Site Directed Spin Labeling and EPR Spectroscopic Studies of Pentameric Ligand-Gated Ion Channels
Summary
This article describes methods for site-directed spin labeling and reconstitution of pentameric ligand-gated channels for Electron Paramagnetic Resonance studies. This protocol can be adapted for any membrane protein. The reconstitution method described here can also be used for patch-clamp measurements of macroscopic and single-channel currents in a defined lipid system.
Abstract
Ion channel gating is a stimulus-driven orchestration of protein motions that leads to transitions between closed, open, and desensitized states. Fundamental to these transitions is the intrinsic flexibility of the protein, which is critically modulated by membrane lipid-composition. To better understand the structural basis of channel function, it is necessary to study protein dynamics in a physiological membrane environment. Electron Paramagnetic Resonance (EPR) spectroscopy is an important tool to characterize conformational transitions between functional states. In comparison to NMR and X-ray crystallography, the information obtained from EPR is intrinsically of lower resolution. However, unlike in other techniques, in EPR there is no upper-limit to the molecular weight of the protein, the sample requirements are significantly lower, and more importantly the protein is not constrained by the crystal lattice forces. Therefore, EPR is uniquely suited for studying large protein complexes and proteins in reconstituted systems. In this article, we will discuss general protocols for site-directed spin labeling and membrane reconstitution using a prokaryotic proton-gated pentameric Ligand-Gated Ion Channel (pLGIC) from Gloeobacter violaceus (GLIC) as an example. A combination of steady-state Continuous Wave (CW) and Pulsed (Double Electron Electron Resonance-DEER) EPR approaches will be described that will enable a complete quantitative characterization of channel dynamics.
Introduction
Over the last decade, the structural understanding of pentameric ligand-gated ion channels (pLGIC) has grown in leaps and bounds, owing to multitudes of high-resolution structures of several members of the family. Key factors that led to the current advancements in the field include, the discovery of prokaryotic pLGIC channels,1-3 major progresses in eukaryotic membrane protein expression,4-6 and tremendous breakthroughs in structure determination approaches.7 These structures provide a clear consensus on the overall conservation of the three-dimensional architecture of pLGIC. However, two major areas that seem to trail behind are the functional characterization of these channel preparations and the mechanistic description of channel function.
Gating conformational changes are complex and occur over a 60 Å distance along the length of the channel and these transitions are extensively modulated by membrane lipids. In particular, negative lipids, cholesterol, and phospholipids have been shown to modulate the function of pLGIC8-11. While the precise role of these lipid constituents in channel function remains unknown, a complete molecular understanding of gating would require studying these channels in their native environment. Site-Directed Spin Labeling (SDSL) and Electron Paramagnetic Resonance (EPR) spectroscopy are the techniques of choice for studying protein dynamics in reconstituted systems. EPR spectroscopy is not limited by the molecular size (as is NMR) or the optical property of the sample (as is fluorescence spectroscopy), and thereby allows measurements of full-length constructs reconstituted in native lipid conditions. The technique is extremely sensitive and has relatively low sample requirements (in the pico-mole range). Both these aspects make the technique well suited for studying large membrane proteins that are difficult to express in over milligram quantities.
The use of EPR spectroscopy in combination with site-directed spin labeling was developed by Wayne Hubbell and colleagues, and has been adapted for studying a range of protein types.12-24 EPR data have been used to investigate secondary structures, changes in the protein conformation, membrane-insertion depths, and protein-protein/protein-ligand interactions.
The method involves cysteine substitution at positions of interest by site-directed mutagenesis. To ensure site-specific labeling, it is necessary to substitute native cysteines with another amino acid (e.g., serine) to create a cysteine-less template. By far, the most popular spin label is a thiol-specific MTSL: (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate that attaches to the protein through a disulfide bond bridge. Due to its high specificity, relatively small size (slightly larger than tryptophan), and flexibility of the linker region, this spin label has been shown to have excellent reactivity even with a buried cysteine. Furthermore, to maximize reactivity, the labeling reaction of the protein is carried out in the detergent-solubilized form. After separation of the excess free spin-label by size exclusion chromatography, the protein is reconstituted into liposomes or bilayer-mimicking systems of defined lipid composition. In general, cysteine mutagenesis is well tolerated in most parts of the protein, and the relatively small size of the spin-probe causes minimal perturbation to the secondary and tertiary structures. To ensure that the modification retained wild type functions, the labeled and reconstituted channels can be studied by patch-clamp measurements.
