Biomimetic Replication of Root Surface Microstructure using Alteration of Soft Lithography
Instructor Prep
concepts
Student Protocol
1. Growing the plants and root preparation
- Option 1: Prepare adventitious roots from stem.
- Take a rooting tray for growing plants.
- Fill the tray with soil.
- Add one seed of M82 tomato cultivar to each cell in the tray.
- Cover the seeds with a little soil.
- Water the tray from the bottom with a dropper as the water fills the bottom of the tray and the soil absorbs water.
- Add 2 mL of fertilizer per 1 L of water to the bottom of the tray once a week.
- Grow in a growing chamber at 25 °C.
- Use lighting conditions of 9 h light (7:00-16:00) alternated with 15 h of darkness.
- After 3 weeks remove the plant from the soil.
- Cut the root system from the plant at the point of interaction with the stem.
- Put the rootless plant in a beaker filled with water.
- After a few days, cut the adventitious roots that emerge from the stem and use them for replication.
- Option 2: Prepare seed germinating roots.
- Wet a Petri dish sized filter paper with water.
- Put several M82 seeds (no more than 10) on the paper, inside a Petri dish.
- Incubate the plate at 25 °C.
- Hydrate the paper every day.
- After germinated roots are long enough (approximately 5 days), remove the seeds and use the roots for replication.
2. Preparation of the root negative replica from polyurethane
- To generate negative replica solution, add 9.49 g of diurethane dimethacrylate to a 20 mL vial.
- Add 1.45 mL of ethyl methacrylate to the vial.
- Stir at room temperature (RT) until the solution looks clear and becomes homogeneous.
NOTE: Approximately 2 h is sufficient to reach a homogeneous solution. - Add 3 mL of the plasticizer, diethyl phthalate, and stir for 1 h at RT.
NOTE: Diethyl phthalate is miscible in acrylate monomer. - Add 300 µL of the photo initiator, 2-hydroxy-2-methylpropiophenone, and stir overnight at RT. Continue stirring until all bubbles are removed.
NOTE: The protocol can be paused here. The solution can be kept at RT.
- To generate the negative replica of the root, take a clean glass slide and pour 1 mL of the negative replica solution on it.
- Place 2‒3 roots over the solution. Do not allow the roots to be fully covered by the solution.
- Keep the slide under 8 W ultra violet (UV) lamp for 8‒10 min. Do not keep the solution under UV light for too long.
NOTE: It is important not to keep the solution under the UV light for too long as it makes the polyurethane too hard, making it impossible to remove the root. - Switch off the UV lamp, remove the replica from the glass slide and put it in a Petri dish filled with ethanol, to remove unreacted monomer.
- To obtain the negative replica, remove the root from the replica very slowly using forceps.
3. Prepare the root positive replica from PDMS.
- To generate the mixture for the positive replica, place 10 g of dimethyl siloxane in a paper cup.
- Add 1 g of curing agent and mix thoroughly.
- Keep the mixture in a desiccator under vacuum for 2 h to remove air bubbles.
- To generate the positive replica, place the polyurethane negative replica in a Petri dish.
- Pour the PDMS mixture on top of the negative replica.
- Apply vacuum for 2 h to assure coverage of the microstructure.
- Keep the Petri dish overnight at RT.
- Separate the cured positive replica from the negative replica by hand.
4. Prepare the root positive replica from ethyl cellulose.
- To generate the ethyl cellulose solution, put 1.32 ml of diethyl pthalate as a plasticizer in a 100 ml cup.
- Add 20 mL of ethanol and stir at RT for 2 h.
- Add 3.3 g of ethyl cellulose and stir overnight.
- To generate the positive replica, place the polyurethane negative replica in a Petri dish.
- Pour the ethyl cellulose solution on top of the negative replica.
- Keep the Petri dish overnight at RT under the hood.
- Remove the positive replica from the negative replica very slowly by forceps.
