Loop-mediated Isothermal Amplification (LAMP) Assays for the Species-specific Detection of Eimeria that Infect Chickens
Instructor Prep
concepts
Student Protocol
1. Template Preparation
NOTE: Any genomic DNA template suspected to contain DNA derived from one of the seven Eimeria species which infect chickens can be used as template for LAMP-based Eimeria species identification. Intestinal tissue samples for field diagnostic analysis should be collected during routine post-mortem as described here.
- Choose the section (or sections) of intestine to be tested. See Table 1 for a guide to sample site selection and the Eimeria species most likely to be present18 and Figure 1 for the species-specific range of distribution and location of the sampling sites.
NOTE: This is of key importance since Eimeria are notably host site specific. Each species that infects chickens is defined by the intestinal region that it targets19. The decision may be influenced by previous experience of the farm, interest in one or more specific Eimeria species or other diagnostic indicators6. - Excise 5 cm or longer lengths of the intestinal section(s) selected to test for Eimeria using sterile scissors or a scalpel. Optionally, store the samples for subsequent analysis in a fixative such as for example in RNAlater18 or 95% ethanol.
NOTE: If storing in ethanol the sample should be washed thoroughly in sterile tris-ethylenediaminetetraacetic acid (TE) buffer prior to use. - Cut the sample open longitudinally, remove most intestinal contents (if present) and scrape cells from the mucosal layer free using either the edge of a sterile glass microscope slide or an ethanol/flame sterilised scissor blade. Optionally for a pooled sample include cells from all four of the species-specific intestinal sites in a single tube.
- Put the scraped material into a sterile 1.5 ml screw-top microcentrifuge tube containing 100 µl sterile TE buffer including 10% (w/v) Chelex 100 resin.
- Shake each sample vigorously for 1 min. Ensure that the screw top is firmly closed and then incubate in a boiling water bath for 10 min.
- After boiling allow each sample to cool at the ambient temperature for 1-2 min.
- Centrifuge each sample using a microcentrifuge at top speed (e.g., ~10,000 x g) for 1 min.
- Collect 2 µl of the resulting supernatant to be template in each LAMP assay to be undertaken. Optionally, pool more than one intestinal site in a single tube to provide a multi-site assay.
2. Eimeria LAMP Primer Preparation (Pre-assay)
- Prepare Eimeria LAMP primer stocks adequate for 100 assays:
- Reconstitute each lyophilised Eimeria LAMP primer by adding molecular grade water to a concentration of 100 µM (as specified by the manufacturer). If not specified, calculate the volume of molecular grade water required using the physical and molecular weights of each primer.
- Pipette 60 µl molecular grade water into a separate 0.5 ml flip-top microcentrifuge tube for each Eimeria species to be assayed.
- Add primers FIP, BIP, F3, B3, LF and LB specific to the target Eimeria species to the water using the volumes shown in Table 2, creating a series of seven species-specific primer mixes.
- Briefly vortex mix the primer solution, then pulse microfuge and freeze until required.
- Prepare a LAMP reaction mastermix for each Eimeria species to be assayed. Multiply the volumes shown in Table 3 by the number of samples and add three to a positive control, negative control and pipetting spare. Pipette into a 0.5 or 1.5 ml flip-top microcentrifuge tube.
3. Eimeria LAMP Assay
- Pipette 23 µl Eimeria species-specific Bst DNA polymerase/LAMP mastermix into a 0.5 ml microcentrifuge tube.
- Add 2 µl DNA template (prepared in section 1), making a final reaction volume of 25 µl.
- Add 2 µl Eimeria species-specific genomic DNA to one reaction (positive control). Add 2 µl molecular grade water to reaction (negative control).
NOTE: If species-specific genomic DNA is not available a previous positive LAMP or standard PCR product may be used instead. - Incubate in a water bath or heat block at 62oC for 30 min. Optionally, de-activate the Bst DNA polymerase by heating to 80oC for 10 min if the reaction is not going to be read immediately.
4. LAMP Assay Read-out
- At the conclusion of the incubation assess the colour of each reaction by eye under indoor light. Negative results appear pink to violet in colour, positive results appear sky blue20.
- Optionally, confirm the LAMP assay result in a laboratory by mixing 5 µl LAMP reaction product with 1 µl DNA gel loading buffer for agarose gel electrophoresis using a 2% agarose gel in 1 x Tris/Borate/EDTA (TBE) buffer, pre-stained using a nucleic acid stain (5 µl per 50 ml agarose). Add 5 µl of a 1Kb molecular size DNA ladder to lane 1 of the gel to allow fragment size calculation.
