes with 100 µL of fixative: 4% formaldehyde in PEM (100 mM piperazine-N,N'-bis(2-ethanesulfonic acid) [PIPES], 1 mM EGTA, 1 mM MgSO4, pH 7.0) (see Table of Materials) and place on ice.
NOTE: As many as 20 brains of a single genotype and condition can be processed in a single tube. - Prepare a dissection plate with one small pool (~100 µL) of 70% ethanol and three small pools of phosphate-buffered saline (PBS; 100 mM of K2HPO4, 140 mM of NaCl, pH 7.0).
- Anesthetize ~10 control or experimental flies on a CO2 pad that has been sanitized with 70% ethanol.
- Separate the head from the trunk of each fly using a scalpel.
- Collect the heads with a paintbrush wetted in 70% ethanol and place heads for 2-5 min in the ethanol pool on the dissection plate.
NOTE: This slightly dehydrates the brains and makes them easier to dissect away from the head cuticle. - Transfer the heads to a ~100 µL pool of PBS and dissect out the brains, moving each brain to a clean ~100 µL pool of PBS. Perform this using two pairs of Watchmakers forceps to open the back of the head cuticle and holding the cuticle with one pair of forceps while gently prying the brain out of the cuticle using the closed tip of the second pair of forceps.
- Transfer the dissected brains to a microcentrifuge tube containing 100 µL of the fixing solution using a P200 pipettor equipped with a plastic tip that has been cut and beveled to permit entry of the brains.
- Fix for 20-25 min at room temperature. Carefully remove the fix with either a P200 or a glass pipette.
- Wash the fixed brains four times with 1 mL of 'PT' (PBS plus 0.1% Triton X-100), allowing the brains to settle for several minutes between each wash.
NOTE: If brains do not settle rapidly between the washes, they are likely to have still fat body and/or trachea attached. - Block samples in 1 mL of PBS plus 0.1% Triton X-100 and 2% bovine serum albumin (PBT) for ~1 h at room temperature.
- Remove the blocking solution and incubate samples with 100 µL of primary antibody solution overnight at 4 °C in PBT. The primary antibodies used in this study are rabbit anti-PH3 (1:500) and mouse anti-Fasiclin II (1:20) (see Table of Materials).
- Wash samples five times with 1 mL of PT, allowing the brains to settle for several minutes between each wash.
- Remove the final wash and incubate in 100 µL of secondary antibody solution overnight at 4 °C. The secondary antibodies used here are anti-rabbit 568 (1:400) and anti-mouse Cy5 (1:100) (see Table of Materials).
- Wash samples five times with 1 mL of PT, allowing the brains to settle for several minutes between each wash. During the final wash, add 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; 1:100 of a 10 µM solution) for 10 min to stain the nuclei. Remove the wash, leaving 50-100 µL in each tube.
- Prepare microscope slides by placing a single, self-adhesive reinforcement label on the middle of each slide and a 50 µL droplet of anti-fade mounting media in the center of each label (see Table of Materials). The reinforcement label holds the coverslip slightly above the slides and prevents the mounted brains from becoming overly flattened.
- Carefully transfer the brains from each microcentrifuge tube to a prepared slide with as little wash buffer as possible using a P200 equipped with a cut and beveled plastic tip. As many as 10 brains can be mounted on a single slide.
- Use forceps to gently reposition the brains before placing a coverslip on each slide and sealing the slide with nail polish. Orient the brains with either posterior side up or anterior side up but should not be touching one another.
NOTE: Because it is difficult to obtain high-quality confocal images through an entire brain, some brains should be mounted in each orientation. - Store prepared slides flat and in the dark at 4 °C until imaging. For longer-term storage (up to 1 year), slides may be stored at -20 °C.
5. Confocal imaging
NOTE: Image brains using a laser-scanning confocal microscope with excitation lasers and emission filter cubes appropriate to DAPI and the fluorescent secondary antibodies (i.e., 405 nm, 488 nm, and 568 nm, 633 nm, respectively).
- Turn on the power of the microscope, lasers, controller, and computer.
- Open the Acquisition software.
- In the Acquisition mode, select up to four channels and set the parameter for sequential scanning of the desired channels. Increase the laser power for each channel to 5-10%.
- Place a slide on the microscope.
- Choose a brain to be imaged using the epifluorescence attachment (see Table of Materials).
- In the Acquisition mode, select 1024 x 1024 pixels as the frame dimension.
- In the Acquisition mode, select the Z-stack option, indicate Z-stack should be taken at 2 µm sections, and focus through the sample, selecting the top and bottom focal planes to be imaged.