Biomimetic Replication of Root Surface Microstructure using Alteration of Soft Lithography
Learning Objectives
To form the root surface microstructure replication, a root must be chosen for molding. We grow tomato plants in soil, making the use of the natural root from the root system extremely challenging. Removal of soil from the root system can be difficult and additionally, the root system roots are fragile and can break upon molding attempts. We therefore suggest to first use more rigid roots, to establish the protocol in the lab. The formation of such roots is described in Figure 1A. The plant root system is removed after the plant was grown for 3 weeks and the rootless plant is placed in water for about a week until adventitious roots emerge from the stem. Those roots can be used for replication during the protocol establishment. Once the protocol has been well established, a more realistic root surface structure is desired. Here we suggest avoiding roots grown in soil as the full removal of soil in extremely challenging. Instead we suggest the use of germinating roots, supplying valuable information on the root surface microstructure of a genetically specific plant. The growth of such roots is described in Figure 1B. The seeds are placed on a wet filter paper and incubated at 25 °C. After approximately 5 days, during which the filter paper is kept moist, the germinated roots are long enough for replication. Those roots are more fragile than the previously suggested roots and require more delicate care.
The production of the root surface microstructure replica is a two-step process. In the first step the natural root is being molded into a polyurethane based mold (the negative replica). The advantage of this step is that all materials for the polyurethane mold are being prepared and the root is placed on top of the prepared solution at the very end for a 10 min exposure to UV. As a result, the biological tissue is not exposed to harsh conditions for too long and can be gently handled at the end of the process. If all protocol steps are followed, a good negative replica is generated. This replica will show the cell structure of the root surface as well as holes representing the location of the root hairs (Figure 2A). If some critical steps in the protocol are not being followed, the procedure will fail. One such step is the placement of the root on the polyurethane solution prior to curing. The root must be placed very gently to avoid the submergence of it in the polyurethane solution. Such submergence, of any part of the root, will cause the entrapment of the root in the hard polymer with no ability to remove it. If such an event occurs, the root will remain within the negative replica after it is cured (Figure 2B). Another crucial step is regarding the curing time by UV light. The recommended curing time is 8‒10 min. Going past 10 min will result in an extremely hard polyurethane mold, making it impossible to remove the root without breaking it within the polyurethane mold. The breakage of the root can sometimes be visible to the naked eye, e.g., when a large piece is broken (Figure 2C, top, marked with purple arrows). However, sometimes small root pieces are left in the material which are difficult to spot by the naked eye and a microscope has to be used (Figure 2C, bottom, marked with purple arrows). We recommend carefully examining the polyurethane negative replica with a microscope prior to the continuation of the protocol to make sure no residual root is present.
Once the polyurethane negative replica is prepared; many materials can be used for the preparation of the positive replica. The preparation of the positive replica, using the polyurethane negative replica as a mold, is straight forward and depends completely on the quality of the polyurethane negative replica. To generate the positive replica we have used both PDMS—as it is well known in the field of soft lithography (Figure 3A)—and ethyl cellulose as a material that better mimics the properties of the root surface which is mostly composed of cellulose (Figure 3B). The SEM image of the PDMS replica shows the root hairs very clearly. The hairs are in the elongation zone, where they begin to emerge. Hence, the length of root hairs varies along the root surface as they become longer, much like in the natural root (Figure 3A). Ethyl cellulose generates harder and less flexible film than PDMS. Hence, the removal of it from the negative mold requires more care. However, some hairs and the surface microstructure are visible under the light microscope (Figure 3B). We used those two materials to generate the positive replica, however, any material that can form a film will be a good candidate for the positive replica, using the polyurethane negative replica.

Figure 1: Tomato plant roots for replication. (A) A tomato (M82) plant is grown at 25 °C with 9 h of light and 15 h darkness. After 3 weeks, the plant is removed from the soil and the root system is cut off. The rootless plant is put in water until adventitious roots emerge from the stem after about a week. These roots do not show the exact structure as the roots from the root system, but they represent a good model. Those roots are less fragile than the root system roots and hence are preferred to work with when establishing the technique in the lab. (B) Tomato (M82) seeds are put on a wet filter paper in a Petri dish and incubated at 25 °C. The paper is hydrated every day and the seeds are germinating. The roots are growing and after approximately 5 days are long enough to be used for replication. These roots are gentler and should be used once the method is well established. Please click here to view a larger version of this figure.