Loop-mediated Isothermal Amplification (LAMP) Assays for the Species-specific Detection of Eimeria that Infect Chickens
Learning Objectives
Assay validation
During validation each Eimeria species-specific LAMP assay was tested using a panel of pure DNA samples representing all seven Eimeria species that infect the chicken, as well as chicken genomic DNA as a host control. Agarose gel electrophoresis was used to resolve each assay and demonstrated absolute species specificity with no host cross-reactivity18. Next, a ten-fold serial dilution series prepared using purified Eimeria tenella genomic DNA revealed an assay sensitivity limit of between one and ten genome copies18. No upper limit was determined with positive results achieved up to and including the highest concentration (100,000 genome copies)18.
Application with field samples
Samples collected for Eimeria testing are likely to be derived from chickens found dead, culled as a consequence of poor health or culled for sentinel health surveillance, indicating a likely sample size of between one and three when part of a routine. Testing three birds collected from a US broiler farm as part of a surveillance programme yielded three sets of intestinal samples. Targeted application of the species-specific LAMP assays, prioritising the preferred intestinal sites for each Eimeria species (Table 1), allowed visual identification of eimerian infection in all birds tested using hydroxynaphthol blue as an indicator (Figure 2A). The colour achieved with a negative LAMP reaction when using hydroxynaphthol blue can range from violet to pink, but is always distinct from the blue achieved by a positive result. Confirmation by agarose gel electrophoresis provided comparable results (Figure 2B). During field application the user may choose to apply the full screen against all seven species, or target only those species prioritised as important or known to be circulating on the farm or surrounding area.
The failure of PCR-based approaches to become established as diagnostics for the occurrence of Eimeria emphasises the requirement for simplicity in any new test. While LAMP offers simpler preparation and processing than PCR, the requirement to test multiple intestinal sites per bird remains discouraging. Production of a single pooled DNA sample per bird, which can then be tested with one or more LAMP assays, is likely to be more appealing. Processing one pooled sample per bird, representing material collected from each of the specific intestinal sites described in Table 1 and pooled prior to DNA preparation, for testing with all seven LAMP assays provided the same result as when each intestinal site was processed separately (Figure 2 compared with Figure 3).
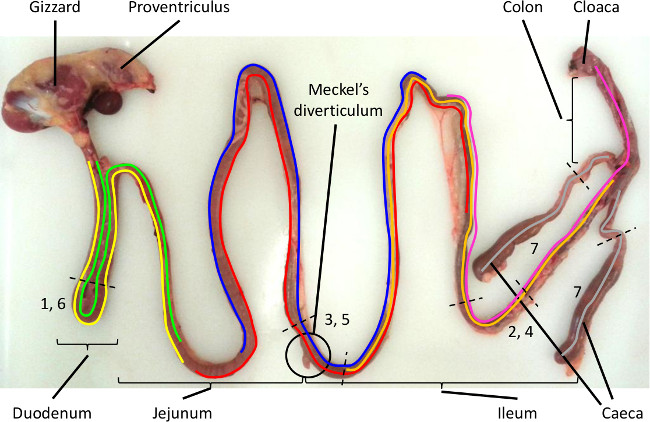
Figure 1. Intestinal sampling sites for LAMP detection of Eimeria species parasites that infect chickens. The intestinal regions targeted by each Eimeria species is highlighted by the coloured lines, with the preferred sites of sampling indicated by the number between the dotted black lines (E. acervulina: yellow/1, E. brunetti: pink/2, E. maxima: blue/3, E. mitis: orange/4, E. necatrix: red/5, E. praecox: green/6 and E. tenella: grey/7). Please click here to view a larger version of this figure.
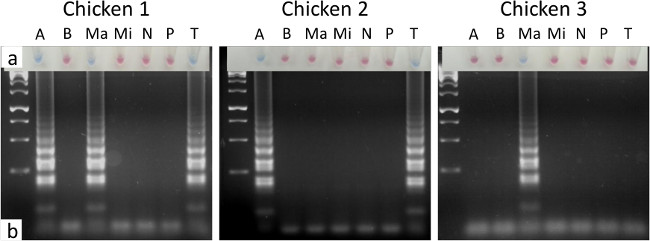
Figure 2. LAMP diagnosis of eimerian infection from three commercial broiler chickens. LAMP reactions resolved using (A) hydroxynaphthol blue, where a sky blue reaction was positive and a violet to pink reaction was negative, and (B) agarose gel electrophoresis. The intestinal sites sampled were as shown in Table 1 for each parasite species. A = E. acervulina, B = E. brunetti, Ma = E. maxima, Mi = E. mitis, N = E. necatrix, P = E. praecox and T = E. tenella. Lane 1 contained the GeneRuler 1Kb DNA ladder. Please click here to view a larger version of this figure.