- Collect Z-stack images of entire brains using a 20x objective and specific brain regions such as the mushroom body using a 60x objective.
6. Data analysis
- Quantify the proliferating cells and the numbers of perma-twin clones manually and/or using the image analysis software (see Table of Materials). When using software, select regions of interest (ROIs) with areas of at least 10 µm.
Representative Results
PTBI stimulates cell proliferation
To determine the extent of neurogenesis after a central brain PTBI, the proliferative response was measured in young adult males collected and injured within 6 h of eclosion. A significant increase in proliferation was observed 24 h post-injury using anti-phosphohistone 3 (PH3), a marker for cells actively undergoing mitosis. Approximately 3 PH3+ cells in control central brains and 11 PH3+ cells in the injured central brains are observed 24 h post-PTBI (Figure 2A-D). The majority of the dividing cells are located near the injury site. A second assay for cell division was used to quantify the cumulative cell proliferation from a single injury and to assess the extent to which the newly created cells survived. 5-ethynyl-2'-deoxyuridine (EdU) is a thymidine analog that can be incorporated into newly synthesized DNA and permanently label cells that have undergone DNA synthesis. Flies were given a 4-day pulse of EdU, followed by a 3-day chase. This revealed that the labeled cells were viable and survived at least 3 days after proliferation. By 7 days, there were an average of 2 EdU+ cells in control central brains and an average of 11 EdU+ cells in the injured central brains, respectively (Figure 2E). This is similar to the results obtained 24 h post-injury using the PH3 antibody. When cell proliferation is measured at 14 days, the uninjured controls averaged 1 EdU+ cell per central brain, while the injured brains averaged 29 EdU+ cells (Figure 2E), demonstrating that cell proliferation continues at least into the second week following a PTBI.
Cell proliferation is age-dependent
The greatest proliferative response in the central brain was observed within the first 24 h after eclosion (Figure 3). By 7 days post-eclosion, a penetrating injury still causes a significant increase in proliferation, with an average of 6 PH3+cells per central brain. Still, by 14 days post-eclosion, the ability for cells to divide following PTBI decreases significantly to 1 dividing cell, similar to that of control brains (Figure 3). Thus, the potential for cell proliferation post-PTBI is age-dependent.
Newly created neurons can project to correct target areas
To evaluate neural regeneration post-PTBI, the perma-twin labeling system15 was used. Perma-twin lineage tracing permanently labels dividing cells and their progeny with a green fluorescent protein (GFP) or red fluorescent protein (RFP)15. More perma-twin clones were detected in injured samples, at 2 days and 2 weeks, than in controls (Figure 4A-E). Notably, there were new mushroom body neurons in ~50% of the PTBI brains 2 weeks post-injury (Figure 4N). These new neurons projected their dendrites appropriately to the mushroom body calyx and their axons appropriately to the mushroom body lobes (Figure 4D,F,G). This indicates that the newly created cells may be functional neurons involved in the repair of the damaged mushroom bodies. Other areas of the brain that appeared to regenerate include the ellipsoid body (EB) (Figure 4H,I), the antennal lobes (AL) (Figure 4J,K), and the lateral horn (LH) (Figure 4L,M) which possessed large clones approximately 26%, 26%, and 20% of the time, respectively (Figure 4N). These results underscore the utility of this system for the investigation of adult neurogenesis. A proposed model for the sequence of events following PTBI and leading to the generation of new neurons is shown in Figure 5.
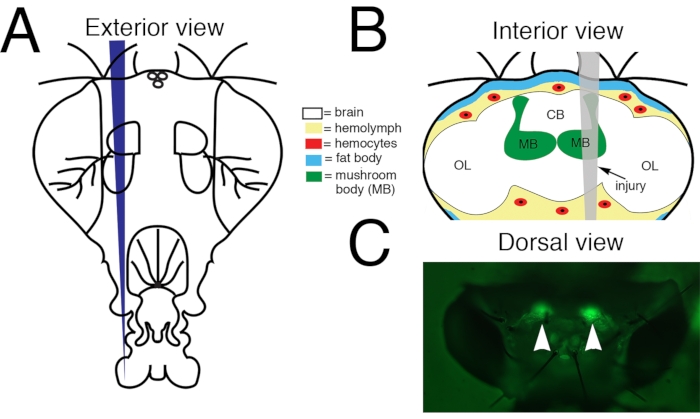
Figure 1: Penetrating Traumatic Brain Injury (PTBI) to the adult Drosophila central brain. (A) Schematic of the exterior of an adult fly head. This is a frontal view. Thus, the right side of the animal is to the viewer's left. (B) Schematic of the interior of an adult Drosophila head with the injury trajectory indicated in grey. This is a posterior view. Thus, in this image and subsequent figures, the right side of the brain is to the right. Central brain PTBI impacts multiple brain structures, including the mushroom body (green) and tissues outside the brain, including the fat body (blue) and hemocytes (red). CB = central brain region. OL= optic lobe region. (C) Dorsal view of a live adult head in which mushroom bodies (arrowheads) are labeled with a green fluorescent protein (GFP). This is the 'standard genotype' (see text for details). The PTBI protocol reproducibly results in injury to the mushroom bodies. This Figure has been adapted from Reference16. Please click here to view a larger version of this figure.