The labeled-functional protein is then subjected to spectroscopic measurements, which essentially provide three main types of information:12,14,15,20,22,23,25-27 spin-probe dynamics by lineshape analysis; accessibility of the probe to paramagnetic relaxation agents; and distance distribution.27 EPR distances are measured by two different approaches. The first is based on the Continuous Wave (CW) technique, where spectral broadening arising from dipolar interactions between spin-labels (in the 8 – 20 Å distance range) is used to determine distance.28,29 The second is a pulsed-EPR method where distance measurements can be extended up to 70 Å.30-34 In Double Electron Electron Resonance (DEER), oscillations in the spin-echo amplitude are analyzed to determine distances and distance distributions. Here the spin echo is modulated at the frequency of the dipolar interaction. Together, these parameters are used to determine protein topology, secondary structural elements, and protein-conformational changes.
Protocol
1. Site-Directed Mutagenesis and Cys Mutations
- Cloning and Mutagenesis
NOTE: GLIC wild type (wt) 35 has a single-native cysteine (C27), which is mutated to serine to create a cysteine-less background. Cysteine mutations are introduced on the cysteine-less background by site-directed mutagenesis using primers that carry a cysteine codon at the desired position36.- Mix 5 μl of 10x reaction buffer, 1 μl of 100 ng/μl cysteine-less GLIC template DNA35, 0.5 μl each of 40 μM forward and reverse primer36, 1 μl of dNTPs (100 mM), and 41 μl of deionized water. Pipette the sample a few times to mix it thoroughly, spin (6,000 rpm) and finally add 1 μl of DNA polymerase.
- Set the thermo-cycler to the following protocol:
- Denaturation at 95 ºC for 4 min.
- Denaturation at 95 ºC for 30 sec.
- Annealing at 55 ºC for 1 min.
- Elongation at 68 ºC for 10 min (roughly 1 min per kb plasmid length).
- Repeat steps 1.1.2.2 – 1.1.2.4 for 18 cycles.
- A final elongation at 68 ºC for 10 min.
- Holding temperature at 4 ºC
NOTE: Annealing temperature is calculated based on the primer Tm.
- Add 1 μl of DpnI to the reaction mixture, mix thoroughly by pipetting, and spin down (6,000 rpm) for 1 min. Incubate for 2 hr at 37 °C.
- Transformation
- Thaw E. coli competent cells on ice. Aliquot 30 μl of cells into a 14 ml sterile transformation tube and add 1 μl of digested DNA to the cells. Mix by tapping on the side of the tube. Incubate for 20 min on ice.
- Place the tube in a 42 ºC water bath for 45 sec and then place it back on ice for 2 min. Add 400 μl sterile SOC media and shake the cells in an incubator at 37 ºC for 1 hr.
- Plate 100 μl of cells on LB plates containing kanamycin (50 μM) and 1% glucose. Incubate the plates O/N at 37 ºC.
- Harvest a single colony from the plate and inoculate in 5 ml O/N cultures containing LB/kanamycin media. Incubate the culture O/N (~16 hr) at 37 ºC in an incubator shaker.
- Extract DNA from the O/N culture by minipreparation37. Quantify the yield of DNA using a spectrophotometer by measuring the absorbance at 260 nm wavelength. The concentration should be ~100 – 200 ng/μl. Confirm the presence of mutation by standard recombinant DNA sequencing strategies 38,39.
2. GLIC Expression and Purification
- Transformation
- Following the steps described in section 1.2, transform 30 μl of C43 competent cells with 1 μl (~100 – 200 ng/μl) of the plasmid containing the gene coding for GLIC along with a Human Rhinovirus (HRV)-3C protease cleavage site (For the plasmid and gene sequence, refer to the supplemental text) 35 (from step 1.2.5).
- Plate 100 μl of cells on LB-agar plates containing 1% glucose and 50 μg/ml kanamycin, and incubate them O/N (~16 hr) at 37 ºC in an incubator.
- Check visually for colonies and ensure that they are not over-grown or show the presence of satellite colonies. Promptly seal the plates with parafilm and store them at 4 ºC.
- Setting up O/N-cultures and Preparation of growth media
- Pick a single isolated colony using loop inoculating wire and transfer into a 100 ml sterile flask containing 20 ml of Luria Broth (LB) media supplemented with sterile 1% glucose and 50 μg/ml kanamycin. Incubate them O/N (~16 hr) at 37 ºC in a shaker at 250 rpm.
