Visualizing Single-Stranded DNA Foci in the G1 Phase of the Cell Cycle
Summary
The following protocol presents the detection of single-stranded DNA foci in the G1 phase of the cell cycle utilizing cell cycle synchronization followed by RPA2 immunofluorescent staining.
Abstract
DNA has dedicated cellular repair pathways capable of coping with lesions that could arise from both endogenous and/or exogenous sources. DNA repair necessitates collaboration between numerous proteins, responsible for covering a broad range of tasks from recognizing and signaling the presence of a DNA lesion to physically repairing it. During this process, tracks of single-stranded DNA (ssDNA) are often created, which are eventually filled by DNA polymerases. The nature of these ssDNA tracks (in terms of both length and number), along with the polymerase recruited to fill these gaps, are repair pathway-specific. The visualization of these ssDNA tracks can help us understand the complicated dynamics of DNA repair mechanisms.
This protocol provides a detailed method for the preparation of G1 synchronized cells to measure ssDNA foci formation upon genotoxic stress. Using an easy-to-utilize immunofluorescence approach, we visualize ssDNA by staining for RPA2, a component of the heterotrimeric replication protein A complex (RPA). RPA2 binds to and stabilizes ssDNA intermediates that arise upon genotoxic stress or replication to control DNA repair and DNA damage checkpoint activation. 5-Ethynyl-2′-deoxyuridine (EdU) staining is used to visualize DNA replication to exclude any S phase cells. This protocol provides an alternative approach to the conventional, non-denaturing 5-bromo-2′-deoxyuridine (BrdU)-based assays and is better suited for the detection of ssDNA foci outside the S phase.
Introduction
To sustain life, cells constantly survey and repair DNA to maintain their genomic integrity. Cells may accumulate various types of DNA damage due to both endogenous (e.g., oxidation, alkylation, deamination, replication errors) and exogenous (e.g., UV, ionizing irradiation) sources of DNA stressors. Failure to repair these lesions results in either apoptosis, cell cycle arrest, or senescence and can lead to diseases1. DNA lesions could be addressed by any of the following main DNA repair pathways: DR (direct reversal repair), which mainly repairs alkylated bases2; BER (base excision repair), which targets non-bulky DNA bases errors and single-stranded DNA breaks (SSBs)3; NER (nucleotide excision repair) correcting bulky, helix-distorting DNA lesions4; MMR (mismatch repair) mainly targeting DNA mismatches, insertion/deletion loops (IDLs), and certain base damages5; NHEJ (non-homologous end joining) and HRR (homologous recombination repair) that are both active at double-stranded DNA breaks (DSBs)6; and TLS (translesion synthesis), which is a DNA lesion bypass mechanism7. Though these pathways have distinct substrate specificities, there are certain overlaps amongst them to ensure redundancy for efficient repair. Understanding the action of the different DNA repair pathways in various cell cycle phases is crucial as these DNA repair factors could serve as essential targets for therapeutic approaches to treat cancer, aging, and neurological disorders8,9.
Single-stranded DNA (ssDNA) is generated throughout the cell cycle due to the repair of DNA lesions generated by both endogenous and exogenous DNA-damaging agents. Upon genotoxic stress, ssDNA is generated abundantly in the S and G2 phases where HRR and MMR have their highest activity and when the replication machinery stalls or collapses when encountering DNA lesions6,10,11. Other DNA repair pathways (e.g., NHEJ/microhomology-mediated end joining (MMEJ)/single-strand annealing [SSA]) also generate ssDNA during DSB repair12. These ssDNA tracks usually arise from DNA resection, carried out by exonucleases such as EXO1, DNA2, and CtIP during HR and MMR, endonucleases such as XPF and XPG during NER, or through the combined action of POLB and FEN1 during BER4,13,14,15,16,17,18,19. Due to the work of the replication machinery, ssDNA tracks are also generated when the DNA helicases unwind DNA in front of the PCNA-bound replicative polymerases20. In contrast, in the G1 phase, the lack of HRR and DNA replication and the limited activity of MMR reduces the extent of ssDNA tracks generated and are therefore more challenging to detect10,11,21.
Cellular ssDNA tracks are highly sensitive structures that must be protected to avoid the formation of DSBs. This is achieved by coating the ssDNA tracks with RPA. RPA is an abundant heterotrimeric protein complex composed of multiple subunits (RPA1, RPA2, and RPA3, also referred to as RPA70, RPA32, and RPA14, respectively), which are ubiquitously expressed throughout the cell cycle22. Each RPA subunit contains a DNA-binding domain (DBD), capable of interacting with 4-6 nucleotides, and the combined subunits form a stable trimerization core. Altogether, RPA binds to approximately 20-30 nucleotides with sub-nanomolar affinity23,24.
Conventional methods use immunofluorescence (IF) microscopy to visualize ssDNA foci by labeling 5-bromo-2'-deoxyuridine (BrdU) incorporated into genomic DNA using BrdU antibodies25. This approach relies on the fact that BrdU antibodies can only detect BrdU in exposed ssDNA25. Although this approach is straightforward, it also displays certain limitations. For instance, cells are pretreated to incorporate BrdU before the start of the experiment, which is time-consuming and can interfere with downstream effectors. Therefore, BrdU-based ssDNA detection is limited to replicating cells and cannot be used for quiescent cells. This excludes the application of this method to study DNA repair in non-replicating cells despite its importance in several diseases such as cancer and neurodegeneration5,26. Additionally, because the structures of BrdU and EdU are very similar, most BrdU antibodies display crossreactivity towards EdU, which must be considered when aiming for dual labeling experiments27. RPA staining has been previously utilized to show ssDNA foci mainly in S phase cells; however, some papers have also successfully used it outside the S phase28,29,30,31,32,33,34,35. The following protocol efficiently utilizes the properties of RPA, allowing the visualization of ssDNA foci following DNA damage in the G1 phase of the cell cycle (although it can be used in all cell cycle phases).
Protocol
1. Maintenance of hTERT-immortalized retinal pigment epithelial cells (RPE1)
- Maintain RPE1 cell lines in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Hi-FBS) and 100 µg/mL Penicillin-Streptomycin (referred to as culturing medium from now on) in a humidified incubator with 5% CO2 at 37 °C. For routine culturing, grow RPE1 cells in a 15 cm tissue culture-treated dish and split when reaching 80-90% confluency (~16-18 × 106 cells per 15 cm dish).
- When splitting, remove the medium and rinse the cells with 10 mL of 1x phosphate-buffered saline (PBS).
- Add 3 mL of 0.05% Trypsin-EDTA to cover the entire surface of the dish. Keep the cells at 37 °C with the trypsin until they detach.