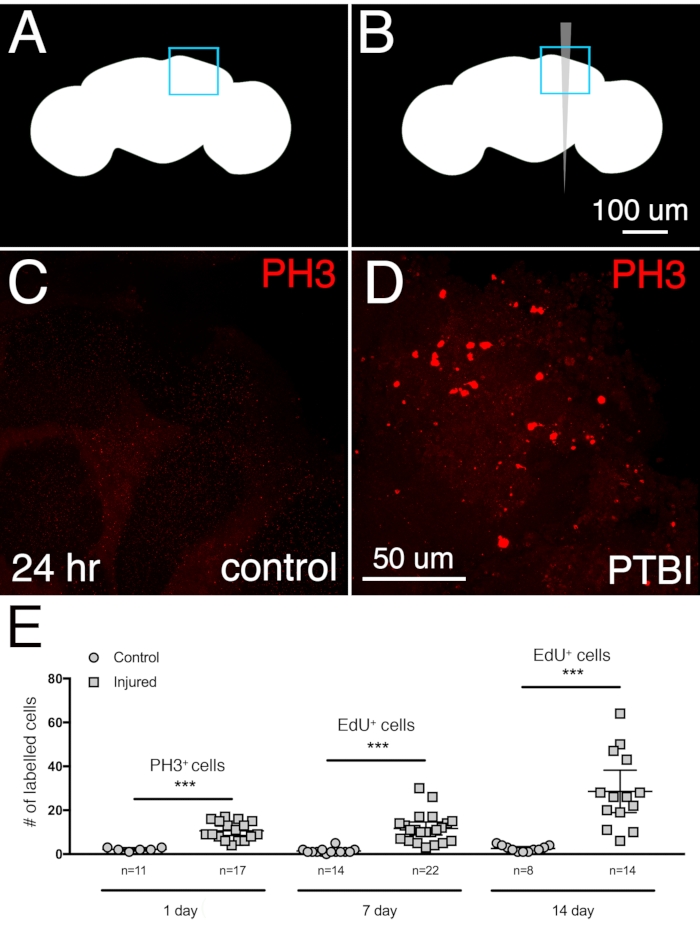
Figure 2: PTBI stimulates cell proliferation. Uninjured control (A) and PTBI (B) schematics. The blue boxes in the upper right corners indicate the brain regions shown at higher magnification in panels (C) and (D). (C,D) PH3 antibody (red) was used to assay cell proliferation 24 h after injury. In control brains (C), there are few PH3+ cells and none near the MB. However, in PTBI brains (D), there are PH3+ cells near the MB.(E) Quantification of proliferating cells. The numbers reflect proliferating cells throughout entire brains, not only in the vicinity of the mushroom body. At 24 h, uninjured control brains had an average of 3 PH3+ cells/brain (n = 11 brains, 28 cells), while 24 h post-PTBI, brains had an average of 11 PH3+ cells/brain (n = 17 brains, 181 cells). At 7 days, uninjured controls have few EdU+ cells, with an average of 2 EdU+ cells/brain (n = 15 brains, 24 cells), while 7-day post-PTBI brains had an average of 11 EdU+ cells/brain (n = 22 brains, 238 cells). At 14 days, uninjured controls have an average of 1 EdU+ cell/brain (n = 8 brains,11 cells), while 14-day post-PTBI brains have an average of 29 EdU+ cells/brain (n = 14 brains, 400 cells). For this set of experiments, young adult males within 6 h of eclosion were used. Unpaired t-tests of control and PTBI samples at the 3-time points yield values of p<0.0001, p<0.0001, and p<0.0002, respectively. Error bars reflect the standard deviation (SD). This Figure has been adapted from Reference16. Please click here to view a larger version of this figure.