- Autoclave 900 ml Terrific Broth (TB) media (See Media and Buffer Composition) in 2.8 L Fernbach glass flasks for the bacterial culture in steam cycle at 121 ºC for 20 min.
- Setting up large growth cultures
- Add 100 ml of sterile potassium phosphate buffer (See Media and Buffer Composition) to the autoclaved TB media. Add sterile glucose (0.2% final concentration), kanamycin (50 μg/ml), and 10 ml of the O/N culture. Incubate at 37 ºC in a shaker at 250 rpm.
- Check the optical density at 600 nm wavelength (OD600) using densitometer or a spectrophotometer every hour until it reaches 0.5 (~2 hr). At this point reduce the speed to 150 rpm and lower the temperature to 18 ºC. Monitor the OD600 every 30 min until it reaches 0.8 (~1 hr).
- Add 50 ml of glycerol and continue shaking the culture until the OD600 reaches 1.0 – 1.2 (~15 – 30 min).
NOTE: Under these conditions, it takes about an hour for the culture to reach 18 ºC from 37 ºC. It is important that glycerol is added to the culture after it has cooled to 18 ºC. - Induce the cells with 200 μM IPTG (isopropyl-thio-β-galactoside) and reset the shaker back to 250 rpm and incubate further for ~16 hr at 18 ºC.
- Protein Purification
- Ensure that the OD600 at the end of 16 hr is ~16 – 18. Harvest the cells in 1.0 L centrifuge bottles by spinning at 8,500 x g, 4.0 ºC for 15 min. Decant the supernatant and weigh the bottle with the cell pellet. Calculate the weight of the pellet by subtracting weight of the empty bottle from the total weight.
NOTE: The expected cell pellet weight is ~30 – 35 g/L. Although, it is to be noted that there may be variability in the final OD600 and cell pellet weight across mutants. - Re-suspend the bacterial pellet in 150 ml of ice-cold Buffer A (100 mM NaCl, 20 mM Tris, pH 7.4) per 1.0 L of culture. Add DNase I (100 μg/ml) and protease inhibitors (phenylmethyl sulfonyl fluoride (1 mM), leupeptin (2.0 μg/ml), aprotinin (2.0 μg/ml), and pepstatin A (1 μg/ml)).
- Homogenize the cells by passing them through a cell disrupter in a 4.0 ºC cold room. Repeat the process three times so that most of the bacteria is lysed. Centrifuge the cells at 14,000 x g for 15 min and pipette the supernatant out and transfer into a clean centrifuge tube. Discard the pellet, which contains un-lysed cells and inclusion bodies.
- Centrifuge the supernatant at 168,000 x g for 1 hr. Carefully decant the supernatant without disturbing the membrane pellet.
- Pool the membrane pellet from 1 L of culture and re-suspend it in Buffer A (supplemented with 10% glycerol) to a final volume of 50 ml. Add 0.5 g of n-Docecyl-β-D-Maltopyranoside (DDM) to the membrane suspension and nutate for 2 hr at 4.0 ºC.
- After membrane solubilization, remove cell debris from the lysate by centrifugation at 168,000 x g for 1hr. In the meantime prepare amylose resin. Transfer 3 ml of resin using a pipette into an empty polypropylene chromatography column. Wash the resin by passing 10 bed volumes of water 3 times and then with 10 bed volumes of Buffer A containing 0.5 mM DDM.
- After centrifugation, gently take the supernatant out using a pipette and transfer to a clean 50 ml conical tube. For batch-wise binding, add pre-equilibrated amylose resin to the conical tube. Bind the extracted protein to amylose resin by nutating the mixture for 2 hr at 4.0 ºC.
- Pass the entire slurry through a chromatography column and collect the flow-through. Then pass the entire collected flow-through over the resin a second time to maximize binding.
- Pass 10 bed volumes of buffer A with 0.5 mM DDM, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) and 1 mM ethylenediaminetetraacetic acid (EDTA) to remove unbound proteins.
- Elute the GLIC protein with 10 ml of Buffer A containing 40 mM maltose in addition to 0.5 mM DDM and 0.05 mM TCEP. TCEP prevents oxidation of cysteine side-chains. Collect the entire elute.
- Concentrate the eluted protein using centrifugal concentrator (Molecular Weight Cut-off = 50 KDa) to 4 – 6 mg/ml and add HRV-3C protease (200 μg per 10 mg protein) and incubate O/N at 4.0 ºC.
NOTE: Ascertain the completion of protease digestion by running the samples on SDS PAGE gel (See step 2.5.5 and Figure 1).
