A Practical and Novel Method to Extract Genomic DNA from Blood Collection Kits for Plasma Protein Preservation
Instructor Prep
concepts
Student Protocol
1. Sample Collection
- Collect blood samples from individuals with proper informed consent. For each individual, draw 4 to 5 ml of blood into a P100 tube (BD Diagnostics, Franklin Lake, NY, USA). For comparison purposes, simultaneously collect blood in an EDTA tube.
- The BD P100 tubes contain 15.8 ml spray-dried K2EDTA and a lyophilized proprietary broad spectrum cocktail of protease inhibitors to prevent coagulation and stabilize the plasma proteins. They also contain a mechanical separator. After collecting the whole blood, keep both tubes at room temperature for 60 min to overnight until processing occurs.
- Centrifuge the BD P100 tubes at 2,500 g for fifteen minutes at room temperature. Transfer the plasma collected above the mechanical separator into micro centrifuge tubes.
- Store the P100 tubes, containing the blood compacted below the separator, at -80 °C until you are ready to begin the DNA extraction.
2.1 From blood of P100 tube (P100_DNA)
- The P100 tubes are stored at -80 °C after plasma extraction. Within 3 to 4 months, retrieve the P100 tube containing the compacted blood from the freezer and thaw at 37 ° in a water bath for 5 min (Figure 1A). Remove any residual plasma that sits above the separator. Add 2 ml of 17% sucrose solution (gradient solution tested in house – unpublished data) in the P100 tubes above the separator (Figure 1B). Centrifuge the tubes at room temperature for 20 min at 2,500 g; this process pushes the mechanical separator to the top of the tube and yields three layers of solution including the buffy coat (Figure 1C). Manually extract the separator using a steriled hooked metal paperclip (Figure 1D).
- After retrieving the mechanical separator from each tube (Figure 1D), remove and discard the top sucrose layer. Transfer the middle layer, representing the buffy coat (BC) layer, into a sterile 15 ml conical tube. Add Phosphate Buffer Saline (PBS) to increase the buffy coat volume to 3 ml. Extract the DNA from the buffy coat using reagents from the Qiagen Puregene Blood Kit as per the manufacturer’s recommendation; except a centrifugation speed of 2,500 g (instead of 2,000 g).
- To lyse and remove the red blood cells (RBC), add 9 ml of RBC lysis to 3 ml of buffy coat. Invert each tube several times and incubate at room temperature for 10 min with occasional inverting during incubation. Spin each mixture down at 2,500 g for 5 min and discard the supernatant. Treat the remaining pellet in each tube with 3 ml of cell lysis solution and vortex for 15 sec. Treat the lysate with 1 ml of protein precipitation solution and vortex for 15 sec.
- Centrifuge at 2,500 g for 5 min and transfer the supernatant into a fresh 15 ml conical tube containing 3 ml of 100% isopropanol. Invert the new tubes 10 times and centrifuge at 2,500 g for 3 min. Discard the supernatant and add 1 ml of 70% ethanol. Invert the conical tubes a few times to dislodge and wash the DNA pellet. Centrifuge at 2,500 g for 1 min. Allow the pellet to dry for 3 min (place the tube upside down) at room temperature and then suspend the DNA with 250 ml of rehydration solution at 37 °C for 10 min and transfer to a pre-labeled 1.7 ml microtube. Store the tubes at -80 °C until use.
2.2 From Blood of EDTA Ttube (EDTA_DNA)
- The whole blood for routine immunohematology testing is collected in plastic vaccutainer tubes spray-coated with 10.8 mg of K2EDTA and stored at -80 °C. Because the tubes do not contain a mechanical separator, the DNA extraction does not include the steps involving the mechanical separator (2.1.1 and 2.1.2).
- Within 3 to 4 months of blood storage, the DNA is extracted using the KingFisher Flex (Thermo Fisher Scientific, Waltham, MA, USA). It is an automation DNA extraction system based on magnetic particle technology and consists of cell lysis / DNA binding, several washing steps and DNA elution.
- The kit used in the DNA extraction is the KingFisher Flex 24 DNA Blood kit (Qiagen, Germany).
3. DNA Quantity and Quality Assessment
The quantity, quality and integrity of the genomic DNA were assessed by a combination of method that includes picogreen, spectroscopy and electrophoresis.
- The concentration of the genomic DNA is determined by Nanodrop and picogreen quantification.
- The purity is determined from the 260/280 ratio measurement on the Nanodrop spectrophotometer.
- The sample (500 ng) is also loaded on 1% agarose gel to assess the integrity (degradation) of the DNA.