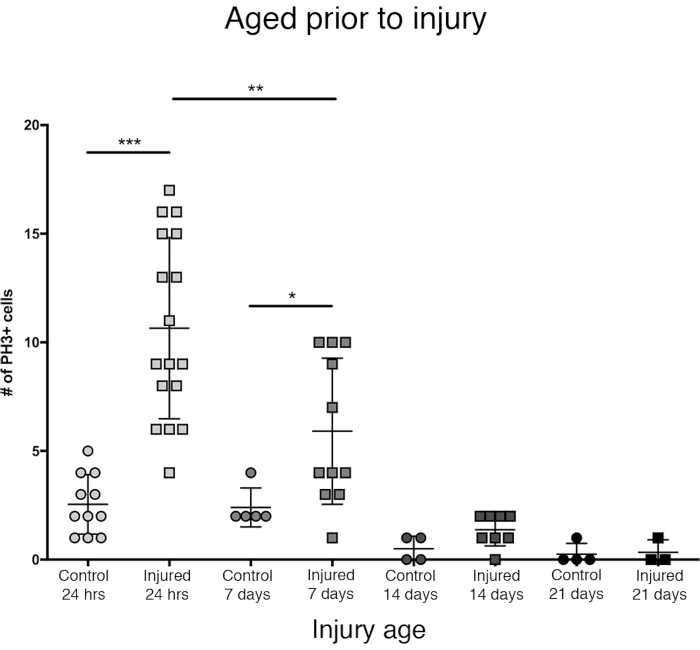
Figure 3: The proliferative response to PTBI decreases with age. To explore whether age impacts the amount of cell proliferation that occurs post-injury, newly eclosed adult males were compared to animals aged to 7 days, 14 days, and 28 days before PTBI, using anti-PH3 to assay cell proliferation 24 h after injury. Flies injured within 6 h of eclosion had an average of 11 PH3+ cells/brain (n = 17 brains, 182 cells) compared to an average of 3 PH3+ cells/brain in age-matched controls (n = 11 brains, 28 cells). Flies aged to 7 days, then subjected to PTBI, had an average of 6 PH3+ cells/brain (n = 11 brains, 65 cells) compared to age-matched controls with an average of 2 PH3+ cells/brain (n = 5 brains, 12 cells). When flies were aged to 14 days before PTBI and assayed 24 h later, there was an average of 1 PH3+ cell/brain (n = 8 brains, 11 cells) similar to age-matched controls, which also averaged 1 PH3+ cell/brain (n = 4 brains, 2 cells). 28-day uninjured control (n = 4, 1 cell) and PTBI (n = 3, 1 cell) flies both averaged 0 PH3+cells/brain. Unpaired t-tests for PTBI to control comparisons at these 4-time points are p<0.0001, p<0.04, p<0.07, and p<0.84, respectively. This Figure has been adapted from Reference16. Please click here to view a larger version of this figure.
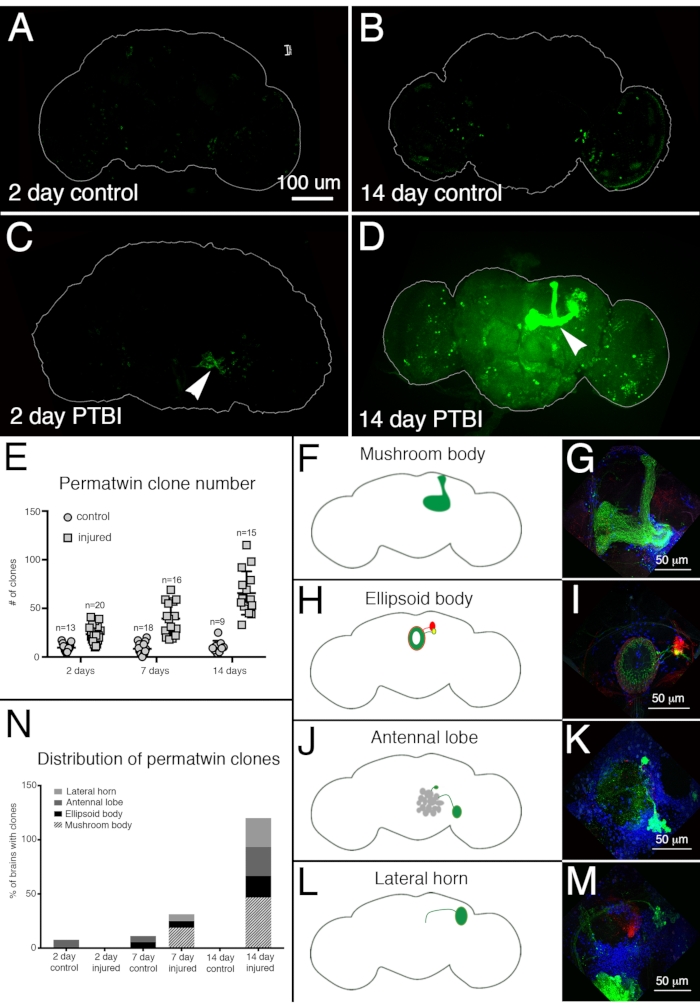
Figure 4: Perma-twin lineage tracing demonstrates brain regeneration and appropriate targeting of axons following PTBI. The perma-twin lineage-tracing system15 was utilized to analyze neurogenesis after PTBI. This system permanently labels dividing cells and progeny with a green fluorescent protein (GFP) or red fluorescent protein (RFP). Flies were reared at 17 °C to keep the system off during development. F1 males carrying perma-twin transgenes were collected upon eclosion, then injured and placed at 30 °C to recover for 2 or 14 days. (A) In 2-day uninjured controls, there are some GFP+ cells scattered throughout the brain. (B) At 14 days, there are relatively few GFP+ cells present in the control central brain. (C) In comparison, 2-day injured brains have more GFP+ cells that tend to cluster near the injury (arrowhead). (D) At 14 days post-injury, there are large clones near the site of injury. Some of these clones have axons that project along the mushroom body tracts (arrowhead). Only the GFP channel is shown here; there were similar RFP+ clones in the PTBI samples. (E) The number of clones increases over time post-PTBI.Control uninjured brains (n = 13) have an average of 10 clones at 2 days, while 2-day PTBI brains (n = 20) have an average of 23 clones (p<0.00002). At 7 days, control brains had an average of 9 clones per brain (n = 18), while 7-day PTBI brains had an average of 