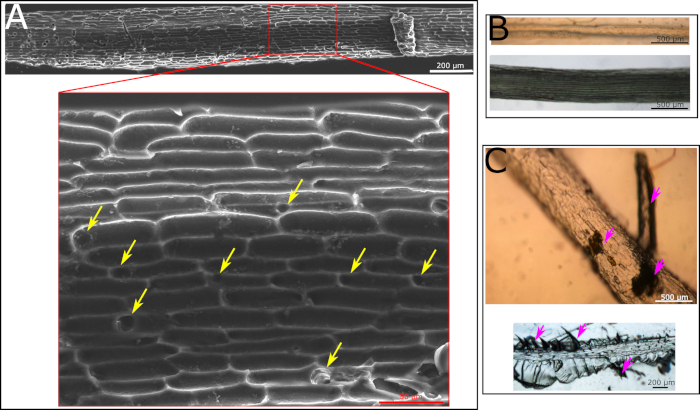
Figure 2: Microscopy images of polyurethane negative replica. (A) SEM image of polyurethane negative replica made according to a protocol following all the steps. Cell structure is clearly visible. Yellow arrows point at holes formed by the hairs in the root. (B) Light microscopy images of polyurethane negative replica with a root inside of it as it was fully covered with the solution and the removal of it was impossible. The polyurethane negative was cured with the root inside. The root is visible by eye and using light microscopy. It is impossible to remove this root from the cured replica. (C) Light microscopy images of polyurethane negative replica that was kept under UV light for too long. As a result, root could not be fully removed from the polymer with either large particles visible by eye (top image, marked with purple arrows) or small fractions visible only by microscope (lower image, marked with purple arrows). Please click here to view a larger version of this figure.
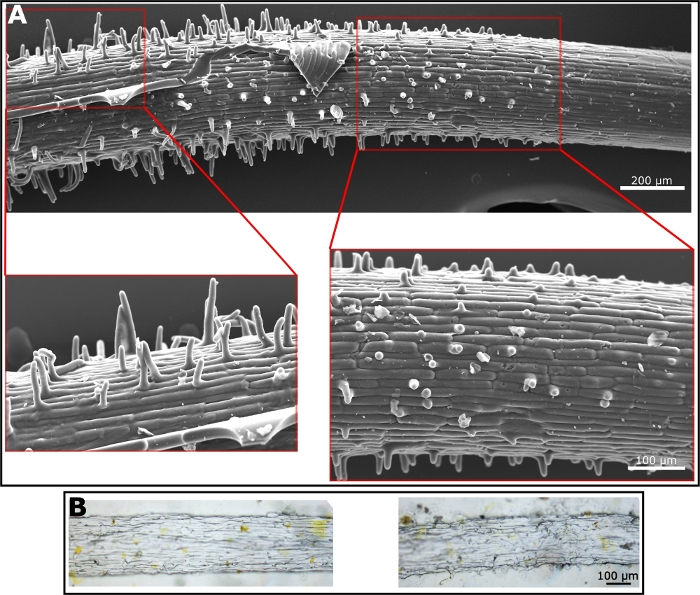
Figure 3: Microscope images of positive replica. (A) SEM micrograph of a positive replica made from PDMS. Enlargement shows root hairs. (B) Light microscopy images of a positive replica made from ethyl cellulose. Hairs are shown in the images on the right while surface texture is visible in the image on the left. Please click here to view a larger version of this figure.