- After trypsinization, resuspend the cells with culturing medium and spin them down at 150 × g for 5 min at room temperature (RT, 22-25 °C). Remove the supernatant and gently resuspend the cells in 10 mL of culturing medium.
- Seed 1.6-1.8 × 106 cells into a new 15 cm dish (~1 mL of the cell suspension).
NOTE: All tissue culture work should be done under BSL-2 safety levels. The incubation time for trypsinization depends on cell confluency. Usually, the process takes 2-3 min to complete for a 90% confluent plate. Cells should be screened for Mycoplasma contamination on a regular basis with commercially available kits (see examples in the Table of Materials).
2. siRNA knocking down of the gene of interest (GOI)
- Seed 1.0 × 106 RPE1 cells into a 10 cm tissue culture treated plate with 10 mL of culturing medium on the day before the transfection.
- On the day of the transfection, complex the siRNA. For a 10 cm plate, use a final concentration of 20 nM siRPA2 and 12 µL of lipid-based transfection reagent in 500 µL of low-serum transfection medium. Gently mix all the components by flicking the tube and incubate at RT (22-25 °C) for 5 min.
- Add the complexed siRNA mixture to the cells dropwise and incubate the cells with the siRNA for 48 h.
3. Synchronization of RPE1 cells into G0 phase
- Trypsinize the RPE1 cells from step 2.3 as outlined in section 1 (~2 × 106 cells).
- Transfer the cell suspension into 15 mL centrifuge tubes and centrifuge it at 150 × g, RT (22-25 °C) for 5 min.
- Remove the supernatant and resuspend the cells in 12 mL of PBS. Centrifuge the cells at 150 × g at RT (22-25 °C) for 5 min. Repeat the removal of the supernatant and centrifugation twice.
- Resuspend the cells in 10 mL of serum-free DMEM supplemented with 100 μg/mL Penicillin-Streptomycin, 1 mM Sodium Pyruvate, 15 mM HEPES, and plate them onto a 10 cm tissue culture dish.
NOTE: If the cells tend to clump, resuspend them in just 1 mL of serum-free DMEM and pipette them up and down 5x using a P1000 tip to dislodge the clumps before diluting the suspension up to a final volume of 10 mL. - After 24 h of serum starvation, introduce the second round of silencing using the same procedure as described in section 2 by adding the complexed siRNA to the serum-starving cells.
- Keep the RPE1 cells in serum-free DMEM for 72 h before proceeding to G1 release.
4. Coverslip coating and releasing cells into G1 phase
- Sterilize tweezers with 70% ethanol and place a single glass coverslip (12 mm in diameter and #1.5 thickness [0.17 mm]) in a well of a 24-well plate.
- Dilute the vitronectin coating matrix with PBS to obtain a final concentration of 10 µg/mL. Add 500 μL of the vitronectin solution into each well containing the coverslips and incubate for 1 h at RT.
- Remove the coating solution and wash the coverslips with 1 mL of PBS.
- Detach the serum-starved RPE1 cells from the 10 cm tissue cultured treated plate using 1 mL of 0.05% Trypsin after a PBS wash for 1 min at 37 °C.
NOTE: Cells detach much faster after serum starvation. Use caution when washing the cells with PBS and use short trypsinization times. - To inactivate the trypsin, resuspend the RPE1 cells in a total of 6 mL of culturing medium. Remove the inactivated trypsin by spinning down the cells using 150 × g at RT (22-25 °C) for 5 min.
- Resuspend the cells in 1 mL of culturing medium and measure the cell number.
- Seed 4 × 104 RPE1 cells onto the coated coverslip in a total of 500 μL of culturing medium.
NOTE: Make sure cell viability is above 90% before proceeding to downstream steps. Cell viability could be quickly assessed by trypan blue staining during the cell counting step. - After 6 h of plating the cells into the culture medium, G0-released cells will be in early G1 phase. Perform experiments in G1 in this 6-12 h window before the cells start entering S phase.
- Before introducing DNA damage, pulse the cells with 10 μM 5-ethynyl-2'-deoxyuridine (EdU) for 30 min at 37 °C, diluted in culturing medium.
- Remove the medium containing EdU and chase the cells with 10 μM thymidine for 10 min at 37 °C to prevent remaining EdU incorporation during DNA damage induction.
- Remove the medium with thymidine and treat the cells with 250 μM H2O2 for 1 h, diluted in culturing medium.
5. Immunofluorescence staining of ssDNA
- Wash cells once with 1 mL of RT (22-25 °C) PBS to remove the medium and serum components.
NOTE: Be gentle when washing the cells to avoid detachment and drying. Do not process many wells at the same time. - Pre-extraction: Incubate the washed cells in 1 mL of CSK extraction buffer (Table 1) for 5 min at RT (22-25 °C).
NOTE: CSK pre-extraction removes all the non-chromatin-bound proteins, including soluble RPA2.
CAUTION: Triton X-100 is harmful if swallowed and may cause skin irritation and eye damage. - Remove the CSK buffer from the cells and fix them directly by adding 0.5 mL of 3.6% paraformaldehyde solution (in PBS) containing 0.05% Triton X-100 for 10 min at RT (22-25 °C).
CAUTION: It is important to prepare 3.6% PFA from 32% PFA stock freshly. Paraformaldehyde may cause serious eye damage, skin irritation, and respiratory irritation. - Wash the cells once with 1 mL of PBS containing 0.05% Triton X-100 to remove the PFA.
- Further permeabilize cells using 1 mL of PBS containing 0.5% Triton X-100 for 15 min at RT (22-25 °C).
- EdU click-IT reaction to visualize replicating cells (S phase)
- Remove the permeabilization solution and wash the cells 2x using 1 mL of blocking buffer (Table 1).
CAUTION: Bovine serum albumin (BSA) may cause respiratory irritation. - Add 1 mL of blocking buffer (Table 1) and gently rock the coverslip-containing plate for 10 min at RT (22-25 °C).
- Remove the blocking buffer and add 500 μL of click reaction cocktail containing picolyl azide 647 (Table 1). Incubate the coverslips for 30 min at RT (22-25 °C) using gentle rocking and perform downstream incubations in the dark.
NOTE: When using BrdU antibodies, use double the amount (1 mL) and time (60 min) for the click-reaction as recommended by the manufacturer to ensure that the reaction is saturated and the incorporated EdU is labeled. This limits the cross-reactivity of the BrdU antibodies27.
- Remove the permeabilization solution and wash the cells 2x using 1 mL of blocking buffer (Table 1).
- Remove the click reaction mixture and wash the cells 2x with PBS with 0.05% Triton X-100 for 10 min at RT (22-25 °C) (Figure 1 and Figure 2).
- Add 1 mL of blocking buffer and incubate at RT (22-25 °C) for 30 min. Alternatively, keep the cells in blocking buffer at 4 °C overnight.