39 clones per brain (n = 16) (p-value<0.00000002). This is significantly more than the number of clones seen at 2 days post-injury (p-value<0.0009). In 14-day control brains, there is an average of 10 clones per brain, which is not significantly different from the 2-day and 7-day controls. However, at 14 days post-PTBI, there is an average of 66 GFP+ clones, which is significantly more than either age-matched controls (p<0.0000003) or 2-day post-PTBI brains (p-value<0.0001). Error bars reflect SD. (F-M) PTBI stimulates clone formation in multiple regions in the brain. Panels on the left side are schematics of brain regions where large clones were found 14 days post-PTBI (A, H, J, L). Panels on the right show high magnifications of representative brains (G, I, K, M). Many 14-day brains had clones that projected to particular target areas. These included the mushroom body (MB) (F,G), the ellipsoid body (EB) (H,I), the antennal lobe (AL) (J,K), and the lateral horn (LH) (L,M). (N) Both clone number and clone size increase with time post-PTBI.The proportions of brain regions with large clones were calculated at 2, 7, and 14 days in controls and injured brains. At 2 days, ~8% of control brains (n = 13) showed AL clones, while there were no AL clones in 2-day injured brains (n = 20). In 7-day control brains (n = 18), 6% had AL and 6% had EB clones. At 7 days post-PTBI (n = 16), 6% of brains also had AL clones, 6% had EB clones, and 19% had large MB clones. At 14 days, control brains (n = 9) did not exhibit any specific areas with clones, while 47% of PTBI brains (n = 15) had MB clones, 20% of PTBI brains had AL clones, and 27% of PTBI brains had EB clones, and 27% had LH clones. This Figure has been adapted from Reference16. Please click here to view a larger version of this figure.
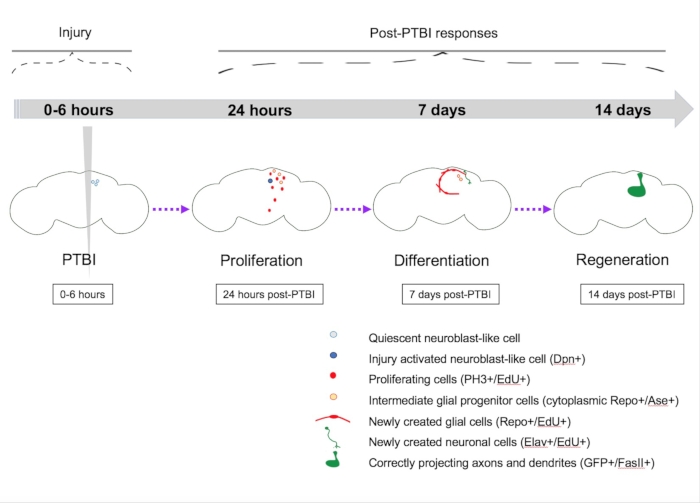
Figure 5: Summary model for regeneration following penetrating traumatic brain injury (PTBI). In young adult Drosophila, there are quiescent neuroblast-like cells within the central brain that lack expression of canonical neuroblast genes. By 24 h post-PTBI, the quiescent neuroblast-like cells are activated, express neuroblast genes, and have begun to proliferate. At both 4 h and 24 h post-PTBI, there is a wave of cell death16. At 7 days, the proliferation rate is still high, and many of the new cells have adopted mature cell identities, becoming neurons or glia. At 14 days post-PTBI, large clones of new neurons with axons and dendrites correctly projecting to their respective target areas. Locomotor defects are also restored by 14 days, suggesting that adult Drosophila can regenerate functionally and structurally. This Figure has been adapted from Reference16. Please click here to view a larger version of this figure.
