Identification of Novel Genes Associated with Alginate Production in Pseudomonas aeruginosa Using Mini-himar1 Mariner Transposon-mediated Mutagenesis
Summary
Here we describe a protocol using the mini-himar1 mariner transposon-mediated mutagenesis for generating a high-density insertion mutant library to screen, isolate and identify novel alginate regulators in the prototypic Pseudomonas aeruginosa strain PAO1.
Abstract
Pseudomonas aeruginosa is a Gram-negative, environmental bacterium with versatile metabolic capabilities. P. aeruginosa is an opportunistic bacterial pathogen which establishes chronic pulmonary infections in patients with cystic fibrosis (CF). The overproduction of a capsular polysaccharide called alginate, also known as mucoidy, promotes the formation of mucoid biofilms which are more resistant than planktonic cells to antibiotic chemotherapy and host defenses. Additionally, the conversion from the nonmucoid to mucoid phenotype is a clinical marker for the onset of chronic infection in CF. Alginate overproduction by P. aeruginosa is an endergonic process which heavily taxes cellular energy. Therefore, alginate production is highly regulated in P. aeruginosa. To better understand alginate regulation, we describe a protocol using the mini-himar1 transposon mutagenesis for the identification of novel alginate regulators in a prototypic strain PAO1. The procedure consists of two basic steps. First, we transferred the mini-himar1 transposon (pFAC) from host E. coli SM10/λpir into recipient P. aeruginosa PAO1 via biparental conjugation to create a high-density insertion mutant library, which were selected on Pseudomonas isolation agar plates supplemented with gentamycin. Secondly, we screened and isolated the mucoid colonies to map the insertion site through inverse PCR using DNA primers pointing outward from the gentamycin cassette and DNA sequencing. Using this protocol, we have identified two novel alginate regulators, mucE (PA4033) and kinB (PA5484), in strain PAO1 with a wild-type mucA encoding the anti-sigma factor MucA for the master alginate regulator AlgU (AlgT, σ22). This high-throughput mutagenesis protocol can be modified for the identification of other virulence-related genes causing change in colony morphology.
Introduction
The ability of the opportunistic, Gram-negative pathogen Pseudomonas aeruginosa to overproduce alginate is a major factor in its ability to establish a biofilm. The overproduction of alginate is a phenotype often referred to as mucoidy. The isolation of mucoid colonies from the sputa of individuals afflicted with cystic fibrosis (CF) is indicative of a chronic infection, and is directly associated with an overall decline in the patient's health1. Currently, it is understood that the regulation and production of alginate in P. aeruginosa primarily occurs at two operons. The first is the alginate biosynthetic operon, which contains 12 genes (algD–alg8–alg44–algK–algE–algG–algX–algL–algI–algJ–algF–algA) that are responsible for the synthesis and export of the alginate polymer across the periplasm to the extracellular environment2-5. The second operon is a cluster of genes beginning with the alternative sigma factor algU/T and continuing with mucA, mucB, and mucD. AlgU/T is a positive regulator, while mucAB-D are classified as negative regulators of alginate production6-8. Additionally, transcriptional regulators, such as AlgB, AlgQ, AlgR, and RpoN, as well as post-transcriptional and post-translational modification by catabolite repression control, kinase activity (KinB) and intramembrane proteolysis, have also been shown to be involved in alginate regulation9-14.