- Ensure that the OD600 at the end of 16 hr is ~16 – 18. Harvest the cells in 1.0 L centrifuge bottles by spinning at 8,500 x g, 4.0 ºC for 15 min. Decant the supernatant and weigh the bottle with the cell pellet. Calculate the weight of the pellet by subtracting weight of the empty bottle from the total weight.
- Site-Directed Spin-labeling
- Make about 100 μl of 200 mM stock of (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) MTSL 40 in DMSO and store the stock at -20 ºC in 25 μl aliquots.
- Use the protease-digested sample for spin-labeling with MTSL. Add 10-fold molar excess of MTSL spin-label (GLIC monomer: MTSL in 1:10 molar ratio). Incubate on ice for 1 hr. Add a second shot of spin label at a 5-fold molar excess ratio and incubate for another hour.
NOTE: For buried positions (with low spin labeling efficiency, described below), a 30 times molar excess of spin label is added and the protein is incubated for 6 hr. - Pass the sample through a size-exclusion Fast Protein Liquid Chromatography (FPLC) column that is pre-equilibrated with Buffer A and 0.5 mM DDM to separate the cleaved MBP and the excess free spin-label from GLIC pentamers. Collect 1 ml fractions for a total of 30 ml elution. Pool the fractions containing GLIC pentamers (Figure 1). Follow the manufacturer's instruction to run the FPLC.
- Determine the concentration of the sample using a spectrophotometer by measuring the absorbance at 280 nm wavelength. Molar extinction coefficient and the estimated molecular weight of GLIC monomer are 49,850 M-1 cm-1 and 36,380 Da, respectively. Concentrate the protein solution using a centrifugal concentrator (Molecular Weight Cut-off = 50 KDa) to a final concentration of ~8-10 mg/ml and place it on ice. The sample is now ready for reconstitution.
- As an additional quality check to ensure that the preparation is biochemically homogenous, run a reducing (1.0% β-ME) SDS-PAGE gel.
NOTE: The commassie stained gel should show a single band at ~33 kDa corresponding to the GLIC monomer (Figure 1). - For the spin-labeled mutants suspected of exhibiting dipolar broadening, under-label the samples in the presence of diamagnetic label 41. Incubate the eluted-protein with 0.1-fold MTSL and incubate on ice for 1.0 hr. Afterwards, add 20-fold molar excess of diamagnetic label (1-acetoxy-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methane thiosulfonate.
NOTE: The diamagnetic reagent used here is iso-steric to the paramagnetic label with an identical labeling chemistry. - Incubate the sample on ice for 2 hr. Pass the sample through a size exclusion chromatography column that is pre-equilibrated with Buffer A and 0.5 mM DDM as in Step 2.5.3.
- Efficiency of spin labeling
- Spin-labeling efficiency is defined as the ratio of the spin-label concentration to the concentration of cysteine side-chain (GLIC-monomer). To determine the efficiency of spin-labeling of the sample, the integrated intensity of the sample will be compared with a standard of known concentration using a calibration plot. Make solutions of an EPR standard (such as 2,2,6,6-tetramethyl-1-piperidinyloxyl: TEMPO) in a range of concentrations (10 – 250 μM) by dissolving in water.
- Measure the EPR signal for each of these solutions and determine the area under the curve (double integration of the EPR signal) for each concentration (as described in Steps 6.1). Generate a calibration curve by plotting the measured area as a function of the TEMPO concentration and fitting with a straight line.
NOTE: The area under a derivative EPR signal is a better description of the spectral intensity than the peak to peak amplitude. Integrating the first-derivative EPR signal will give the absorption spectrum, a second integration will then give the area under the absorption spectrum (which is proportional to the spin concentration). Ensure a constant baseline both in the low-field and high field region of the spectrum. - Measure the concentration of the spin-labeled protein in the detergent using a spectrophotometer (as in step 2.5.4). Now measure the EPR signal of the sample and then determine the area of the double integral of the EPR signal following steps in section 6.1. By using the calibration plot, determine the concentration of the label which corresponds to the measured EPR signal. Calculate the spin labeling efficiency as the molar ratio of spin label and protein concentration.
3. GLIC Reconstitution in Asolectin Membranes
- Liposome Preparation
- Rinse a clean 25 ml round bottom flask with 5 ml of chloroform and dry the flask in a stream of N2 gas in the fume hood. Transfer 10 mg of asolectin (soybean polar extract 25 mg/ml stock in chloroform) into the flask and dry the lipids under a continuous stream of N2 gas.