4. Genotyping Assays and Quality Control
- Five P100_DNA and their corresponding EDTA_DNA samples are randomly selected for further analysis using the custom Immunochip (Immuno DNA Analysis BeadChip). Immunochip is an Infinium genotyping chip containing 196,524 polymorphisms and is designed for immunogenetic studies5.
- For each sample, 200 ng of DNA is amplified, fragmented, precipitated and resuspended in the appropriate hybridization buffer.
- The denatured samples are hybridized on Immunochips at 48 °C for a minimum of 16 hr.
- After hybridization, the Immunochips are processed for single-base extension reactions and stained.
- The immunochips are then imaged using Illumina Hiscan Bead array reader and the normalized bead intensity data is loaded into the Illumina GenomeStudio software and converted into genotypes. Genotypes are called using the autocalling algorithm of GenomeStudio. The quality control involves exclusion of single nucleotide polymorphisms (SNPs) with a call rate <95%.
- The SNP call rate is used as a measure of the immunochip assay efficiency. It is calculated as the percentage of the total number of assays on the chip (196,524 SNPs encoded on each chip) that achieve sufficient signal intensity and quality to receive an automated genotype assignment. High quality DNA (260/280 ratio of 1.8 or higher and absence of degradation) is expected to achieve at least the same call rate as the HapMap control DNA included in the immunochip assay. Quality control involves exclusion of SNPs with a call rate <95% in each data set.
A Practical and Novel Method to Extract Genomic DNA from Blood Collection Kits for Plasma Protein Preservation
Learning Objectives
DNA quality assessment and concentration measurement
The yields of P100_DNAs were significantly lower than those of EDTA_DNAs (Tables 1 and 2). The DNA purity represented by its 260/280 ratio value is similar for both P100_DNA and EDTA_DNA sample set (Tables 1 and 2). The quality of the DNA is fairly uniform within each set of sample although some degradation is seen in the EDTA_DNA samples, as indicated by light smear (Figure 2). The EDTA_DNAs that showed more signs of degradation also showed reduced DNA concentration.
Genotyping
The genotyping call rate for both EDTA_DNAs and P100_DNAs were compared. They yielded similar call rates and were both comparable to the internal HapMap control DNA (Table 3).
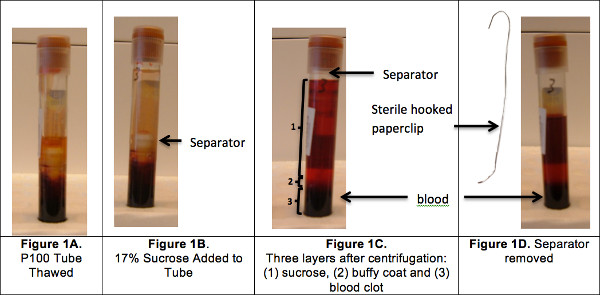
Figure 1. Buffy coat isolation.
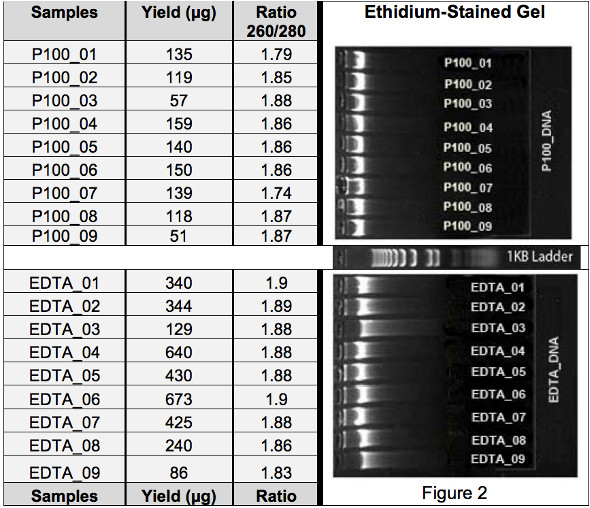
Table 1. DNA sample yield (PicoGreen) and ratio (nanodrop).
| DNA Yield | DNA Purity | |||
| Samples | P100 | EDTA | P100 | EDTA |
| 1 | 135 | 340 | 1.79 | 1.9 |
| 2 | 119 | 344 | 1.85 | 1.89 |
| 3 | 57 | 129 | 1.88 | 1.88 |
| 4 | 159 | 640 | 1.86 | 1.88 |
| 5 | 140 | 430 | 1.86 | 1.88 |
| 6 | 150 | 673 | 1.86 | 1.9 |
| 7 | 139 | 425 | 1.74 | 1.88 |
| 8 | 118 | 240 | 1.87 | 1.86 |
| 9 | 51 | 86 | 1.87 | 1.83 |
| p value | p = 0.00221 | p = 0.09836 | ||
Table 2. Comparison of yield and purity (two-tailed, Paired Sample t-test).