The mini-himar1 transposon vector known as pFAC was originally created in Dr. Mekalanos' lab at Harvard Medical School15. The pFAC plasmid consists of a transposable element flanked by two inverted repeats of 27 bps and a gentamycin resistance cassette in the middle (aacC1: 534 bp), a gene encoding the hyperactive mariner transposase16, and a gene encoding β-lactamase (bla) (Figure 1) . Sequence information for the transposable element of pFAC is available at the GenBank accession number: DQ36630013. In pFAC, there is a multiple cloning site (MCS) behind the aacC1 gene which is used for the identification of the chromosomal insertion site using inverse PCR. One major advantage with using the mini-himar1 transposon is no specific host factors are required for transposition (mutagenesis). Additionally, there is high abundance of the TA dinucleotide insertion sites found throughout the genome of P. aeruginosa. For example, TA dinucleotide insertion sites occurs 94,404 and 100,229 times in the PAO1 (6,264,404 bps, GenBank accession number NC_002516.2) and PA14 (6,537,648 bps; GenBank access number NC_008463) genomes, respectively. Because of the abundance of TA dinucleotide in the genome, mini-himar1 transposon can cause the high-density and random mutagenesis, which is particularly suitable for the analysis of virulence genes and the genes that are highly regulated. Theoretically, the mini-himar1 transposon can insert into any nonessential gene within the genome of P. aeruginosa. This provides approximately 18 TA insertion sites per open reading frame in the PAO1 genome.
Here, we describe a protocol using mini-himar1 transposon-mediated mutagenesis to identify novel regulators of mucoidy in P. aeruginosa. More specifically, we biparentally conjugated the pFAC vector containing a mini-himar1 transposon from E. coli SM10/λpir into the nonmucoid prototrophic strain PAO1. After the transposon is integrated in the genome, the recipient strain is cultured on Pseudomonas isolation agar (PIA) containing triclosan which inhibits the growth of E. coli. Thus, a library of mutants of P. aeruginosa can be selected by the growth on PIA plate supplemented with gentamycin and the presence of the mucoid phenotype. Note, a true transposon-mediated vs. an integration mutant will have a gentamycin resistant and carbenicillin-sensitive phenotype. In this study, approximately 80,000 insertion mutants of PAO1 were isolated through four separate conjugations. We then screened for mucoid isolates, and determined the site of insertion by restriction enzyme digestion, ligation and inverse PCR. We performed Southern blot analysis using the gentamycin resistance cassette as a probe to see the number of insertion per genome. We determined that more than 90% of mutants obtained per conjugation using this protocol had only a single copy of the himar1 in the genome and displayed the gentamycin resistant and carbenicillin-sensitive phenotype. A total of 32 mucoid isolates were identified, 22 of them were mapped to different loci of the P. aeruginosa PAO1 chromosome. This rate of insertion provides adequate coverage to identify several novel regulators of alginate overproduction.
Protocol
1. Preparation of Bacterial Strains and Biparental Conjugation
- Inoculate E. coli SM10/λpir/pFAC in 5 ml of Luria Broth (LB) supplemented with 15 µg/ml of gentamycin and place in a shaking incubator overnight at 37 °C.
- Inoculate P. aeruginosa strain PAO1 in 5 ml of LB, and place in a shaking incubator overnight at 42 °C.
- Measure OD600 of overnight cultures and mix equal amounts of PAO1 and E. coli pFAC such that the final volume is between 1-1.4 ml.
- Centrifuge mixture at 6,000 x g for 5 min.
- Meanwhile, dry LB plates for conjugation: leave plates open in incubator at 37 °C for 10-15 min.
- Remove all but 50 µl of supernatant from cell mixture.
- Resuspend the cell pellet in the remaining 50 µl of supernatant and pipette onto a dry LB plate in one compact droplet.
- Carefully place the plate in a fume hood to dry (IMPORTANT Note: Do not let cells spread over the plate, this will significantly decrease the efficiency of the conjugation).
- Once the droplet is dry, invert the LB plate and incubate at 37 °C for 4-6 hr.
- While the cell mixture is incubating, prepare large (150 O.D. x 15 mm) PIA plates with 300 µg/ml of gentamycin. Before use, remove any residual moisture by placing in an incubator at 37 °C for 15-20 min.
- After the incubation of the cell mixture is complete, collect the cells using a sterile inoculation loop and in a microcentrifuge tube containing 1 ml LB and vortex, or pipette, to mix thoroughly.