- Apply primary antibody (anti-RPA2 rat, 1:1,000 dilution) for 2 h at RT (22-25 °C) in 250-500 μL of blocking buffer with gentle rocking.
- Wash the cells 2x with PBS containing 0.05% Triton X-100 to quickly remove most of the antibody solution.
- Continue washing the cells for 3 x 10 min with blocking buffer at RT (22-25 °C).
- Apply secondary antibody (anti-rat Alexa-488, 1:1,000 dilution) in 250-500 μL of blocking buffer at RT (22-25 °C) for 2 h with gentle rocking.
- Wash the cells with blocking buffer 2x to quickly remove most of the secondary antibody. Continue washing the cells for 3 x 10 min with PBS containing 0.05% Triton X-100 at RT (22-25 °C).
- To counterstain the nuclei, wash cells once with PBS containing 0.05% Triton X-100 and 1 μg/mL 4',6-diamidino-2-phenylindole (DAPI) for 10 min at RT (22-25 °C). Wash cells once with PBS for 5 min at RT (22-25 °C).
- Mount the cover glass onto microscope slides using 10 μL of mounting medium/coverslip. Dip the coverslips into distilled water before mounting to get rid of any salt crystals. Image the slides on the following day and store them at 4 °C for weeks (Figure 3).
6. Image acquisition and quantification
- To capture images, use any available epifluorescent microscope equipped with routine filter sets to image DAPI, FITC, and Cy5 channels with at least 60-63x magnification, high numeric aperture, and oil objectives to visualize nuclear foci.
NOTE: Optimal DAPI excitation is ~359 nm; Alexa 488 excitation is ~488 nm; while Alexa 647 excitation is ~647 nm. - For image analysis, open image files in Fiji/ImageJ.
- Make nuclear masks using the DAPI staining (Figure 4A-F and Supplementary Video S1).
- Open the DAPI image.
- Select Process | Enhance Contrast and set saturated pixel to 0.35.
- Click Process | Binary | Convert to Mask. Choose Binary | Fill Holes and click on Analyze | Analyze Particles. Set the size to 10-Infinity.
- In ROI manager, click on Show all.
- Finding RPA2 foci in the nucleus (Figure 4G,H and Supplementary Video S1)
- Open the RPA2 image.
- Choose Process | Find Maxima. Set the prominence to a value that highlights the RPA2 foci (between 500 and 750), separating it from the background.
- Finally, click the Measure button in ROI Manager.
- Calculate the total number of nuclear ssDNA foci by dividing the value in the RawinDen column by 255 (the maximum value of pixel intensity in each foci).
- Perform statistical analysis using the preferred statistical software tool.
NOTE: Exclude all EdU-positive cells and improperly segmented DAPI masks from the analysis.
- Make nuclear masks using the DAPI staining (Figure 4A-F and Supplementary Video S1).
Representative Results
To overcome the limitations of detecting ssDNA in G1, we utilized RPA2, which enhances both the specificity and intensity of ssDNA foci detection35. To achieve precise cell synchronization, we used RPE1 cells that can be efficiently serum-starved and synchronized into G0 phase. They can then be induced to re-enter the cell cycle by the addition of serum following serum deprivation. To confirm the synchronization efficiency, we labeled the cells with EdU and their DNA content with propidium-iodide. We further gathered qualitative and quantitative results via flow cytometry (Supplementary Figure S1A). The dot-plots show that after 72 h of serum starvation, ~98% of the cells are in G0 phase. Following the addition of serum-containing media for 6 h, cells reenter the cell cycle (as seen by the increase in p27 levels in Figure 1A), having ~97% of cells in G1, while only having <1% cells in S phase, <2% cells in G2 phase (Supplementary Figure S1A). After 20-28 h of addition of serum to the cells, they gradually pass through S phase, as shown by the flow cytometry plots (Supplementary Figure S1A). This cell synchronization protocol gives a ~97% pure G1 population (6 h post serum addition following 72 h of serum starvation). To further validate the synchronization efficiency, we compared the expression of cell cycle markers following serum release using western blotting (Figure 1A and Supplementary Figure S1B) and in parallel, performed an EdU incorporation assay to visualize DNA replication. The EdU staining also highlights synchronization efficiency and the lack of DNA replication in G1 phase (Figure 1B,C).
Conventional methods to detect ssDNA in mammalian cells rely on the detection of BrdU in ssDNA. Figure 2A demonstrates that upon H2O2 and neocarzinostatin (NCS) treatment, the BrdU foci were detectable only in S phase cells, while no ssDNA foci were detectable in non-S phase cells. The BrdU antibody staining also showed a noticeable nucleolar background staining that could be detected in all the nuclei, independent of the cell cycle stage or treatments applied. Using the EdU click protocol described here, we could not detect co-localizing EdU and BrdU foci, which is evident in the untreated samples of Figure 2A. To completely rule out any BrdU signal that arose from cross-reactivity, we avoided EdU labeling and rather used cyclin A2 as an S-G2 marker. However, cyclin A2 staining did not allow CSK pre-extraction, and under this condition, we did not see any BrdU foci, even after genotoxic stress (Supplementary Figure S2A). This highlights the fact that CSK pre-extraction is necessary for anti-BrdU-based ssDNA staining. As a control, we tested BrdU antibody staining under denaturing conditions. This opens the DNA to expose the incorporated BrdU, which reveals that BrdU was uniformly incorporated (Supplementary Figure S2B).
In contrast, RPA2 staining shows NCS- and H2O2-dependent foci formation not only in the S phase but also in other cell cycle phases (Figure 2B). As a control, we also treated the cells with HU, which only causes ssDNA accumulation in cells undergoing replication. As expected, we only detected a signal increase upon HU treatment with the RPA2 antibody in EdU-positive cells, highlighting the specificity of this approach. The RPA2 antibody can also detect naturally occurring ssDNA formation during replication in the absence of exogenous genotoxic stress (Figure 2B). The highly sensitive nature of the RPA2 antibody prompted us to try to utilize it in the G1 phase where conventional BrdU staining failed to detect any signal upon genotoxic stress (Supplementary Figure S2C). Figure 3A shows that the formation of ssDNA foci upon H2O2 treatment was detectable when using an anti-RPA2 antibody, even in G1. There was a significant increase in the number of RPA2 foci in these nuclei upon H2O2 treatment (Figure 3B). These foci were specific to RPA2 as silencing of RPA2 abolished the IF signal (Figure 3A,B). Figure 3C and Supplementary Figure S1C show the efficiency of RPA2 silencing in these cells. Compared to conventional methods, RPA2-based detection of ssDNA is highly sensitive, and its application can therefore be extended to G1 phase cells.