Discussion
Although penetrating injuries to the adult Drosophila brain have been described previously15,17,18, these injuries focused on the optic lobes and not the central brain. Further, detailed instructions for how to carry out the injuries are thus far lacking. This protocol describes a model for penetrating injury to the adult Drosophila central brain that reproduces statistically significant evidence for adult neurogenesis after PTBI.
The reproducibility of this PTBI protocol is due, in part, to the mushroom body as the injury target region. The mushroom body is large, consisting of ~2200 neurons with complex dendrite and axon arbors in large and highly stereotyped arrays18. The cell bodies of mushroom body neurons lie near the brain's surface and can be visualized through the head cuticle using the expression of green fluorescent protein (GFP) (Figure 1C). Mushroom body precursors are the last neural stem cells to undergo apoptosis during development13,12,19. Thus, many mushroom body neurons are pretty young at the time of eclosion. This led to the hypothesis that the mushroom body might have more mitotic potential than other brain regions16. In addition, the mushroom body is critical for learning and memory18. This allows one to ask whether PTBI-triggered neurogenesis leads to functional recovery.
Other factors that contribute to the reproducibility of the results include using outcrossed flies of consistent genotypes, performing crosses in the same direction each time, precisely controlling the rearing and aging temperatures, and analyzing males and females separately. Using F1 flies from an outcross reduces the probability of analyzing brains homozygous for spontaneous mutations. The standard cross of y[1] w[1];UAS-mCD8-GFP;; OK107-GAL4 adult females to y[1] w[1] adult male flies results in F1 progeny of the genotype y[1] w[1];UAS-mCD8-GFP/+;; OK107-GAL4/+. OK107-GAL4 is expressed in all intrinsic neurons of the mushroom body and drives expression of the membrane-bound reporter UAS-mCD8-GFP permitting visualization of mushroom bodies and their projections. For the perma-twin crosses, crosses must remain at 17 °C at all times to keep the lineage tracing system switched off. This ensures that no dividing cells are labeled during development and that only adult-born neurons and glia are labeled. To this end, the fly room can also be maintained at 17 °C. Although the initial description of the perma-twin system15 recommended rearing flies at 18 °C, this can lead to significant background labeling.
For consistency, it also is recommended to keep the control uninjured flies on the CO2 pad as one carries out the PTBI. This ensures that both sets of flies have identical anesthetic exposure. In addition, it is desirable for reproducibility to completely penetrate the head. However, care must be taken not to bend the tip of the pin against the pad, making it unusable for future injuries. For skilled practitioners, there is little unintended harm to PTBI flies. Nonetheless, pressing too hard on the thorax to stabilize the fly during injury can be lethal. One way to assess the extent of the unintended injury is to quantify the mortality of PTBI flies 24 h post-injury. For unilaterally injured flies, this can be 50% or higher for beginners. Therefore, to ensure that observed outcomes are due to PTBI and not to unintended injury, it is advised that beginners practice administering PTBI on ~20 flies daily over several weeks and do not analyze the resulting brains until the 24 h mortality is consistently <10%.
To quantify the amount of proliferation stimulated by central brain PTBI, both anti-phosphohistone H3 (PH3) immunostaining and 5-ethynyl-2´-deoxyuridine (EdU) incorporation can be employed. Anti-PH3 labels cells before and throughout metaphase, limiting detection to only a fraction of the actively dividing cells. Thus, anti-PH3 staining provides only a partial glimpse of proliferation. EdU is a thymidine analog that can be incorporated into newly synthesized DNA. By feeding flies EdU before and after injury, it is possible to gain a more complete picture of the cells that are either dividing or have divided following the injury. The fact that any cells that divide are permanently marked is helpful both for the identification of slowly cycling cells and assay the survival of cells after initial proliferation. For unclear reasons, but maybe due to limited permeability of the blood-brain barrier, EdU labeling is inefficient and under-reports cell proliferation in the adult brain. This is evidenced by the similar numbers of PH3+ and EdU+ cells in both control and experimental brains at 24 h post-PTBI and by observing that only a subset of new cells in perma-twin clones incorporate EdU16. For maximal labeling, it is essential to pre-feed the flies with EdU because injured flies do not resume feeding for several hours post-PTBI. Feeding was assessed by adding food coloring to the EdU solution and monitoring the amount of dye in the gut through the abdominal cuticle16.
