Solid Plate-based Dietary Restriction in Caenorhabditis elegans
Instructor Prep
concepts
Student Protocol
1. Establishing a calibration curve for the estimation of bacteria concentrations
This section describes a method for the estimation of bacteria concentration for any given bacteria strain (e.g. OP50, HT115) that will be used as a food source for Dietary restriction experiments in C. elegans. Separate calibration curves should be established for each bacteria strain used.
- Inoculate a single colony of E. coli OP50 bacteria picked from a fresh streak of bacteria on LB agar in 3 mL liquid LB broth.
- Place OP50 culture in 37 °C shaker and allow bacteria to grow overnight.
- Prepare serial 2-fold dilutions of the bacterial suspension from the overnight culture (2x, 4x, 8x, 16x, 32x).
- Measure the absorbance (optical density) of each dilution at 600 nm by spectrophotometer in triplicate.
- Further dilute each bacterial suspension 1,000,000-fold (1000-fold dilution twice).
- Spread 0.4 mL of bacterial suspension of each dilution evenly on a 100 mm LB agar plate.
- Place LB plates with OP50 in a 37 °C incubator for 18-24 hours.
- Count the number of bacterial colonies grown on each plate. The colony counts are used to calculate cfu (colony forming unit) per mL for each sample.
- The relationship between measured OD600 values and OP50 bacteria concentrations can be established by plotting OD600 as a function of bacterial concentration (cfu/mL) and performing a linear regression analysis (Figure 1).
Solutions:
| Luria Broth (LB), 1L: | |
| 10 g | Bacto-tryptone |
| 5 g | Yeast Extract |
| 10 g | NaCl |
Add deionized water to 1L.
Autoclave and store at room temperature.
2. Preparation of solid plate-based dietary restriction (sDR) plates
This section describes the preparation of plates for use in solid-plate based dietary restriction (sDR) experiments. Normally, we consider a bacteria concentration of 1 x 1011 cfu/mL as the ad libitum feeding condition and 1 x 108 cfu/mL as the optimal DR feeding condition for this method. However, 4-6 different bacteria concentrations may be needed to establish a complete dose-dependent relationship for sDR and its responses.
- Inoculate a single colony of E. Coli OP50 bacteria in 3 mL liquid LB broth.
- Place the OP50 culture in 37 °C shaker and allow the bacteria to grow for 6-8 hours.
- Transfer the OP50 culture to a new flask containing 500 mL LB broth, and place the OP50 culture back in 37 °C shaker overnight.
- Measure the OD600 of the OP50 culture to determine the bacteria concentration (the concentration is calculated using the calibration curve generated in Part 1). Appropriate dilutions may be needed for an accurate measurement of absorbance.
- Pellet OP50 bacteria by centrifugation at 1,500 g for 20 minutes.
- Resuspend bacteria to a concentration of 1 x 1012 cfu/mL.
- Prepare 10 mL bacteria culture in six different concentrations: 1 x 1012, 1011, 1010, 109, 108, 107 cfu/mL.
- Pipette 0.2 mL of bacteria culture into the center of a 60 mm solidified NGM plate. Avoid touching the surface of the plate with pipette tip, as nicks on the surface allow worms to burrow into the agar plates.
- Place plates in 37 °C incubator for 1 hour.
- Apply 10μL of 100mg/mL carbenicillin and 10 μL of 50mg/mL kanamycin to each plate to arrest growth of the bacteria.
- Store plates with different concentrations of antibiotic-killed bacteria at 4 °C for future use (up to one month). It is important to prepare and store enough plates for the entire planned experiment at least 1-2 days ahead. Using the same batch of plates for the entire experiments is preferable.
- To verify the viability of the bacteria, scrape some bacteria out of the sDR plates and spread it on an empty NGM plate with carbenicillin and kanamycin. Place the plate in 37 °C incubator for 6-8 hours and then check for growth of the bacteria (no bacteria growth should be observed).
Solutions
| Nematode Growth Media (NGM), 1L: | |
| 3 g | NaCl |
| 2.5 g | Bacto-Peptone |
| 20g | Agar |
Autoclave for 40 minutes and let cool to 65 °C, then add:
| 1 mL | 1 M MgSO4 |
| 1 mL | 1 M CaCl2 |
| 1 mL | 5 mg/mL Cholesterol |
| 25 mL | 1M Potassium Phosphate, pH 6.0 |
Immediately pour liquid NGM onto 60 mm x 15 mm petri dishes in 10 mL aliquots
3. Setting up sDR experiments
For most of the sDR-related studies, such as life span analysis and fertility profiling, preparation of an age-synchronized population of worms is essential. For other studies, simply transfer worms from a regular OP50 plate to an sDR plate to initiate the dietary restriction. In this section, a protocol to set up a lifespan analysis for sDR animals is described as an example.
- Use a platinum worm picker to transfer 20-30 reproductively active adults onto several regular NGM plates with OP50. Depending on the number of eggs required for the experiments, more adults may be needed.
- Leave the plates at 20°C for 4 hours to allow egg-laying.
- Remove the adults from the plate. Plates should be visually inspected for eggs.
- Leave the plates at 20°C and allow the progeny to develop into the Late-L4/day 1 adult stage. Usually, this takes 3 days for N2 wild types worms at 20 °C.
- Transfer a suitable number of day 1 adults onto sDR plate to initiate the dietary restriction.
- For a lifespan analysis, set up a total of 6-8 plates with 10-20 worms per plate for each sDR condition.