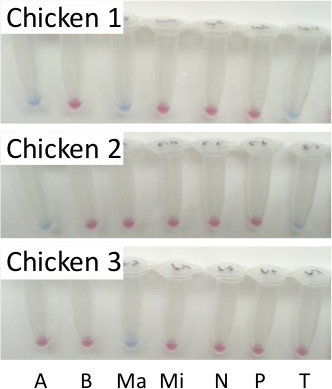
Figure 3. LAMP diagnosis of eimerian infection using pooled samples from three separate commercial broiler chickens. LAMP reactions resolved using hydroxynaphthol blue, where a sky blue reaction was positive and violet to pink reaction was negative. A = E. acervulina, B = E. brunetti, Ma = E. maxima, Mi = E. mitis, N = E. necatrix, P = E. praecox and T = E. tenella. Please click here to view a larger version of this figure.
| Sample site | Eimeria species assay (most likely) |
| Duodenum (D) | E. acervulina, E. praecox |
| Jejunum/ileum* (J/I) | E. maxima, E. necatrix |
| Caeca (C) | E. necatrix, E. tenella |
| Terminal ileum (TI) | E. brunetti, E. mitis |
| Pooled sample (P) | E. acervulina, E. brunetti, E. maxima, E. mitis, E. necatrix, E. praecox, E. tenella |
Table 1. Intestinal region-specific selection of candidate Eimeria species assays. The choice of region to be sampled varies for each Eimeria species as illustrated in Figure 1. Pooled samples include material collected from all four specific sites which were then combined for DNA preparation.
| Primer* | Stock concentration (µM) | Volume (µl) |
| Water | – | 60 |
| Forward Inner Primer (FIP) | 100 | 40 |
| Backward Inner Primer (BIP) | 100 | 40 |
| Forward Outer Primer (F3) | 100 | 10 |
| Backward Outer Primer (B3) | 100 | 10 |
| Loop Forward (LF) | 100 | 20 |
| Loop Backward (LB) | 100 | 20 |
| Total | 200 |
Table 2. Preparation of a LAMP primer premix. The components and proportions required to prepare a primer premix for LAMP. Volumes shown are for 100 LAMP reactions. *Primer identities as shown in the Materials and Barkway et al (2011)18.
| Stock concn | Final reaction concn | Volume per reaction(µl) | |
| DDW | – | – | 10.1 |
| ThermoPol buffer | 10 x | 1 x | 2.5 |
| MgSO4 | 100 mM | 2 mM | 0.5 |
| Primer mix* | Table 2 | 2.5 | |
| dNTPs | 25 mM | 400 uM | 0.4 |
| Betaine | 5 M | 1 M | 5 |
| Hydroxynaphthol blue | 3 mM | 120 µM | 1 |
| Bst DNA polymerase | 8,000 U/ml | 8 U | 1 |
| Total | 23 | ||
Table 3. Preparation of a LAMP reaction mastermix. *Eimeria species-specific.
List of Materials
| Name | Company | Catalogue number | Comments |
| RNAlater | Ambion | AM7024 | |
| Ethanol | VWR Chemicals | 20821.321 | Caution, highly flammable |
| 100 x Tris-EDTA (TE) buffer concentrate | Sigma-Aldrich | T9285 | |
| Chelex 100 resin | Bio-Rad | 142-1253 | |
| Molecular grade water | Invitrogen | 10977035 | |
| E. acervulina F3 | Sigma-Aldrich | VC00021 | *CCTAACATTTCGCTTCACGGAC |
| E. acervulina B3 | Sigma-Aldrich | VC00021 | *ATGAGCAAGTGGAACACCTTG |
| E. acervulina FIP | Sigma-Aldrich | VC00021 | *AGAGCACAGTGGCAGTGC-AGCAGACAGCATGGCTTACCT |
| E. acervulina BIP | Sigma-Aldrich | VC00021 | *GAAGACCCTCTGAAGAACGGA-CCTTCTCACCGCTTACCGG |
| E. acervulina LB | Sigma-Aldrich | VC00021 | *TAAGGTTACACCCGTGGAGG |
| E. acervulina LF | Sigma-Aldrich | VC00021 | *GCCATGCACAAAGCGACTT |
| E. brunetti F3 | Sigma-Aldrich | VC00021 | *GGCCATCAAGTTCCATGAGC |
| E. brunetti B3 | Sigma-Aldrich | VC00021 | *TCAACCTCCTGAGTGTGGTT |
| E. brunetti FIP | Sigma-Aldrich | VC00021 | *GAAAATGCCTTCGTAGCTGCT-GCTGGGTACGGAGCGTCTT |
| E. brunetti BIP | Sigma-Aldrich | VC00021 | *TACTTCCTAGGATCCATCCTCGC-AGTTTCGCTGCCGCCTC |
| E. brunetti LB | Sigma-Aldrich | VC00021 | *GAAACGCTCGAACATGGC |
| E. brunetti LF | Sigma-Aldrich | VC00021 | *CTTCTCCACAGACCCAGAGGT |
| E. maxima F3 | Sigma-Aldrich | VC00021 | *ACTACGGAAAAGTGCGTAGCT |
| E. maxima B3 | Sigma-Aldrich | VC00021 | *CCTTCCTCCCTTCTGAAAACTG |
| E. maxima FIP | Sigma-Aldrich | VC00021 | *GAGTCACTGCTGATGTACCAAA AG-GAACTATGCCGCTTTCCCCTG |