NOTE: The choice of lipids for reconstitution is based on findings from experiments in section 4. - When the chloroform has evaporated, place the flask in a vacuum for 1 hr to ensure complete drying. Add 1 ml of reconstitution Buffer A to the dried lipid and vortex the flask vigorously to get the lipid pellet into suspension.
- To prepare small unilamellar vesicles, sonicate the lipid suspension in a cold bath sonicator until the vesicle solution becomes more or less translucent and no clumps are observed (roughly 10 – 15 min). To this mixture, add DDM to a final concentration of 4 mM and incubate at RT for 30 min.
- Rinse a clean 25 ml round bottom flask with 5 ml of chloroform and dry the flask in a stream of N2 gas in the fume hood. Transfer 10 mg of asolectin (soybean polar extract 25 mg/ml stock in chloroform) into the flask and dry the lipids under a continuous stream of N2 gas.
- Reconstitution
- Add the purified protein from step 2.5.4 to the lipid mixture.
NOTE: The protein-to-lipid ratio depends upon the type of experiment. For macroscopic current measurements, a 1:10,000 ratio is used. For EPR studies, reconstitution is done at a higher protein to lipid ratio (1:2,500) to maximize the signal. It is previously shown that at these concentrations, no lateral aggregation is observed42.
NOTE: Typical working amount of GLIC for EPR is ~1 mg, although based on the position probed, lower amounts would work as well. - Incubate the sample at 4 ºC for 1 hr with gentle rotation then dilute the sample to 15 ml with Buffer A.
NOTE: This step brings the detergent concentration below the critical micellar concentration (CMC). - Remove the residual detergent in the suspension by adding polystyrene beads (with hydrophobic pores that trap detergent). Add 80 mg of clean the beads and incubate the liposome suspension O/N on a nutator at 4 ºC.
- To prepare the beads for use, weigh ~100 mg and transfer them to a 50 ml conical tube. Add 10 ml of methanol, shake vigorously, spin down the beads at 8,000 x g for 2 min, and decant the methanol. Repeat this process of methanol wash three times. Repeat the same process with 30 ml of deionized water, washing three times.
- The next day, transfer the liposome to a 25 ml ultracentrifuge tube using a column filter to remove the beads and further dilute the sample to 25 ml. Centrifuge the samples at 168,000 x g for 2 hr. Decant the supernatant and re-suspend the pellet using 100 μl of Buffer B.
- Add the purified protein from step 2.5.4 to the lipid mixture.
4. Determine Optimal Lipid Composition for Reconstitution by FRET
- Use a fluorescence resonance energy transfer (FRET)-based assay to determine the membrane composition that maintains the protein monodisperse.
- Purify wt GLIC (with the native cysteine, C27) by following steps described in section 2. Label the protein at a 10:1 (label/monomer) molar ratio with either fluorescein-maleimide or tetramethylrhodamine-maleimide using steps in section 2.5.
- Following steps in section 3.2 for reconstitution, prepare three different samples for each lipid composition to be tested; First, fluorescein-labeled GLIC in a molar ratio of 1: 1,500 (pentamer: lipid). Second, rhodamine-labeled GLIC in a molar ratio of 1: 1,500 (pentamer: lipid). Third, mix detergent solubilized fluorescein and rhodamine labeled GLIC (1:1 molar ratio). Reconstitute this mixture in 1: 750 (pentamer: lipid) ratio.
NOTE: We used three lipid systems: asolectin, POPC:POPG (3:1 molar ratio), and E. coli polar extract. The protein to lipid reconstitution ratio in the FRET experiment is higher than that used for EPR measurements (1:750 in comparison to 1: 1,500 used for EPR studies). - Dilute the liposome suspension (to reduce the light scattering signal; ~5 μl in 995 μl of Buffer A) then pipette the sample into a quartz cuvette and place it in a fluorimeter.
- Set the excitation wavelength at 490 (which corresponds to the absorption maxima for the donor:fluorescein). Using manufacturer's instructions, record the emission spectra from 500 nm to 660 nm.