- Score the viability of each worm every 2 days until all the worms have died. It is critical that worms be transferred to fresh sDR plates every 12-48 hours to avoid complete deprivation of food.
4. Representative Results:
Shown in Figure 1 is a plot of a representative calibration curve for the estimation of OP50 concentration in relation to OD600. The equation generated by liner regression analysis can be used for all future calculations of OP50 bacteria concentrations. Shown in Figure 2 is an example of assaying the effect of chronic sDR treatments on mean lifespan of different worm strains. The blue line represents a normal dose-dependent response of wild-type worms to sDR. The red line indicates that the lifespan effect of sDR is partially suppressed by drr-2 o.e., while daf-16(mu86) has no effect on sDR-induced lifespan extension. Detail method for performing lifespan analysis in C. elegans is described in previous literature 1-3.
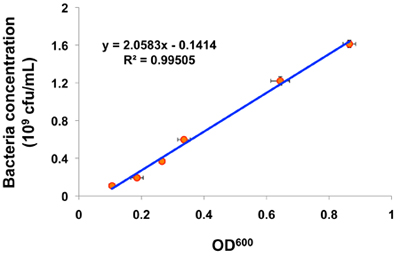
Figure 1. Representative result of a calibration curve for the estimation of E. Coli OP50 concentration. OP50 concentration is plotted as a function of measured OD600 values. Error bars = S.D. Overnight bacterial culture was diluted 1.5x, 2x, 4x, 8x, 16x, 32x before absorbance measurement, and further diluted 1,000x twice before cfu measurement.
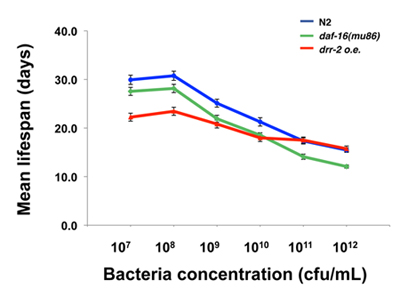
Figure 2. Representative result of a C. elegans lifespan experiment comparing wild type N2 animals (blue) with two different mutants (green and red) in response to sDR (Figure from Ching et al. 4). Error bars = S.E.M.
Solid Plate-based Dietary Restriction in Caenorhabditis elegans
Learning Objectives
List of Materials
| Name of the reagent | Company | Catalogue number | Comments |
| Carbenicillin | Duchefa Biochemie | C0109.25 | Dissolved in water. |
| Kanamycin | Roche | 10106801001 | Dissolved in water. |
| Bacto-peptone | Becton, Dickinson and company | DF0118072 | |
| Bacto-tryptone | Becton, Dickinson and company | DF0123075 | |
| Bacto-yeast extract | Becton, Dickinson and company | DF0127071 | |
| Agar | Becton, Dickinson and company | DF00145170 | |
| Sterile culture tube | USA Scientific | 1485-0810 | |
| Spectrophotometer | Amersham Biosciences | Gene Quant pro | Amersham is now part of GE Healthcare. |
| Dissecting Microscope | Zeiss | Stemi 2000 | |
| Centrifuge | Beckman Coulter | Accuspin FR |
Lab Prep
Reduction of food intake without malnutrition or starvation is known to increase lifespan and delay the onset of various age-related diseases in a wide range of species, including mammals. It also causes a decrease in body weight and fertility, as well as lower levels of plasma glucose, insulin, and IGF-1 in these animals. This treatment is often referred to as dietary restriction (DR) or caloric restriction (CR). The nematode Caenorhabditis elegans has emerged as an important model organism for studying the biology of aging. Both environmental and genetic manipulations have been used to model DR and have shown to extend lifespan in C. elegans. However, many of the reported DR studies in C. elegans were done by propagating animals in liquid media, while most of the genetic studies in the aging field were done on the standard solid agar in petri plates. Here we present a DR protocol using standard solid NGM agar-based plate with killed bacteria.
Reduction of food intake without malnutrition or starvation is known to increase lifespan and delay the onset of various age-related diseases in a wide range of species, including mammals. It also causes a decrease in body weight and fertility, as well as lower levels of plasma glucose, insulin, and IGF-1 in these animals. This treatment is often referred to as dietary restriction (DR) or caloric restriction (CR). The nematode Caenorhabditis elegans has emerged as an important model organism for studying the biology of aging. Both environmental and genetic manipulations have been used to model DR and have shown to extend lifespan in C. elegans. However, many of the reported DR studies in C. elegans were done by propagating animals in liquid media, while most of the genetic studies in the aging field were done on the standard solid agar in petri plates. Here we present a DR protocol using standard solid NGM agar-based plate with killed bacteria.
Procedure
Reduction of food intake without malnutrition or starvation is known to increase lifespan and delay the onset of various age-related diseases in a wide range of species, including mammals. It also causes a decrease in body weight and fertility, as well as lower levels of plasma glucose, insulin, and IGF-1 in these animals. This treatment is often referred to as dietary restriction (DR) or caloric restriction (CR). The nematode Caenorhabditis elegans has emerged as an important model organism for studying the biology of aging. Both environmental and genetic manipulations have been used to model DR and have shown to extend lifespan in C. elegans. However, many of the reported DR studies in C. elegans were done by propagating animals in liquid media, while most of the genetic studies in the aging field were done on the standard solid agar in petri plates. Here we present a DR protocol using standard solid NGM agar-based plate with killed bacteria.