| Genotyping Cell Rate | ||
| Samples | P100 | EDTA |
| 1 | 97.9 | 97.5 |
| 2 | 97.9 | 97.8 |
| 4 | 97.4 | 97.5 |
| 7 | 97.5 | 98.2 |
| 8 | 97.9 | 98 |
| P value | p = 0.67970 | |
Table 3. Genotyping call rate (two-tailed, Paired Sample t-test).
| Samples | Layers | Yield (μg) | Ratio (260/280) |
| P100_06 | Buffy coat | 150 | 1.86 |
| RBC | 4.13 | 1.73 | |
| P100_07 | Buffy coat | 139 | 1.74 |
| RBC | 1.00 | 1.70 | |
| P100_08 | Buffy coat | 118 | 1.87 |
| RBC | 3.72 | 1.78 | |
| P100_09 | Buffy coat | 51 | 1.87 |
| RBC | 0.23 | 0.98 |
Appendix. Evaluation of the amount of DNA found in red blood cell (RBC) layer.
List of Materials
| Name of the reagent/equipment | Company | Catalogue number |
| BD P100 tubes | BD Diagnostics | 366448 |
| EDTA tubes | BD Diagnostics | 367863 |
| Sucrose | Sigma | S9378 |
| Paper clip | Office product | |
| 15 ml tube | Corning | 430052 |
| Red blood cells (RBC) | Qiagen | 158389 (kit) |
| Phosphate Buffer Saline (PBS) | Thermo Scientific | SH30256.01 |
| BioSprint 96 DNA Blood Kit (48) | Qiagen | 940054 |
| 1.7 ml microtubes | Axygen | MCT-175-C |
| Human Immuno DNA Analysis BeadChip Kit | Illumina | WG-352-1001 |
| Bead array reader | Illumina | NA |
| GenomeStudio Software | Illumina | N/A |
Lab Prep
Laboratory tests can be done on the cellular or fluid portions of the blood. The use of different blood collection tubes determines the portion of the blood that can be analyzed (whole blood, plasma or serum). Laboratories involved in studying the genetic basis of human disorders rely on anticoagulated whole blood collected in EDTA-containing vacutainer as the source of DNA for genetic / genomic analysis. Because most clinical laboratories perform biochemical, serologic and viral testing as a first step in phenotypic outcome investigation, anticoagulated blood is also collected in heparin-containing tube (plasma tube). Therefore when DNA and plasma are needed for simultaneous and parallel analyses of both genomic and proteomic data, it is customary to collect blood in both EDTA and heparin tubes. If blood could be collected in a single tube and serve as a source for both plasma and DNA, that method would be considered an advancement to existing methods. The use of the compacted blood after plasma extraction represents an alternative source for genomic DNA, thus minimizing the amount of blood samples processed and reducing the number of samples required from each patient. This would ultimately save time and resources.
The BD P100 blood collection system for plasma protein preservation were created as an improved method over previous plasma or serum collection tubes1, to stabilize the protein content of blood, enabling better protein biomarker discovery and proteomics experimentation from human blood. The BD P100 tubes contain 15.8 ml of spray-dried K2EDTA and a lyophilized proprietary broad spectrum cocktail of protease inhibitors to prevent coagulation and stabilize the plasma proteins. They also include a mechanical separator, which provides a physical barrier between plasma and cell pellets after centrifugation. Few methods have been devised to extract DNA from clotted blood samples collected in old plasma tubes2-4. Challenges from these methods were mainly associated with the type of separator inside the tubes (gel separator) and included difficulty in recovering the clotted blood, the inconvenience of fragmenting or dispersing the clot, and obstruction of the clot extraction by the separation gel.
We present the first method that extracts and purifies genomic DNA from blood drawn in the new BD P100 tubes. We compare the quality of the DNA sample from P100 tubes to that from EDTA tubes. Our approach is simple and efficient. It involves four major steps as follows: 1) the use of a plasma BD P100 (BD Diagnostics, Sparks, MD, USA) tube with mechanical separator for blood collection, 2) the removal of the mechanical separator using a combination of sucrose and a sterile paperclip metallic hook, 3) the separation of the buffy coat layer containing the white cells and 4) the isolation of the genomic DNA from the buffy coat using a regular commercial DNA extraction kit or a similar standard protocol.