- Spread the cells evenly onto the large PIA plates contain 300 µg/ml gentamycin. (IMPORTANT: For this step, it is best to add increasing volumes of the cell mixture to separate plates (e.g. Plate 1: 10 µl, Plate 2: 50 µl, Plate 3: 100 µl, Plate 4: 500 µl, etc.).
- Incubate overnight at 37 °C.
2. Detection and Isolation of Mucoid Colonies
- Remove plates from incubator and examine for mucoid colonies.
- Isolate a single mucoid colony and streak for isolation on small plates (100 O.D. x 10 mm) with PIA and 300 µg/ml of gentamycin.
- Incubate overnight at 37 °C.
- Repeat step 2.2 on the previous overnight culture.
- Repeat steps 2.2-2.4 two more times to determine stability of the mucoid isolate.
- After final isolate step, inoculate 4.5 ml of LB with a single mucoid colony and incubate in shaker overnight at 37 °C.
- Next day, label 3 microcentrifuge tubes for each mucoid mutant.
- Prepare two 1.25 ml aliquots of overnight culture into 2 tubes for genomic DNA extraction and identification of the transposon insertion site (see Protocol 3).
- Additionally, archive each mucoid mutant by pipetting 1 ml overnight culture into an appropriately-labeled cryovial containing an equal volume of 10% skim milk. Store at -86 °C.
3. gDNA Restriction Digestion and Ligation
- Extract gDNA from the mucoid mutant using any preferred method (e.g. phenol-chloroform, spin column, etc.).
- Determine the concentration of the gDNA, digest a total of 2 µg with 1 µl of the restriction endonuclease SalI, 0.5 µl of bovine serum albumin, 5 µl of SalI enzyme buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM Dithiothrietol (DTT), pH 7.9 at room temperature), and the appropriate volume of dH2O to achieve a total volume of 50 µl.
- Digest DNA overnight at 37 °C. (IMPORTANT: Depending on the efficiency of the restriction enzyme, it may be necessary to add an additional 1 µl to the overnight digest and incubate at 37 °C for 1-2 hr. This is in addition to the overnight digest.)
- Purify the digested DNA using any preferred method (e.g. agarose gel purification, spin columns) and elute with 25 µl of buffer.
- Ligate the digested DNA by mixing 11 µl of purified DNA with a preferred concentration of ~10-20 ng/µl, 1.5 µl of Ligation Buffer (330 mM Tris-acetate pH 7.5, 660 mM potassium acetate, 100 mM magnesium acetate, 5 mM DTT), 1.5 µl of 100 mM ATP and 1 µl DNA ligase.
- Incubate the mixture for 15 min at room temperature, and then inactivate the enzyme by incubating for 15 min at 70 °C. (Note: Incubating the mixture at room temperature for longer may increase the success of the ligation).
4. Inverse PCR (iPCR) and Sequence Analysis
- Perform iPCR on ligation product using the following forward and reverse primers and thermocycling conditions:
-Forward Primer (Gm3OUT): GGGCATACGGGAAGAAGTGA
-Reverse Primer (Gm5OUT): GACTGCCCTGCTGCGTAACA
Thermocycler Conditions:
1. 94 °C for 1 min.
2. 94 °C for 1 min.
3. 58 °C for 2 min.
4. 72 °C for 2 min.
5. 72 °C for 8 min.
6. 10 °C hold.
Repeat steps 2-4 for 34 cycles.
(NOTE: Amplified iPCR products can be stored at 4 °C.) - Perform agarose gel electrophoresis to confirm successful iPCR amplification.
- Perform sequencing on the iPCR product using the Gm3OUT and Gm5OUT primers. The insertion site and the orientation of himar1 are mapped out.