It is to be noted that while we have provided a brain dissection protocol in step 4, alternative techniques may be used. Several of these are available in previously published protocols20,21,22. Drosophila melanogaster offers a low-cost model with powerful genetic and molecular tools that can be used to study the mechanisms underlying regeneration of multiple tissues, including the gut and components of the nervous system. A novel and reproducible injury model that can be used to study the response to brain injury is outlined here. Data obtained using these protocols support the idea that the adult Drosophila central brain retains the proliferative ability, generating new neurons in response to injury. These observations warrant further investigation of both the extent of adult neurogenesis and its underlying molecular mechanisms. Once the components involved in neural regeneration are identified in this system, we can convert our knowledge of adult Drosophila neurogenesis to humans.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are grateful to Stacey Rimkus and Becky Katzenberger for technical assistance and to Eduardo Moreno for sharing the perma-twin stocks. We would like to thank Barry Ganetzky and David Wassarman for lively discussions that undoubtedly improved science and Kent Mok, Cayla Guerra, and Bailey Spiegelberg for their contributions to the laboratory. The FasII antibodies were developed by Corey Goodman and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Most of the Drosophila strains used in this study were obtained from the Bloomington Drosophila Stock Center (BDSC; NIH P40OD018537). This work was supported by NIH T32 GM007133 (KLC); NIH NS090190 (GBF); NIH NS102698 (GBF); the University of Wisconsin Graduate School (GBF); and the UW-Madison Women in Science and Engineering Leadership Institute (WISELI) (GBF).
Materials
| #11 disposable scalpels | Santa Cruz Biotechnology | sc-395923 | used for separating Drosophila heads from trunks prior to brain dissection |
| 150 mm diameter black Sylgard dishes | Dow | 1696157 | made in the laboratory with reagents from Dow; used for brain dissection |
| 18 mm coverslips | any | for mounting brains on microscope slides | |
| 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) | ThermoFisher | D1306 | for immunohistochemistry |
| 70% Ethanol | made from 95% ethanol sourced variously | ||
| anti-mouse Cy5 | Jackson ImmunoResearch | 715-175-151 | for immunohistochemistry |
| anti-rabbit 568 | ThermoFisher | A11036 | for immunohistochemistry |
| bovine serum albumin (BSA) | SIgma Aldrich | A7030 | for immunohistochemisty |
| Clear nail polish | any | for sealing coverslips | |
| Click-It EdU labeling kit | InVitrogen | C10640 | to detect newly synthesized DNA |
| CO2 bubbler | Genesee Scientific | 59-181 | for anesthesia |
| CO2 pad | Genesee Scientific | 59-114 | for anesthesia |
| CO2 regulator and supply | any | for anesthesia | |
| Confocal microscope | any | for imaging fixed, stained and mounted brains | |
| cotton plugs | Genesee Scientific | 51-101 | for EdU labeling |
| Drosophila vials | Genesee Scientific | 32-109 | for EdU labeling |
| Fix buffer (Pipes, EGTA, Magnesium; PEM) | components sourced from various companies | for fixing adult brains; 100 mM piperazine-N,N’-bis(2-ethanesulfonic acid) [PIPES], 1 mM EGTA, 1 mM MgSO4, pH 7.0 | |
| Formaldehyde | Sigma Aldrich | 252549 | for fixing adult brains, added to PEM |
| Grade 3 round Whatman filters, 23 mm round | Tisch Scientific | 1003-323 | for EdU labeling |
| Microfuge tubes | any | for fixing and staining reactions and for storing Minutien pins | |
| Microscope slides | any | for mounting brains | |
| Minutien pins | Fine Science Tools | 26002-10 | for brain injury; 12.5 μm diameter tip and 100 μm diameter rod |
| mouse anti-Fasiclin II | Developmental Studies Hybridoma Bank | 1D4-s | for immunohistochemistry |
| NIGHTSEA stereo microscope fluorescence adaptor | Electron Microscopy Sciences | SFA-GR | fluorescence setup for dissecting microscope |
| P20, P200 and P1000 pipettors and tips | any | for measuring solutions | |
| phosphate buffered saliine (PBS) | components sourced from various companies | for dissecting brains and making immunohistochemistry blocking and washing solutions; 100 mM of K2HPO4, 140 mM of NaCl, pH 7.0 | |