Laboratory tests can be done on the cellular or fluid portions of the blood. The use of different blood collection tubes determines the portion of the blood that can be analyzed (whole blood, plasma or serum). Laboratories involved in studying the genetic basis of human disorders rely on anticoagulated whole blood collected in EDTA-containing vacutainer as the source of DNA for genetic / genomic analysis. Because most clinical laboratories perform biochemical, serologic and viral testing as a first step in phenotypic outcome investigation, anticoagulated blood is also collected in heparin-containing tube (plasma tube). Therefore when DNA and plasma are needed for simultaneous and parallel analyses of both genomic and proteomic data, it is customary to collect blood in both EDTA and heparin tubes. If blood could be collected in a single tube and serve as a source for both plasma and DNA, that method would be considered an advancement to existing methods. The use of the compacted blood after plasma extraction represents an alternative source for genomic DNA, thus minimizing the amount of blood samples processed and reducing the number of samples required from each patient. This would ultimately save time and resources.
The BD P100 blood collection system for plasma protein preservation were created as an improved method over previous plasma or serum collection tubes1, to stabilize the protein content of blood, enabling better protein biomarker discovery and proteomics experimentation from human blood. The BD P100 tubes contain 15.8 ml of spray-dried K2EDTA and a lyophilized proprietary broad spectrum cocktail of protease inhibitors to prevent coagulation and stabilize the plasma proteins. They also include a mechanical separator, which provides a physical barrier between plasma and cell pellets after centrifugation. Few methods have been devised to extract DNA from clotted blood samples collected in old plasma tubes2-4. Challenges from these methods were mainly associated with the type of separator inside the tubes (gel separator) and included difficulty in recovering the clotted blood, the inconvenience of fragmenting or dispersing the clot, and obstruction of the clot extraction by the separation gel.
We present the first method that extracts and purifies genomic DNA from blood drawn in the new BD P100 tubes. We compare the quality of the DNA sample from P100 tubes to that from EDTA tubes. Our approach is simple and efficient. It involves four major steps as follows: 1) the use of a plasma BD P100 (BD Diagnostics, Sparks, MD, USA) tube with mechanical separator for blood collection, 2) the removal of the mechanical separator using a combination of sucrose and a sterile paperclip metallic hook, 3) the separation of the buffy coat layer containing the white cells and 4) the isolation of the genomic DNA from the buffy coat using a regular commercial DNA extraction kit or a similar standard protocol.
Procedure
Laboratory tests can be done on the cellular or fluid portions of the blood. The use of different blood collection tubes determines the portion of the blood that can be analyzed (whole blood, plasma or serum). Laboratories involved in studying the genetic basis of human disorders rely on anticoagulated whole blood collected in EDTA-containing vacutainer as the source of DNA for genetic / genomic analysis. Because most clinical laboratories perform biochemical, serologic and viral testing as a first step in phenotypic outcome investigation, anticoagulated blood is also collected in heparin-containing tube (plasma tube). Therefore when DNA and plasma are needed for simultaneous and parallel analyses of both genomic and proteomic data, it is customary to collect blood in both EDTA and heparin tubes. If blood could be collected in a single tube and serve as a source for both plasma and DNA, that method would be considered an advancement to existing methods. The use of the compacted blood after plasma extraction represents an alternative source for genomic DNA, thus minimizing the amount of blood samples processed and reducing the number of samples required from each patient. This would ultimately save time and resources.
The BD P100 blood collection system for plasma protein preservation were created as an improved method over previous plasma or serum collection tubes1, to stabilize the protein content of blood, enabling better protein biomarker discovery and proteomics experimentation from human blood. The BD P100 tubes contain 15.8 ml of spray-dried K2EDTA and a lyophilized proprietary broad spectrum cocktail of protease inhibitors to prevent coagulation and stabilize the plasma proteins. They also include a mechanical separator, which provides a physical barrier between plasma and cell pellets after centrifugation. Few methods have been devised to extract DNA from clotted blood samples collected in old plasma tubes2-4. Challenges from these methods were mainly associated with the type of separator inside the tubes (gel separator) and included difficulty in recovering the clotted blood, the inconvenience of fragmenting or dispersing the clot, and obstruction of the clot extraction by the separation gel.
We present the first method that extracts and purifies genomic DNA from blood drawn in the new BD P100 tubes. We compare the quality of the DNA sample from P100 tubes to that from EDTA tubes. Our approach is simple and efficient. It involves four major steps as follows: 1) the use of a plasma BD P100 (BD Diagnostics, Sparks, MD, USA) tube with mechanical separator for blood collection, 2) the removal of the mechanical separator using a combination of sucrose and a sterile paperclip metallic hook, 3) the separation of the buffy coat layer containing the white cells and 4) the isolation of the genomic DNA from the buffy coat using a regular commercial DNA extraction kit or a similar standard protocol.