Representative Results
As illustrated in Figure 1, the mini-himar1 mariner transposon vector, pFAC, contains two 27 bp inverted repeats with TA insertion sites flanking the aacC1 gentamycin resistance cassette, with its σ70-dependent promoter, and a multiple cloning site (MCS). Additionally, the pFAC vector contains genes encoding the highly active himar1 transposase, β-lactamase (bla), and the tra transfer operon. The TA insertion sites allow for an efficient integration into the genome of P. aeruginosa strains. Accuracy in identifying and selecting mucoid mutants is critical for the overall success in determining gene loci involved with alginate regulation. Examples of mucoid and nonmucoid isolates are shown in Figure 1.
To more accurately identify mucoid mutants, the completed conjugations should be plated on multiple large plates (150 mm). The cell mixtures should be resuspended in sterile PBS and plated by spreading onto the PIA plates plus gentamycin (300 µg/ml). Dilutions of cell mixture in PBS will be spread on the plates with the goal of having between 1,000-3,000 colonies/plate. Based on the protocol above, each conjugation should produce a bank of ~20,000 mutants over 15-20 large plates. Optimal incubation time for the plates at 37 °C is between 24-36 hr. Due to the overproduction of alginate, the mucoid colony should be clear or white in color and have creamy or slimy appearance. If mucoid colonies cannot be discerned, then incubate for an additional 24-36 hr at room temperature. The mucoid mutants are best isolated by passing three times on a regular PIA plate with gentamycin. During this time, you can determine the stability of the mucoid phenotype. The purified mucoid colony will be used for genomic DNA extraction and identification of the genes by iPCR. iPCR products can be visualized using agarose gel electrophoresis (Figure 2). If successful, there should be a single amplicon at the appropriate size. PAO1 genomic DNA that has undergone restriction digestion and ligation can be used as a negative control. As illustrated in Figure 2, there is an absence of iPCR amplicon in PAO1, however we detect a single amplicon of ~1,400 bp and ~1,000 bp in PAO1-VE2 and PAO1-VE13, respectively. The size of these amplicons corresponds to the insertion of the himar1 mariner transposon into the promoter region of PA4033 (mucE) in PAO1-VE2 and internally in PA5484 (kinB) in PAO1-VE13. DNA sequencing of the iPCR amplicons should be carried out with the GM5OUT primer. When using the Basic Local Alignment Search Tool (BLAST) to analyze the target sequence, the inverted repeat on the 5' end of pFAC and TA dinucleotide mark the site and orientation of the himar1 insertion in the genome of P. aeruginosa (Figure 2).
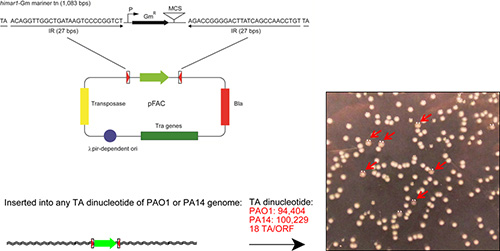
Figure 1. Schematic diagram of mini-himar1 mariner transposon vector, pFAC, and a representation of mucoid mutants. Plasmid pFAC contains a himar1 mariner transposon element with two inverted repeats, and a gentamycin resistance cassette (aacC1) for selection, a gene encoding the hyperactive himar1 transposase, and a conditional replicon. The mini-himar1 mariner transposon can cause high-density insertion in P. aeruginosa because of the abundance of substrate (TA dinucleotide). The number of TA dinucleotides in the genomes of two P. aeruginosa strains, PAO1 and PA14 are shown in red. The red arrows indicate the mucoid mutants identified using this procedure. Please click here to view a larger version of this figure.