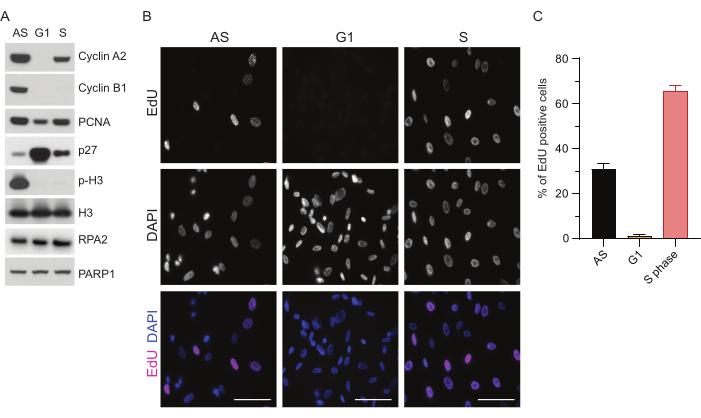
Figure 1: Synchronization efficiency of RPE1 cells following serum starvation. (A) Immunoblots show indicated protein levels in asynchronous, G1, and S phase synchronized RPE1 cells. (B) Representative images show asynchronous, G1, and S phase synchronized RPE1 cells that were exposed to 10 µM EdU for 30 min before fixation and visualized by Click-IT reaction. DAPI was used to counterstain nuclear DNA. Scale bars = 50 µm. (C) Graph shows percentage of EdU-positive cells over the total cell population assessed by DAPI. The error bar represents standard error of mean, and the analyzed numbers of nuclei were the following: AS n = 219, G1 n = 630, S n = 437. Abbreviations: RPE1 = hTERT-immortalized retinal pigment epithelial cells; AS = asynchronous; EdU = 5-ethynyl-2'-deoxyuridine; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
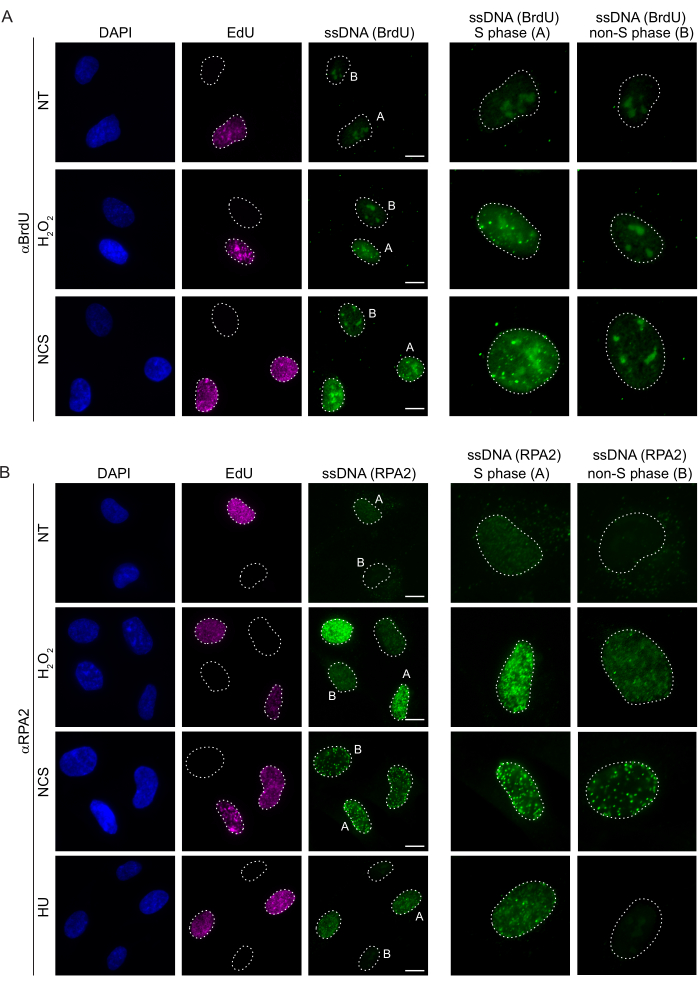
Figure 2: ssDNA detection with either BrdU antibody or RPA2 antibody upon DNA damage. (A) Representative images illustrate ssDNA foci using αBrdU (green), S phase cells are highlighted by EdU (purple), and DAPI was used to counterstain nuclear DNA (blue). RPE1 cells were kept in 10 µM BrdU for 48 h prior to any additional treatment. After 48 h, cells were pulsed with 10 µM EdU for 30 min followed by treatment of H2O2 (250 μM) for 1 h or Neocarzinostatin (0.5 μg/mL) for 4 h. Cells were fixed after CSK pre-extraction. A white dashed line denotes the border of each nucleus. Scale bar = 5 µm. Panels on the right are enlarged images of the indicated S phase or non-S phase nuclei. (B) Representative images illustrate ssDNA foci using αRPA2 antibody (green). S phase cells are highlighted by EdU (purple), and DAPI was used to counterstain nuclear DNA (blue). RPE1 cells were pulsed with 10 µM EdU for 30 min followed by either 1 h H2O2 (250 μM), 4 h of Hydroxyurea (2 mM), or 4 h of NCS (0.5 μg/mL). Cells were fixed after CSK pre-extraction. A white dashed line denotes the border of each nucleus. Scale bar = 10 µm. Panels on the right are enlarged images of the indicated S phase or non-S phase nuclei. Abbreviations: ssDNA = single-stranded DNA; BrdU = 5-bromo-2'-deoxyuridine; DAPI = 4',6-diamidino-2-phenylindole; RPE1 = hTERT-immortalized retinal pigment epithelial cells; EdU = 5-ethynyl-2'-deoxyuridine; NCS = Neocarzinostatin; HU = hydroxyurea. Please click here to view a larger version of this figure.