NOTE: For the fluorescein-labeled GLIC sample, you will see a peak at 518 nm (corresponding to fluorescein emission) with no peak at 572 nm (corresponding to rhodamine emission). For the rhodamine-labeled GLIC sample, you will not see a peak at 518 nm but instead at 572 nm arising from direct excitation of rhodamine at 490 nm. For the sample containing both fluorescein and rhodamine labels, a decrease in fluorescein peak intensity and a corresponding increase in rhodamine peak is a FRET signal that is likely an indicator of lateral aggregation of the protein on the membrane. - Monitor the amplitude of rhodamine emission at 572 nm at various time intervals (from 0 – 24 hr) recording at every hour for the first 6 hr and then after O/N incubation (Figure 2). For each interval, measure the fluorescence intensity at 572 nm (for rhodamine: Ia) and at 518 nm (for fluorescein: Id). Subtract the contribution of direct rhodamine activation (sample 2 in step 4.3) from Ia (Iac). Determine the FRET ratio as Iac/(Iac + Ib). Compare FRET at different time intervals and across different lipid compositions.
NOTE: As a control, perform step 4.5 and 4.6 for detergent solubilized samples and compare the emission spectra to those from reconstituted liposomes. Use an equimolar mixture of labeled protein in detergent (described in step 4.3) and dilute in Buffer A supplemented with 0.5 mM DDM. It is expected that detergent samples will show minimal FRET signal.
5. Functional Measurements by Patch-clamp Recordings
- Prepare proteoliposomes for patch-clamp measurements in exactly the same way as for EPR studies (Section 3.1).
NOTE: However, adjust the protein:lipid ratio based on the type of measurement being made (Single-channel vs Macroscopic recordings). Flash-freeze the proteoliposomes in liquid nitrogen and store at -80 ºC until use. Typically, reconstitute GLIC in 1: 10,000 protein: lipid (molar ratio) for macroscopic currents and 1: 50,000 molar ratio for single-channel recordings. - Thaw liposomes on ice. Rinse a clean glass slide with water and ethanol and dry completely. Place a 10 μl drop of the proteoliposome on the center of the slide and dry O/N in a desiccator at 4 oC under vacuum.
- Re-hydrate the dried proteoliposomes by adding 20 μl of Buffer B (150 mM NaCl, 10 mM HEPES, pH 7.0) and place the slide in a petriplate on top of a moist filter paper. Cover the plate and allow the rehydration to proceed for about 2 hr.
NOTE: This process yields giant multi-lamellar liposomes that are conducive to patch from. - Pull patch pipettes from thin-walled borosilicate glass capillaries (~1 – 2 μm tip diameter) using Manufacturer's recommended setting on the pipette puller. Heat-polish using a fire-forge to a resistance of 1.5 – 2MΩ when filled with Buffer B.
- Fill the recording chamber with Buffer B using a pipette and attach the Ag/AgCl ground electrode. Using a 2 μl pipette tip, gently dislodge the liposomes from the edge and transfer 1 μl of the suspension on the recording chamber. Wait a few minutes for the liposomes to settle to the bottom.
- Fill the patch-pipette with Buffer B using a nonmetallic syringe needle and mount it on the amplifier head-stage. Identify a single-isolated vesicle to patch. Apply a slight positive pressure to prevent the patch-pipette from clogging, and insert the tip into the recording chamber.
- Apply test pulses to measure the pipette resistance and compensate for the pipette voltage offset. Approach the vesicle, and when contact is made, apply a slight negative pressure to gently pull the vesicle against the patch pipette to form a gigaohm seal.
- Monitor the resistance of the pipette during the entire process. Once the gigaohm seal is formed, withdraw the pipette carefully away from the liposome to form an inside-out patch. Turn off the test pulse.
- Set the holding potential to -100 mV, and apply a pH pulse. Low pH was obtained using 10 mM sodium citrate Buffer C (150 mM NaCl, 10 mM citrate, pH 3.0). (Figure 3)
NOTE: Recordings are made at a sampling frequency of 10 kHz for macroscopic and 40 kHz for single channel measurements.
6. EPR Measurements
- Continuous-Wave EPR Spectroscopy
- Transfer the liposome suspension from Step 3.2 into a 200 μl tube and pellet the samples using a centrifuge. Discard the supernatant and sample is ready for EPR measurements.
NOTE: For EPR measurements, it is important to remove as much buffer from the liposome sample as possible. Since aqueous samples absorb the electric part of the microwave resulting in heating and loss of sensitivity. - To carry out buffer exchange, transfer 20 μl of liposome pellet using a pipette to a microfuge tube. Add 180 μl of Buffer D (100 mM NaCl, 10 mM citrate, pH 3.0) and incubate at 42 ºC water bath for 5 min. Centrifuge the sample, remove the supernatant using a pipette and repeat the process three times to ensure complete buffer exchange.