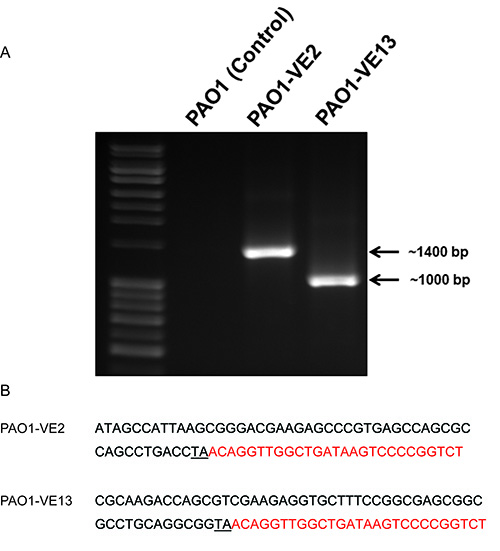
Figure 2. Representative iPCR and sequencing results. A) 1% agarose gel electrophoresis of iPCR amplification using the Forward Gm3OUT and Reverse Gm5OUT primers and a 1 kB ladder: PAO1 (negative control), PAO1-VE2 (1396 bp), and PAO1-VE13 (999 bp). The genomic DNA from three strains of P. aeruginosa was extracted and digested with SalI restriction enzymes followed by self-ligation. The closed circular DNA was used as templates for iPCR as described in the text. B) Comparative sequence analysis using the PAO1 reference genome determines the precise location of the himar1 mariner transposon insertion in PAO1-VE2 and PAO1-VE13. The sequence labeled in red indicates the 5' end of inverted repeat in pFAC. The insertion site of TA was underlined. A BLAST search of these two sequences will map the exact location and orientation of the himar1 transposon within the genome of PAO1.
Discussion
It is important to note that this method can be used in other Pseudomonas species with these alterations: incubate P. fluorescens and P. putida at 30 °C, and P. stutzeri at 42 °C; P. stuzeri should be cultured on LB plates supplemented with 150 µg/mL of gentamycin; on step 1.11, P. stutzeri cells should be transferred into 500 µl of LB instead of 1 ml. Additionally, there are two critical steps for this protocol. First, the recipient strain should be cultured at the appropriate temperature. For example, PAO1 should be incubated at 42 °C to increase the frequency of recombination15. The efficiency of the mutant library was significantly reduced if the recipient strain if not cultured at the correct temperature. Second, iPCR is highly reproducible, but the gDNA must be completely digested to make ensure that a circular covalently-closed DNA can be made through DNA ligation. If this is done correctly, the iPCR product will appear as one band on an agarose gel. If multiple bands are identified, they indicate that there are multiple insertions in the mutant. We found that iPCR results were strongly correlated with our Southern blot results (~100%). Therefore, iPCR can be used in place of Southern blot analysis for the number of insertions per mutant.
The mini-himar1 mariner transposon in pFAC has no transcriptional terminator, and it is regulated via a σ70-dependent promoter driving the expression of the gentamycin cassette. We observed that the position and orientation of the himar1 insertion had varying effects on the regulation of a particular gene. Genes were inactivated when integrated in the middle of a gene. An example of this is the inactivation of the sensory kinase gene kinB (PA5484)which causes a conversion to mucoidy in PAO1 (PAO1-VE13)11. Additionally, it can also up-regulate the expression of the gene, if inserted upstream of the coding region in the same orientation as the gene. An example of this is PAO1-VE2, where the himar1 drives the expression of mucE (PA4033)13. The limitation of this protocol is that only regulatory genes that are nonessential for growth can be identified. Additionally, those mucoid mutants identified using this method may not occur naturally.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Aeronautics and Space Administration West Virginia Space Grant Consortium (NASA WVSGC), Cystic Fibrosis Foundation (CFF-YU11G0) and NIH P20RR016477 and P20GM103434 to the West Virginia IDeA Network for Biomedical Research Excellence. We thank Vonya M. Eisinger for the technical assistance with this work.