| phosphate buffered saline with 0.1% Triton X-100 (PT) | components sourced from various companies | for washing dissected brains | |
| phosphate buffered saline with 0.1% Triton X-100 + 2% bovine serum albumin (PBT) | components sourced from various companies | blocking solution for immunohistochemistry and for diluting antibodies | |
| rabbit anti-PH3 | Santa Cruz Biotechnology, Inc | sc-8656-R | for immunohistochemistry |
| Reinforcement labels | Avery | 5721 | to maintain space between the microscope slide and the coverslip |
| Size 0 paintbrushes | any | to manipulate and stabilize adult Drosophila during injury | |
| Triton X-100 | Sigma Aldrich | 93443 | |
| Two pair of #5 watchmakers forceps | Fine Science Tools | 11255-20 | used to hold the Minutien pins and for brain dissections |
| Vectashield | Vector Laboratories | H-1000 | mounting medium for microscope slides |
References
- . Report to Congress: Traumatic brain injury in the United States Available from: https://www.cdc.gov/traumaticbraininjury/pubs/tbi_report_to_congress.html (1999)
- . NIEHS Available from: https://www.nih.gov/research/supported/health/neurodegenerative/index.cfm (2021)
- Morton, N. V., Wehman, P. Psychosocial and emotional sequelae of individuals with traumatic brain injury: A literature review and recommendations. Brain Injury. 9 (1), 81-92 (2017).
- Bonini, N. M., Berger, S. L. The sustained impact of model organisms-in genetics and epigenetics. Genetics. 205, 1-4 (2017).
- Brace, E. J., DiAntonio, A. Models of axon regeneration in Drosophila. Experimental Neurology. 287, 310-317 (2017).
- Hao, Y., Collins, C. Intrinsic mechanisms for axon regeneration: insights from injured axons in Drosophila. Current Opinion in Genetics & Development. 44, 84-91 (2017).
- Chiang, A. S., et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Current Biology. 21 (1), 1-11 (2011).
- Meinertzhagen, I. A. The organisation of invertebrate brains: cells, synapses and circuits. Acta Zoologica. 91 (1), 64-71 (2010).
- Bellen, H. J., Tong, C., Tsuda, H. 100 years of Drosophila research and its impact on vertebrate neuroscience: A history lesson for the future. Nature Reviews Neuroscience. 11 (7), 514-522 (2010).
- Lessing, D., Bonini, N. M. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nature Reviews Genetics. 10 (6), 359-370 (2009).
- Boone, J. Q., Doe, C. Q. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Developmental Neurobiology. 68 (9), 1185-1195 (2008).
- Ito, K., Hotta, Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Developmental Biology. 149 (1), 134-148 (1992).
- Siegrist, S. E., Haque, N. S., Chen, C. H., Hay, B. A., Hariharan, I. K. Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Current Biology. 20 (7), 643-648 (2010).
- von Trotha, J. W., Egger, B., Brand, A. H. Cell proliferation in the Drosophila adult brain revealed by clonal analysis and bromodeoxyuridine labelling. Neural Development. 4, 9 (2009).
- Fernandez-Hernandez, I., Rhiner, C., Moreno, E. Adult neurogenesis in Drosophila. Cell Reports. 3 (6), 1857-1865 (2013).
- Crocker, K. L., et al. Neurogenesis in the adult Drosophila brain. Genetics. , (2021).
- Plavicki, J., Mader, S., Pueschel, E., Peebles, P., Boekhoff-Falk, G. Homeobox gene distal-less is required for neuronal differentiation and neurite outgrowth in the Drosophila olfactory system. Proceedings of the National Academy of Sciences of the United States of America. 109 (5), 1578-1583 (2012).
- Aso, Y. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 3, 04577 (2014).
- Ito, K., Awano, W., Suzuki, K., Hiromi, Y., Yamamoto, D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 124 (4), 761-771 (1997).
- Tito, A. J., Cheema, S., Jiang, M., Zhang, S. A. Simple one-step dissection protocol for whole-mount preparation of adult Drosophila brains. Journal of Visualized Experiments. (118), e55128 (2016).
- Kelly, S. M., Elchert, A., Kahl, M. Dissection and immunofluorescent staining of mushroom body and photoreceptor neurons in adult Drosophila melanogaster brains. Journal of Visualized Experiments. (129), e56174 (2017).
- Arain, U., Valentino, P., Islam, I. M., Erclik, T. Dissection, immunohistochemistry and mounting of larval and adult Drosophila brains for optic lobe visualization. Journal of Visualized Experiments. (170), e61273 (2021).