NOTE: Alternatively, buffer exchange can be made by subjecting the samples to multiple freeze/thaw cycles. - Gas permeable plastic capillaries are suitable for measurements of both spectral line-shapes and solvent accessibility. Tap the capillary onto the pelleted proteoliposome to draw the sample inside and seal the end with bone wax.
NOTE: Typical sample volumes are ~5 μl and a desirable spin concentration is 150 μM. If the objective is to measure line shapes alone, one can use 0.4 mm OD quartz capillary tubes. If necessary, a pipette could be used for filling the sample inside the capillary. - Operate the spectrometer
- Perform CW-EPR measurements at RT (292 – 297 K) on an X-band spectrometer equipped with a dielectric resonator as per the manufacturers' instruction manual.
- Turn on the water chiller, power supply, and microwave bridge console. Allow the system to thermally equilibrate for about 30 min before the recording. Open the acquisition software.
- Insert the sample tube/capillary into the resonator, make sure that the section of the tube filled with sample is at the center of the resonator cavity. Tune the resonator as per the instruction on the spectrometer manual.
- Record the first derivative absorption spectrum by field sweep at an incident microwave power of 1.0 mW, a modulation frequency of 100 kHz and a sweep width of 100 G. Determine the optimal power for the experiment by carrying out a progressive power saturation experiment where the peak-to-peak height of the first derivative CW EPR signal is measured as a function of the square root of the incident microwave power.
NOTE: At lower microwave powers, the intensity of the signal increases in proportion to the square root of the power as long the equilibrium population of the spin states is unaffected. However, if power is further increased, the spin-lattice relaxation can no longer maintain the equilibrium population difference of the two spin states and the signal amplitude starts to plateau or "saturate." Beyond this point, the signal amplitude may actually decrease with increasing observe power due to population inversion of spin sates. In addition, for broadened spectra with extensive wings where a well-defined flat baseline is not visible, extend the sweep width to 150 – 200 G to prevent area loss. - Start with using a modulation amplitude of 1.0 G.
NOTE: This value is sample-dependent and adjusted to decrease the low frequency noise in the signal. If too large a value is used, it may distort or broaden the signal. - Set the time constant and conversion time to 20.48 msec, sweep time to 20.97 sec, and resolution to 1024 points.
NOTE: The time constant is also based on the sample and is adjusted to filter out high frequency noise. - After the first scan, set the center field at the center of the EPR signal and choose sweep width such that the trace includes the signal and sufficient baseline on either side.
NOTE: Also, to improve the signal/noise ratio, increase the number of scans based on the strength of the EPR signal. Include sweep width and number of points - Measure the accessibility of the spin label to collisional relaxing agents O2 or ethylenediaminediacetic acid (NiEDDA) using power saturation protocols27 described in step 6.1.4.4.
NOTE: Prepare NiEDDA as described previously 26. - First perfuse the sample in the cavity with N2 for 5 min (to flush out O2). Measure the spectra for each microwave power between 5 – 35 dB in steps of 3 dB with a sweep width of 30 G.
- Then perfuse the sample in the cavity with air for 10 min and measure the spectra at different microwave powers (5 – 35 dB in steps of 3 dB) as in the step 6.1.4.9.
- Incubate 20 μl of liposome pellet with 180 μl of 50 mM NiEDDA (in Buffer B or Buffer D) in a 42 ºC water-bath for 5 min. Centrifuge the sample and remove the supernatant using a pipette. Load the sample into the capillary and place it in the resonator cavity. Repeat step 6.1.4.9.
NOTE: The principle behind the experiment is that the interaction with paramagnetic relaxing agents through Heisenberg spin exchange would prevent saturation of the signal and population inversion. Water soluble NiEDDA and lipid soluble molecular O2 are used as reporters for water (both bulk and intraprotein vestibules) and membrane exposed positions on the protein, respectively.
- Transfer the liposome suspension from Step 3.2 into a 200 μl tube and pellet the samples using a centrifuge. Discard the supernatant and sample is ready for EPR measurements.
- Data Analysis:
NOTE: The analysis of EPR data involves the following steps:14,22,23,25-27- Calculate the mobility of the spin probe (ΔHo-1), as the inverse of the central line width of the first derivative absorption spectrum. ΔHo-1 is reflective of the motional freedom or the dynamics of the probe.
- Calculate the accessibility parameter (П), which is an estimate of the accessibility of the probe to other paramagnetic collisional relaxing agents, using steps described below.