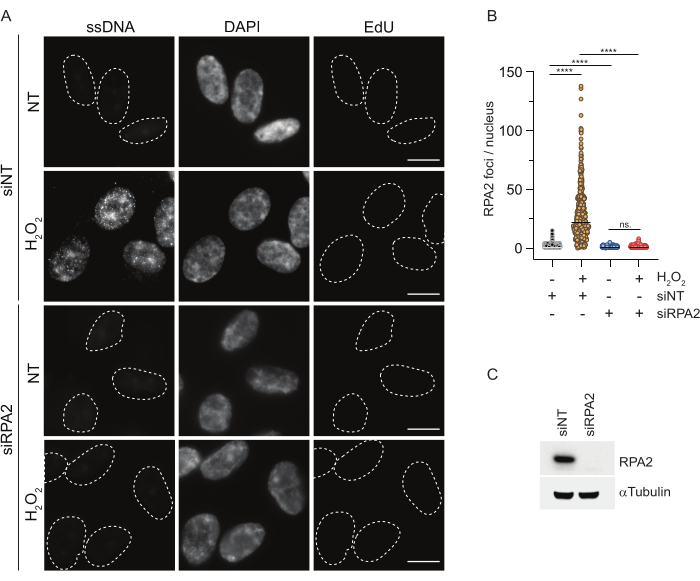
Figure 3: Detection of ssDNA foci in G1 phase using RPA2 antibody. (A) RPE1 cells were transfected with either siRNAs targeting RPA2 or a non-targeting siRNA control, and subsequently synchronized in G1 and pulse-labeled with 10 µM EdU for 30 min before treating them with H2O2 (250 μM) for 1 h where indicated. DAPI was used to counterstain nuclear DNA. Cells were fixed after CSK pre-extraction. A white dashed line denotes the border of each nucleus. Scale bar = 5 µm. (B) The measurements for the number of RPA2 foci/nucleus were carried out from two independent experiments. Only EdU-negative cells were considered during the analysis. Lines represent the mean value on the plots. Non-parametric ANOVA test (Kruskal-Wallis) was performed for statistical analysis. The stars indicate P < 0.0001. The analyzed number of nuclei were the following: siNT no H2O2 n = 513, siNT H2O2 n = 603, siRPA2 no H2O2 n = 266, siRPA2 H2O2 n = 536. (C) The efficiency of the siRNA knockdown is shown in the immunoblotting. Abbreviations: siNT = non-targeting siRNA control; BrdU = 5-bromo-2'-deoxyuridine; DAPI = 4',6-diamidino-2-phenylindole; RPE1 = hTERT-immortalized retinal pigment epithelial cells; EdU = 5-ethynyl-2'-deoxyuridine. Please click here to view a larger version of this figure.
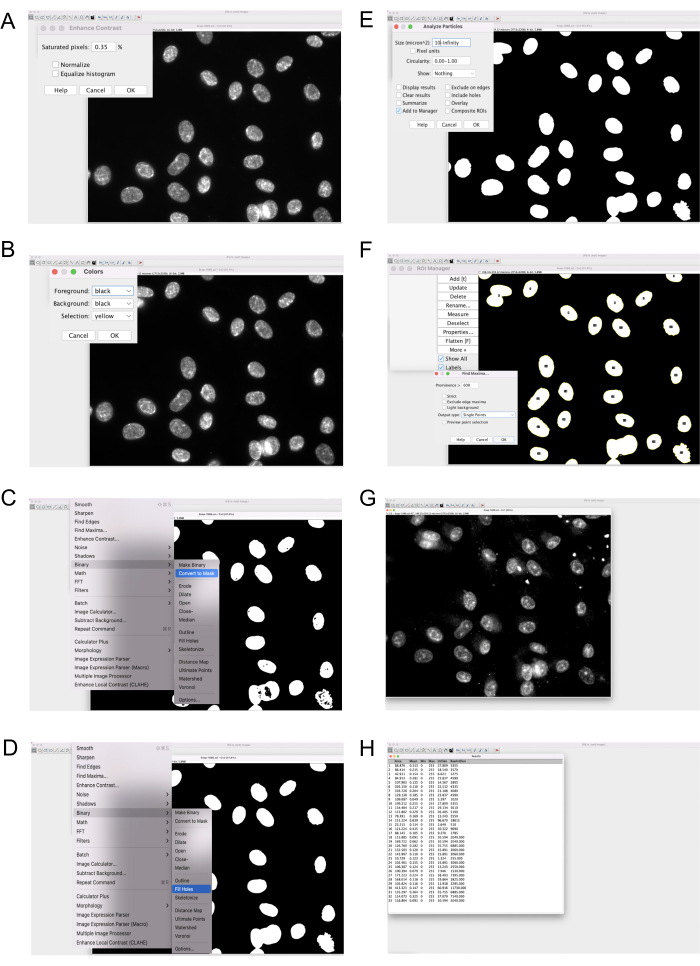
Figure 4: Quantification of ssDNA foci using Fiji. Detailed steps in Fiji showing how to assess RPA2 foci numbers in the nucleus. (A–E) The creation of a nuclear mask using the DAPI channel. (F–H) Thresholding to identify individual nuclear ssDNA foci from the background signal. Abbreviations: ssDNA = single-stranded DNA; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
| Cytoskeletal (CSK) buffer | |
| PIPES pH 7.0 | 10 mM |
| NaCl | 100 mM |
| EDTA pH 8 | 1 mM |
| MgCl2 | 3 mM |
| D-sucrose | 300 mM |
| Triton X-100 | 0.20% |
| Phosphatase inhibitor cocktail | 1 tablet per 10 mL |
| Protease inhibitor cocktail | 1 tablet per 10 mL |
| diluted in ddH2O | n/a |
| Washing buffer | |
| Triton X-100 | 0.05% |
| diluted in PBS | n/a |
| Permeabilization buffer | |
| Triton X-100 | 0.50% |
| diluted in PBS | n/a |
| Fixation solution | |
| Paraformaldehyde | 3.60% |
| Triton X-100 | 0.05% |
| diluted in PBS | n/a |
| Blocking buffer | |
| Bovine serum albumin (BSA) | 5% |
| Triton X-100 | 0.10% |
| diluted in PBS | n/a |
| Click-iT Plus reaction cocktail | |
| 1x Click-iT reaction buffer | 435 mL |
| Alexa Fluor PCA solution | 5 mL |
| CuSO4-copper protectant premix | 10 mL |
| 1x Click-iT buffer additive | 50 mL |
| Total volume | 500 mL |
Table 1: Composition of the buffers used in this protocol.
Supplementary Figure S1. (A) RPE1 cells were synchronized to G0 phase using serum starvation for 72 h and subsequently released into different cell cycle phases by reintroducing serum. Dot plots show cells in G0/G1, S, or G2/M phases, where hours indicate the time after the re-addition of serum following serum starvation. Graph on the right shows the percentage of G0/G1, S, and G2/M cells in each condition. FACS analysis was carried out using a commercially available cell proliferation kit using EdU and propidium iodide according to the manufacturer's recommendations. (B) Uncropped western blot scans for Figure 1. Numbers show molecular weight markers in kDa. PARP1 was used as a loading control and loaded on the gel that was also developed against CCNA2, p27 (further stripped for PCNA), and pH3 (S10) (further stripped for H3) by cutting up the membrane. CCNB1 and RPA2 were loaded onto a separate gel, using the same amount of protein lysate to ensure comparability. (C) Uncropped western blot scans for Figure 3. Numbers show molecular weight markers in kDa. Abbreviation: EdU = 5-ethynyl-2'-deoxyuridine. Please click here to download this File.