List of Materials
| 2-hydroxy-2-methylpropiophenone | Sigma | 405655 | |
| Diethyl phthalate | Across | 114520010 | |
| Diurethane dimetharylate | Sigma | 436909 | |
| Ethyl cellulose | Across | 232705000 | |
| Ethyl methacrylate | Sigma | 234893 | |
| Shaphir Solution | GAT fertilizer | 6-2-4 | |
| Sylgard 184 kit | Polymer-G | 510018400500 |
Lab Prep
Biomimetics is the use of chemistry and material sciences to mimic biological systems, specifically biological structures, to better humankind. Recently, biomimetic surfaces mimicking the microstructure of leaf surface, were used to study the effects of leaf microstructure on leaf-environment interactions. However, no such tool exists for roots. We developed a tool allowing the synthetic mimicry of the root surface microstructure into an artificial surface. We relied on the soft lithography method, known for leaf surface microstructure replication, using a two-step process. The first step is the more challenging one as it involves the biological tissue. Here, we used a different polymer and curing strategy, relying on the strong, rigid, polyurethane, cured by UV for the root molding. This allowed us to achieve a reliable negative image of the root surface microstructure including the delicate, challenging features such as root hairs. We then used this negative image as a template to achieve the root surface microstructure replication using both the well-established polydimethyl siloxane (PDMS) as well as a cellulose derivative, ethyl cellulose, which represents a closer mimic of the root and which can also be degraded by cellulase enzymes secreted by microorganisms. This newly formed platform can be used to study the microstructural effects of the surface in root-microorganism interactions in a similar manner to what has previously been shown in leaves. Additionally, the system enables us to track the microorganism’s locations, relative to surface features, and in the future its activity, in the form of cellulase secretion.
Biomimetics is the use of chemistry and material sciences to mimic biological systems, specifically biological structures, to better humankind. Recently, biomimetic surfaces mimicking the microstructure of leaf surface, were used to study the effects of leaf microstructure on leaf-environment interactions. However, no such tool exists for roots. We developed a tool allowing the synthetic mimicry of the root surface microstructure into an artificial surface. We relied on the soft lithography method, known for leaf surface microstructure replication, using a two-step process. The first step is the more challenging one as it involves the biological tissue. Here, we used a different polymer and curing strategy, relying on the strong, rigid, polyurethane, cured by UV for the root molding. This allowed us to achieve a reliable negative image of the root surface microstructure including the delicate, challenging features such as root hairs. We then used this negative image as a template to achieve the root surface microstructure replication using both the well-established polydimethyl siloxane (PDMS) as well as a cellulose derivative, ethyl cellulose, which represents a closer mimic of the root and which can also be degraded by cellulase enzymes secreted by microorganisms. This newly formed platform can be used to study the microstructural effects of the surface in root-microorganism interactions in a similar manner to what has previously been shown in leaves. Additionally, the system enables us to track the microorganism’s locations, relative to surface features, and in the future its activity, in the form of cellulase secretion.
Procedure
Biomimetics is the use of chemistry and material sciences to mimic biological systems, specifically biological structures, to better humankind. Recently, biomimetic surfaces mimicking the microstructure of leaf surface, were used to study the effects of leaf microstructure on leaf-environment interactions. However, no such tool exists for roots. We developed a tool allowing the synthetic mimicry of the root surface microstructure into an artificial surface. We relied on the soft lithography method, known for leaf surface microstructure replication, using a two-step process. The first step is the more challenging one as it involves the biological tissue. Here, we used a different polymer and curing strategy, relying on the strong, rigid, polyurethane, cured by UV for the root molding. This allowed us to achieve a reliable negative image of the root surface microstructure including the delicate, challenging features such as root hairs. We then used this negative image as a template to achieve the root surface microstructure replication using both the well-established polydimethyl siloxane (PDMS) as well as a cellulose derivative, ethyl cellulose, which represents a closer mimic of the root and which can also be degraded by cellulase enzymes secreted by microorganisms. This newly formed platform can be used to study the microstructural effects of the surface in root-microorganism interactions in a similar manner to what has previously been shown in leaves. Additionally, the system enables us to track the microorganism’s locations, relative to surface features, and in the future its activity, in the form of cellulase secretion.