| E. maxima BIP | Sigma-Aldrich | VC00021 | *AGAATGCGGATTTGTTAGCAGC-AGCAAGTCCAAGGTGTGTGTA |
| E. maxima LB | Sigma-Aldrich | VC00021 | *CAAGCCTACGCGGACATC |
| E. maxima LF | Sigma-Aldrich | VC00021 | *TTATGCAGCTGGGTCAACG |
| E. mitis F3 | Sigma-Aldrich | VC00021 | *ACGATAGCCAAGACACGTAAGG |
| E. mitis B3 | Sigma-Aldrich | VC00021 | *CCCCGTGATAAGAGTAGGAACA |
| E. mitis FIP | Sigma-Aldrich | VC00021 | *CGCGGGTCGTGAGATTTAAATT AT-GGAAGATCAGGACGGGCACT |
| E. mitis BIP | Sigma-Aldrich | VC00021 | *GTTTCAGTTGATGAACAAGCGA GA-TGCGCCTCTAGAATCAAGACG |
| E. mitis LB | Sigma-Aldrich | VC00021 | *TCCATGCATCCCCTTGTT |
| E. mitis LF | Sigma-Aldrich | VC00021 | *CGTGGGCACAGATTGATTC |
| E. necatrix F3 | Sigma-Aldrich | VC00021 | *TGGCTTTCCCGCGTACC |
| E. necatrix B3 | Sigma-Aldrich | VC00021 | *CGGCCCAACACAAAGACTG |
| E. necatrix FIP | Sigma-Aldrich | VC00021 | *CGCTTGAGTTTTAAGCTATGCA CA-GACCCAAGCAGCTCACCAA |
| E. necatrix BIP | Sigma-Aldrich | VC00021 | *CGCCATGCCATTCAATGAACG-*GAGGCATACCGGCGTTGTC |
| E. necatrix LB | Sigma-Aldrich | VC00021 | *GTCTGTAACTTGGGACGTTGT |
| E. necatrix LF | Sigma-Aldrich | VC00021 | *GAACAGCCGGAGCCTCTC |
| E. praecox F3 | Sigma-Aldrich | VC00021 | *GCCCTTGTATGTTGCTGTTTCT |
| E. praecox B3 | Sigma-Aldrich | VC00021 | *GCGCACGAATCTGAATCACAC |
| E. praecox FIP | Sigma-Aldrich | VC00021 | *ATCTCCTCAAAGACTTTCGCGT A-GCGCTTGGCTATATCCATAGG |
| E. praecox BIP | Sigma-Aldrich | VC00021 | *GCTCTCGTGGCATACTTGC-GCCAGGAGCCACTGATTGT |
| E. praecox LB | Sigma-Aldrich | VC00021 | *GAATAGCATTGCCAGGTGG |
| E. praecox LF | Sigma-Aldrich | VC00021 | *GTCCACTGTCATTAATATTGC TGC |
| E. tenella F3 | Sigma-Aldrich | VC00021 | *GCTTGTGAAGGTCAGCGTG |
| E. tenella B3 | Sigma-Aldrich | VC00021 | *GCTGAGTCCATACGTACTTCCT |
| E. tenella FIP | Sigma-Aldrich | VC00021 | *GCCACTGCTATGGAAAGTCAC AC-CATAACTGGCATGCAGGGGT |
| E. tenella BIP | Sigma-Aldrich | VC00021 | *GTTTGGCCCGAAAGTTGTGAA GA-CGTCAGAAATTGCTGCCCAAT |
| E. tenella LB | Sigma-Aldrich | VC00021 | *CGCATGTGCAGTTGAAGACA |
| E. tenella LF | Sigma-Aldrich | VC00021 | *CCAAATGTATCTGCTAGTTATA TTAACAAG |
| 10 x ThermoPol reaction buffer | New England Biolabs | B9004S | |
| MgSO4 | Sigma-Aldrich | M7506 | |
| dNTPs | Promega | U1330 | |
| Betaine solution (5 M) | Sigma-Aldrich | B0300 | |
| Bst polymerase | New England Biolabs | M0275S | |
| Hydroxynaphthol blue | Sigma-Aldrich | 33936 | Dissolved in molecular grade water. |
| UltraPure agarose | Invitrogen | 16500-500 | |
| 10 x Tris/Borate/EDTA (TBE) buffer | Invitrogen | AM9863 | |
| Blue/Orange DNA loading dye (x6) | Promega | G1881 | |
| GeneRuler 1Kb DNA ladder | Thermo Scientific | SM0313 | |
| SafeView nucleic acid stain | NBS Biologicals | NBS-SV |
Lab Prep
Eimeria species parasites, protozoa which cause the enteric disease coccidiosis, pose a serious threat to the production and welfare of chickens. In the absence of effective control clinical coccidiosis can be devastating. Resistance to the chemoprophylactics frequently used to control Eimeria is common and sub-clinical infection is widespread, influencing feed conversion ratios and susceptibility to other pathogens such as Clostridium perfringens. Despite the availability of polymerase chain reaction (PCR)-based tools, diagnosis of Eimeria infection still relies almost entirely on traditional approaches such as lesion scoring and oocyst morphology, but neither is straightforward. Limitations of the existing molecular tools include the requirement for specialist equipment and difficulties accessing DNA as template. In response a