Supplementary Figure S2: (A) Representative images illustrate ssDNA foci using BrdU antibody (green); S phase cells are highlighted by cyclin A2 (red); and DAPI was used to counterstain nuclear DNA (blue). RPE1 cells were kept in 10 µM BrdU for 48 h prior to further treatment. After 48 h, cells were treated with H2O2 (250 μM) for 1 h or Neocarzinostatin (0.5 µg/mL) for 4 h before fixation. A white dashed line denotes the border of each nucleus. Scale bar = 5 µm. (B) BrdU staining of RPE1 cells with and without denaturing condition. Asynchronous RPE1 cells were pre-treated with 10 µM BrdU for 48 h. Scale bar = 10 µm. (C) The measurements for the number of BrdU foci/nucleus were carried out from two independent experiments in G1 synchronized RPE1 cells. Only EdU-negative cells were considered during the analysis. Lines represent the mean value on the plots. Non-parametric ANOVA test (Kruskal-Wallis) was performed for statistical analysis. The 'ns' indicates non-significant difference. The analyzed number of nuclei were the following: NT n = 52, NCS n = 105, H2O2 n = 82. Abbreviations: siNT = non-targeting siRNA control; BrdU = 5-bromo-2'-deoxyuridine; DAPI = 4',6-diamidino-2-phenylindole; RPE1 = hTERT-immortalized retinal pigment epithelial cells; NCS = Neocarzinostatin. Please click here to download this File.
Supplementary Video S1: Screen recording of Fiji-based RPA2 foci analysis. Please click here to download this File.
Discussion
Maintaining a healthy, mycoplasma-free cell culture is critical for all the experiments described above. RPE1 cells have a strong attachment to tissue culture-treated plasticware under normal culturing media; however, their binding characteristics significantly diminish when kept in serum-free conditions. Additionally, to capture high-resolution images of ssDNA foci under a microscope, the cells need to be plated onto a 0.17 mm thick cover glass, which is not hydrophilic enough to support proper attachment of RPE1 cells. Without properly flattened and evenly distributed cells, it is very challenging to visualize individual ssDNA foci. Therefore, it is critical to choose the proper coating material (e.g., vitronectin) and to leave adequate time (6-12 h) for the cells to spread and attach after releasing them into G1 phase.
A challenging part of the protocol is to obtain homogeneous G1 synchronized RPE1 cells. This requires two critical steps. First, for efficient serum starvation, the cells need to be trypsinized, washed thoroughly with PBS, and directly seeded onto new tissue culture dishes using serum-free media. Washing the cells directly in tissue culture dishes to remove the serum will not yield efficient G0 synchronization. Second, when releasing cells into G1 phase, the cells must be trypsinized again and seeded onto fresh tissue culture plates. Similarly, just changing the medium and adding serum-containing culturing medium to the cells will not result in a synchronous G1 entry. Additionally, for proper G1 entry, the seeding density of the cells on the coated cover glasses must be at certain confluency levels. While perfect cell synchronization is generally unachievable, this synchronization protocol described here gives a ~97% pure G1 population. The recommended seeding density for RPE1 on a 12 mm diameter coverslip is ~4 × 104 to acquire a homogeneous field of view for imaging, with approximately 70% confluency. Higher seeding density causes cells to detach and "peel-off" after CSK extraction and will result in a higher background signal during image acquisition.
To reduce any background signal and achieve a favorable signal-to-noise ratio, thorough washing after primary and secondary antibody incubation is essential. Since numerous washing steps are to be applied, it is also essential to prevent the well from drying out during each washing step. We minimize this artifact by applying a minimum of 0.05% Triton X-100 in all the washing and incubation steps. Once the wells dried out, the cells displayed an altered signal-to-noise ratio; this leads to a mosaic-like pattern under the microscope and could interfere with evaluation. Z-stack image acquisition combined with deconvolution can assist in capturing foci in different focal planes to improve the analysis.
Conventional methods rely on the detection of incorporated BrdU under non-denaturing conditions. These methods, however, depend on the pretreatment of the cells with high dosages of BrdU for at least 1-2 days (or time equivalent to a full cell cycle in the cell line used) to ensure uniform genomic incorporation. Undesirably, extensive BrdU incorporation can cause cell cycle interference36. To address these limitations, this method utilizes endogenous RPA2 to detect ssDNA foci. This approach does not require replication-driven BrdU incorporation, it can also be used in post-mitotic cells. Since extensive BrdU incorporation is not needed, this saves time and reduces experimental complexity. By using RPA2 staining to visualize ssDNA, we can use 2′-deoxy-5-ethynyluridine (EdU) and click-chemistry to mark DNA replication while avoiding possible crossreactivity of the BrdU antibodies against EdU27,37,38. Special care must be taken to properly mask the incorporated EdU during the click-reaction so that the BrdU antibodies will not cross-react with EdU27,39.
Finally, an important benefit of utilizing RPA2 instead of BrdU is simply having a superior signal-to-noise ratio when compared to BrdU staining outside S phase. We found that the non-denaturing BrdU staining and its ability to visualize ssDNA is restricted to S phase even in replicating cells (Figure 2). BrdU antibody binds only to the sufficiently exposed BrdU in ssDNA stretches. The binding of repair proteins, including RPA2, to the ssDNA stretches may suppress or hamper the sufficient exposure of BrdU in ssDNA. We also found that CSK pre-extraction is necessary for ssDNA visualization using BrdU antibody. This is possible because the ssDNA tracks are not accessible for the antibody without removing lightly bound protein components from them.
Nonetheless, there are some limitations associated with this protocol. A limitation of using RPA2 for ssDNA detection is the need to optimize the CSK pre-extraction step. Unbound, excess RPA2 must be washed away from the DNA before fixing the cells. On the one hand, underextraction leads to a high background due to the RPA2 protein fraction that is not bound to ssDNA. On the other hand, overextraction will lead to signal loss. For BrdU detection, this is not a variable since BrdU is stably incorporated into the DNA and cannot be washed away by pre-extraction. Therefore, the time of the CSK pre-extraction, the amount of Triton X-100 in the buffer, the volume, and the temperature at which the pre-extraction is performed must be carefully considered. CSK pre-extraction also limits the use of nucleus size to discriminate G0/G1 cells from S/G2 cells.