Materials
| Luria Broth | Difco | 240230 | via Fisher Scientific |
| Pseudomonas isolation agar | Difco | 292710 | via Fisher Scientific |
| Small Plates (100 O.D. x 10 mm) | Fisher Scientific | 08-757-13 | |
| Large Plates (150 O.D. x 15 mm) | Fisher Scientific | 08-757-14 | |
| Glycerol | Fisher Scientific | BP229-4 | |
| Benchtop Shaking Incubator | New Brunswick Scientific | Innova 4080 | shake at 200 rpm |
| Cabinet Incubator | VWR | 1540 | |
| Benchtop Microcentrifuge | Sorvall | 75-003-287 | via Fisher Scientific |
| SmartSpec Plus Spectrophotometer | Bio-Rad | 170-2525 | or preferred method/vendor |
| Diposable Inoculation Loops | Fisher Scientific | 22-363-597 | |
| 1.5 mL Microcentrifuge Tubes | Fisher Scientific | 05-408-129 | |
| 2.0 mL Cryogenic Vials | Corning | 430659 | via Fisher Scientific |
| 15 mL Tubes | Fisher Scientific | 05-539-12 | |
| Skim Milk | Difco | DF0032-17-3 | via Fisher Scientific |
| DNeasy Blood and Tissue (250) | Qiagen | 69506 | or preferred method/vendor |
| QIAquick PCR Purification Kit (250) | Qiagen | 28106 | or preferred method/vendor |
| QIAprep Spin Miniprep Kit (250) | Qiagen | 27106 | or preferred method/vendor |
| FastLink II DNA Ligation Kit | Epicentre Technologies | LK6201H | via Fisher Scientific |
| Accu block Digital Dry Bath | Labnet | NC0205808 | via Fisher Scientific |
| Sal1, restriction endonuclease | New England BioLabs | R0138L | |
| EasyStart Micro 50 | Molecular BioProducts | 6020 | via Fisher Scientific |
| Taq DNA Polymerase | New England BioLabs | M0267L | |
| iCycler, Thermocycler | Bio-Rad | 170-8740 | |
| LE agarose | Genemate | 3120-500 | via Fisher Scientific |
| Gentamycin Sulfate | Fisher Scientific | BP918-1 | |
| 2.0 mL Cryogenic Vials | Corning | 430659 | via Fisher Scientific |
References
- Govan, J. R., Deretic, V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539-574 (1996).
- Chitnis, C. E., Ohman, D. E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8, 583-593 (1993).
- Franklin, M. J., et al. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176, 1821-1830 (1994).
- Franklin, M. J., Ohman, D. E. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184, 3000-3007 (2002).
- Deretic, V., Gill, J. F., Chakrabarty, A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 169, 351-358 (1987).
- Mathee, K., McPherson, C. J., Ohman, D. E. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN).. J. Bacteriol. 179, 3711-3720 (1997).
- Boucher, J. C., Schurr, M. J., Yu, H., Rowen, D. W., Deretic, V. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology. 143, 3473-3480 (1997).
- Martin, D. W., Holloway, B. W., Deretic, V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175, 1153-1164 (1993).
- Damron, F. H., Goldberg, J. B. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol. Microbiol. 84 (4), 595-607 (2012).
- Browne, P., Barret, M., O’Gara, F., Morrissey, J. P. Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 10, 300 (2010).
- Damron, F. H., Qiu, D., Yu, H. D. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 191, 2285-2295 (2009).
- Damron, F. H., et al. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J. Bacteriol. 194, 1317-1330 (2012).
- Qiu, D., Eisinger, V. M., Rowen, D. W., Yu, H. D. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 104, 8107-8112 (2007).
- Schurr, M. J. Which bacterial biofilm exopolysaccharide is preferred, Psl or alginate. J. Bacteriol. 195, 1623-1626 (2013).
- Wong, S. M., Mekalanos, J. J. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 10191-10196 (2000).
- Lampe, D. J., Akerley, B. J., Rubin, E. J., Mekalanos, J. J., Robertson, H. M. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U.S.A. 96, 11428-11433 (1999).