simple field DNA preparation protocol and a panel of species-specific loop-mediated isothermal amplification (LAMP) assays have been developed for the seven Eimeria recognised to infect the chicken. We now provide a detailed protocol describing the preparation of genomic DNA from intestinal tissue collected post-mortem, followed by setup and readout of the LAMP assays. Eimeria species-specific LAMP can be used to monitor parasite occurrence, assessing the efficacy of a farm’s anticoccidial strategy, and to diagnose sub-clinical infection or clinical disease with particular value when expert surveillance is unavailable.
Eimeria species parasites, protozoa which cause the enteric disease coccidiosis, pose a serious threat to the production and welfare of chickens. In the absence of effective control clinical coccidiosis can be devastating. Resistance to the chemoprophylactics frequently used to control Eimeria is common and sub-clinical infection is widespread, influencing feed conversion ratios and susceptibility to other pathogens such as Clostridium perfringens. Despite the availability of polymerase chain reaction (PCR)-based tools, diagnosis of Eimeria infection still relies almost entirely on traditional approaches such as lesion scoring and oocyst morphology, but neither is straightforward. Limitations of the existing molecular tools include the requirement for specialist equipment and difficulties accessing DNA as template. In response a simple field DNA preparation protocol and a panel of species-specific loop-mediated isothermal amplification (LAMP) assays have been developed for the seven Eimeria recognised to infect the chicken. We now provide a detailed protocol describing the preparation of genomic DNA from intestinal tissue collected post-mortem, followed by setup and readout of the LAMP assays. Eimeria species-specific LAMP can be used to monitor parasite occurrence, assessing the efficacy of a farm’s anticoccidial strategy, and to diagnose sub-clinical infection or clinical disease with particular value when expert surveillance is unavailable.
Procedure
Eimeria species parasites, protozoa which cause the enteric disease coccidiosis, pose a serious threat to the production and welfare of chickens. In the absence of effective control clinical coccidiosis can be devastating. Resistance to the chemoprophylactics frequently used to control Eimeria is common and sub-clinical infection is widespread, influencing feed conversion ratios and susceptibility to other pathogens such as Clostridium perfringens. Despite the availability of polymerase chain reaction (PCR)-based tools, diagnosis of Eimeria infection still relies almost entirely on traditional approaches such as lesion scoring and oocyst morphology, but neither is straightforward. Limitations of the existing molecular tools include the requirement for specialist equipment and difficulties accessing DNA as template. In response a simple field DNA preparation protocol and a panel of species-specific loop-mediated isothermal amplification (LAMP) assays have been developed for the seven Eimeria recognised to infect the chicken. We now provide a detailed protocol describing the preparation of genomic DNA from intestinal tissue collected post-mortem, followed by setup and readout of the LAMP assays. Eimeria species-specific LAMP can be used to monitor parasite occurrence, assessing the efficacy of a farm’s anticoccidial strategy, and to diagnose sub-clinical infection or clinical disease with particular value when expert surveillance is unavailable.