Additionally, we cannot exclude the possibility that some of the signal that comes from RPA2 originates from it being bound to other chromatin-binding protein interactors. One also must consider the species specificity of the RPA2 antibody. The antibody used in this protocol can recognize human, mouse, rat, hamster, and monkey RPA2. Another limitation of this approach is that not all cell lines can be serum-starved for G0 synchronization. Most cancer cell lines can bypass cell cycle checkpoints and proliferate even in serum-deprived media. Though serum starvation is beneficial, since it does not cause DNA damage, one must carefully monitor their cell synchronization efficiency to make sure that proper cell cycle phase enrichment is achieved. For cells that do not respond to serum deprivation, other cell synchronization methods must be considered (e.g., mitotic shake off, CDK1 inhibition for G2 arrest, or non-invasive techniques such as centrifugal elutriation). Another possible method is using high-content imaging to measure EdU and nuclear DNA content for cell cycle profiling of asynchronous cells31. One must consider the implications of utilizing alternate synchronization methods to prevent interference with downstream analysis. For instance, the use of double thymidine block or aphidicolin, often used in literature, will result in replication stress and DNA damage40.
The investigation of DNA repair mechanisms continues to be a focal point of discussion in the fields of cancer and cell biology. The protocol presented here offers a valuable approach for the preparation of cells, enabling the visualization and quantitative analysis of ssDNA upon exposure to DNA-damaging agents. Notably, this protocol highlights the utilization of the ssDNA binding protein, RPA2, demonstrating its high specificity to visualize small amounts of ssDNA foci while avoiding unwanted cross-reactivity in all the cell cycle phases. Using RPA2 confers numerous advantages, particularly for researchers aiming to analyze cells in the G1 phase of the cell cycle. This protocol considers several limitations and addresses concerns related to signal interference, undesired background noise, and cross-reactivity when using RPA2 or BrdU staining to detect ssDNA.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Michele Pagano for his support and helpful insights, Ashley Chui and Sharon Kaisari for critically reading the manuscript, and Jeffrey Estrada and Vilma Diaz for their continuous support. This work was supported by a diversity supplement to the National Institutes of Health grant GM136250.
Materials
| Alpha-tubulin antibody | Sigma-Aldrich | T6074 | primary antibody (1:5,000) |
| Axio Observer Inverted Microscope | Zeiss | na | microscope |
| Bis-Tris Plus Mini Protein Gels, 4-12% | Invitrogen | NW04127BOX | Western Blot |
| Bovine Serum Albumin | Jackson ImmunoResearch | 001-000-162 | blocking |
| BrdU (5-Bromo-2'-deoxyuridine) | Sigma-Aldrich | B5002-100MG | nucleotide analogue |
| BrdU antibody BU1/75 | Abcam | ab6326 | primary antibody (1:500) |
| CellAdhere Dilution Buffer | Stemcell Technologies | 07183 | coating reagent |
| Click-iT Plus EdU Flow Cytometry Assay Kits | Invitrogen | C10632 | flow cytomery |
| Click-iT Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor 647 dye | Thermo Fisher Scientific | C10640 | click-reaction kit |
| cOmplete ULTRA Protease inhibitor tablets | Sigma-Aldrich | 5892791001 | reagent |
| Countess 3 Automated cell counter | Thermo Scientific | AMQAX2000 | cell counter |
| Coverslip | neuVitro | GG12PRE | tissue culture |
| Cyclin A2 antibody | Santa Cruz Biotechnology | sc-271682 | primary antibody (1:1,000) for IF and WB |
| Cyclin B1 antibody | Santa Cruz Biotechnology | sc-245 | primary antibody (1:5,000) |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D2650-100ML | vehicle control |
| DMEM, high glucose, with HEPES | Gibco | 12430051 | cell culture medium for RPE cells |
| DPBS, no calcium, no magnesium | Gibco | 14190144 | the PBS used throughout the protocol |
| D-Sucrose | Thermo Fisher Scientific | bp220-1 | reagent |
| Eclipse Ti2 Series Epifluorescent Microscope | Nikon | na | microscope |
| EdU (5-Ethynyl-2'-deoxyuridine) | Thermo Fisher Scientific | C10637 | nucleotide analog |
| Falcon 24-well plate | Corning | 351147 | tissue culture |
| Falcon Cell Culture Dishes 100 mm | Corning | 353003 | tissue culture |
| Fetal Bovine Serum, heat inactivated | Gibco | 16140071 | media supplement |
| Fiji (ImageJ) | NIH | version 1.54f | software and algorithms |
| FxCycle PI/RNase Staining Solution | Invitrogen | F10797 | PI staining |
| Goat anti-mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 | Thermo Fisher Scientific | A21422 | secondary antibody (1:1,000) |
| Goat anti-rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 | Thermo Fisher Scientific | A48262 | secondary antibody (1:1,000) |
| Histone H3 antibody | Abcam | ab1791 | primary antibody (1:10,000) |
| hTERT RPE1 | ATCC | CRL-3216 | cell line |
| Hydrochloric acid | Sigma-Aldrich | H1758-100ML | reagent |
| Hydrogen peroxide 30% soultion | Sigma-Aldrich | H1009-100ML | reagent |
| Hydroxyurea,98% powder | Sigma-Aldrich | H8627-5G | reagent |
| Invitrogen Ultra Pure 0.5 M EDTA pH 8.0 | Thermo Fisher Scientific | 15-575-020 | reagent |
| Lipfectamine RNAiMAX Transfection Reagent | Invitrogen | 13778150 | transfection reagent |
| Magnesium chloride solution 1 M | Sigma-Aldrich | M1028-100ML | reagent |
| MycoFluor | Thermo Fisher | M7006 | Mycoplasma Detection Kit |
| Neocarzinostatin from Streptomyces carzinostaticus | Sigma-Aldrich | N9162-100UG | reagent |
| NuPage MES SDS Running Buffer (20x) | Invitrogen | NP0002 | Western Blot |
| onTARGETplus Human RPA2 siRNA | Dharmacon | L-017058-01-0005 | siRNA |
| p27 antibody | BD Biosciences | 610241 | primary antibody (1:1,000) |
| Paraformaldehyde aqueous solution (32%) | Electron Microscopy Sciences | 50-980-494 | fixative |
| PARP1 antibody | Cell Signaling Technology | 9542S | primary antibody (1:1,000) |
| PCNA antibody | Cell Signaling Technology | 13110S | primary antibody (1:2,000) |
| Penicillin-Streptomycin | Gibco | 15140163 | media supplement |
| pH3 antibody | Cell Signaling Technology | 3377S | primary antibody (1:2,000) |
| PhosSTOP phosphatase inhibitor tablets | Sigma-Aldrich | 4906837001 | reagent |
| PIPES Buffer 0.5 M solution, pH 7.0 | Bioworld | 41620034-1 | reagent |
| Precision Plus Protein Kaleidoscope Prestained Protein Standards | Bio-Rad | 1610395 | Western Blot |
| Prism | GraphPad | version 10 | statistical analysis and graph |
| ProLong Diamond Antifade Mountant | Thermo Scientific | P36961 | mounting media |
| Reduced serum media (Opti-MEM) | Gibco | 31985070 | used for transfection |
| Rpa32/rpa2 antibody (mouse) | EMD Millipore | NA19L | primary antibody (1:1,000) for WB |
| Rpa32/rpa2 antibody (rat) | Cell Signaling Technology | 2208S | primary antibody (1:1,000) for IF |
| Sodium Chloride solution (5 M) | Sigma-Aldrich | S5150 | reagent |
| Sodium Pyruvate (100 mM) | Gibco | 11360070 | media supplement |
| Sodium tetraborate decahydrate | Sigma-Aldrich | B3535-500G | reagent |
| Thermo Scientific Pierce DAPI Nuclear Counterstain | Thermo Scientific | 62248 | nucleic acid stain |
| Thymidine,powder | Sigma-Aldrich | T1985-1G | reagent |
| Triton X-100 aqueous solution (10%) | Sigma-Aldrich | 11332481001 | detergent |
| Trypsin-EDTA (0.5%), no phenol red | Gibco | 1540054 | cell dissociation agent |
| Vitronectin XF | Stemcell Technologies | 07180 | coating reagent |
| ZE5 Cell Analyzer | Bio-Rad | na | flow cytomery |
References
- Hakem, R. DNA-damage repair; the good, the bad, and ugly. EMBO J. 27 (4), 589-605 (2008).
- Gutierrez, R., O’Connor, T. R. DNA direct reversal repair and alkylating agent drug resistance. Cancer Drug Resist. 4 (2), 414-423 (2021).
- Krokan, H. E., Bjoras, M. Base excision repair. Cold Spring Harb Perspect Biol. 5 (4), a012583 (2013).
- Marteijn, J. A., Lans, H., Vermeulen, W., Hoeijmakers, J. H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 15 (7), 465-481 (2014).
- Li, G. M. Mechanisms and functions of DNA mismatch repair. Cell Res. 18 (1), 85-98 (2008).
- Hustedt, N., Durocher, D. The control of DNA repair by the cell cycle. Nat Cell Biol. 19 (1), 1-9 (2016).
- Yang, W., Gao, Y. Translesion and repair DNA polymerases: diverse structure and mechanism. Annu Rev Biochem. 87, 239-261 (2018).
- Bhat, D. S., et al. Therapeutic disruption of RAD52-ssDNA complexation via novel drug-like inhibitors. NAR Cancer. 5 (2), zcad018 (2023).
- Gupta, P., Saha, B., Chattopadhyay, S., Patro, B. S. Pharmacological targeting of differential DNA repair, radio-sensitizes WRN-deficient cancer cells in vitro and in vivo. Biochem Pharmacol. 186, 114450 (2021).
- Pena-Diaz, J., et al. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol Cell. 47 (5), 669-680 (2012).
- Schroering, A. G., Edelbrock, M. A., Richards, T. J., Williams, K. J. The cell cycle and DNA mismatch repair. Exp Cell Res. 313 (2), 292-304 (2007).
- Scully, R., Panday, A., Elango, R., Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 20 (11), 698-714 (2019).
- Escribano-Diaz, C., et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 49 (5), 872-883 (2013).
- Genschel, J., Modrich, P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 12 (5), 1077-1086 (2003).
- Hu, J., et al. Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 288 (29), 20918-20926 (2013).
- Huertas, P., Jackson, S. P. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 284 (14), 9558-9565 (2009).
- Keijzers, G., et al. Human exonuclease 1 (EXO1) regulatory functions in DNA replication with putative roles in cancer. Int J Mol Sci. 20 (1), 74 (2018).
- Symington, L. S. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb Perspect Biol. 6 (8), a016436 (2014).
- Liu, Y., et al. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 280 (5), 3665-3674 (2005).
- Wold, M. S., Kelly, T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 85 (8), 2523-2527 (1988).
- Wienholz, F., Vermeulen, W., Marteijn, J. A. Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Res. 45 (9), e68 (2017).
- Wold, M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 66, 61-92 (1997).
- Chen, R., Wold, M. S. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays. 36 (12), 1156-1161 (2014).
- Kang, Y., et al. Alteration of replication protein A binding mode on single-stranded DNA by NSMF potentiates RPA phosphorylation by ATR kinase. Nucleic Acids Res. 51 (15), 7936-7950 (2023).
- Kilgas, S., Kiltie, A. E., Ramadan, K. Immunofluorescence microscopy-based detection of ssDNA foci by BrdU in mammalian cells. STAR Protoc. 2 (4), 100978 (2021).
- Madabhushi, R., Pan, L., Tsai, L. H. DNA damage and its links to neurodegeneration. Neuron. 83 (2), 266-282 (2014).
- Liboska, R., Ligasova, A., Strunin, D., Rosenberg, I., Koberna, K. Most anti-BrdU antibodies react with 2′-deoxy-5-ethynyluridine — the method for the effective suppression of this cross-reactivity. PLoS One. 7 (12), e51679 (2012).
- Biehs, R., et al. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol Cell. 65 (4), 671-684.e5 (2017).
- Cruz-Garcia, A., Lopez-Saavedra, A., Huertas, P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 9 (2), 451-459 (2014).
- Ercilla, A., et al. Physiological tolerance to ssDNA enables strand uncoupling during DNA replication. Cell Rep. 30 (7), 2416-2429.e7 (2020).
- Lezaja, A., et al. RPA shields inherited DNA lesions for post-mitotic DNA synthesis. Nat Commun. 12 (1), 3827 (2021).
- Mukherjee, B., Tomimatsu, N., Burma, S. Immunofluorescence-based methods to monitor DNA end resection. Methods Mol Biol. 1292, 67-75 (2015).
- Ochs, F., et al. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol. 23 (8), 714-721 (2016).
- Raderschall, E., Golub, E. I., Haaf, T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci U S A. 96 (5), 1921-1926 (1999).
- Forment, J. V., Walker, R. V., Jackson, S. P. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 81 (10), 922-928 (2012).
- Mistrik, M., et al. Cells and stripes: A novel quantitative photo-manipulation technique. Sci Rep. 6, 19567 (2016).
- Aten, J. A., Bakker, P. J., Stap, J., Boschman, G. A., Veenhof, C. H. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem J. 24 (5), 251-259 (1992).
- Podgorny, O., Peunova, N., Park, J. H., Enikolopov, G. Triple S-phase labeling of dividing stem cells. Stem Cell Reports. 10 (2), 615-626 (2018).
- Cappella, P., Gasparri, F., Pulici, M., Moll, J. Cell proliferation method: click chemistry based on BrdU coupling for multiplex antibody staining. Curr Protoc Cytom. Chapter 7, (2008).
- Ligasova, A., Koberna, K. Strengths and weaknesses of cell synchronization protocols based on inhibition of DNA synthesis. Int J Mol Sci. 22 (19), 10759 (2021).