NOTE: Interaction of the spin-probe with paramagnetic relaxing agents enhances the spin-lattice relaxation rate (T1) of the nitroxide by Heisenberg spin exchange.27- Estimate the accessibility parameter (П) from power saturation experiments (6.1.4.8) in which the vertical peak-to-peak amplitude of the central line of the first derivative EPR spectrum is measured at different microwave power.25 Plot the amplitude of the signal as a function of the square-root of the microwave power and fit with the following equation 27.
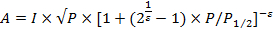
NOTE: Where A is the amplitude of the signal, is a measure of the homogeneity of the saturation of the resonance line (which is usually between 0.5 and 1.5), I is a scaling factor, P is the incident power, and the represents the value at which the first derivative amplitude is half of its unsaturated value.
- Estimate the accessibility parameter (П) from power saturation experiments (6.1.4.8) in which the vertical peak-to-peak amplitude of the central line of the first derivative EPR spectrum is measured at different microwave power.25 Plot the amplitude of the signal as a function of the square-root of the microwave power and fit with the following equation 27.
- Calculate ΔP1/2 as the difference in P1/2 values in the presence (O2 or NiEDDA) and absence (perfused with N2) of paramagnetic relaxing agents. Calculate the accessibility parameter (П) using the equation:
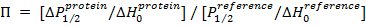
NOTE: Where P1/2reference and ΔHoreference are the half maximal power saturation value and the central line width, respectively, for the reference standard (such as DPPH (2,2-diphenyl-1-picrylhydrazyl))27.
Representative Results
Biochemical Characterization of Spin-labeled GLIC Mutants
Following the above described protocol would typically yield GLIC-MBP fusion protein in the range of 10 – 12 mg/L of culture. Although this value may vary across different mutants, particularly for positions buried within the protein, the yield may be significantly compromised. In these cases, the culture volumes may require scaling up. The cleavage of the fusion construct by HRV3C protease is carried out O/N for convenience. By running the samples on SDS PAGE gel, we found that this cleavage is complete within 30 – 60 min. (Figure 1A). Purified GLIC on size exclusion gel filtration column elutes predominantly as a pentamer with a retention volume of 11.3 ml. The MBP fraction elutes at 15 ml and aggregates elute at the void volume of the column (~7.0 ml). The spin-label comes at the end of the elution (~24 ml) (Figure 1B)
FRET Based Assay for Monitoring Aggregation of GLIC Reconstituted in Various Membrane Lipids
FRET-assay is used to evaluate aggregation of GLIC pentamers in reconstituted vesicles (came together within a distance <50 Å on the membrane). Figure 2 shows data from GLIC wt (with C27, labeled with either Fluorescein or tetramethylrhodamine) and reconstituted in E.coli polar lipids, POPC:POPG (3:1), POPE:POPG (3:1) or asolectin. Fluorescence measurements show that GLIC reconstituted in asolectin showed no FRET and even freeze/thawing the sample (conditions known to promote aggregation) produced minimal change in the FRET levels43. For this reason, asolectin is the lipid mixture of choice for functional and spectroscopic measurements used in these studies.
Macroscopic Behavior of GLIC in Proteoliposomes
Functional properties of purified GLIC are characterized by patch-clamp measurement in reconstituted proteoliposomes. Representative macroscopic current recording from wt GLIC from an inside-out patch in response to a pH-jump is shown in Figure 3. At -100 mV holding potential, switching to pH 3.0 buffer produces rapidly activating inward currents (10 – 100 msec) that decay to about 10% of the peak amplitude within seconds43 (1 – 3 sec, Figure 3). Currents recover from this macroscopic decay or desensitization by switching back to neutral pH. While channels in the inside-out patch were sensitive to changes in the bath pH, there was no effect of pH change in the pipette solution (data not shown) suggesting that GLIC was predominantly oriented in one direction such that in the patch the extracellular domain faced the bath/solution exchanger.
Structural Rearrangements During Channel Activation
At RT, the spin-probe can adopt multiple rotameric conformations, and the spectral line shape is sensitive not only to the motion of spin-label side-chains relative to the peptide backbone, but also to backbone fluctuations and to global protein motions. Therefore, changes in EPR lineshapes can prove to be an excellent diagnostic tool to monitor protein conformational changes. Figure 4 shows spectra at position L241(17)R1 in the extracellular hydrophobic region of the pore-lining M2 region (position highlighted as black circles in Figure 4, Inset) at pH 7.0 and pH 3.0. The spectra show differences under the two conditions, both in the signal amplitude and the extent of motional broadening. As with functional measurements42, the conformational changes reported by the EPR signals are also fully reversible. Spectral broadening is affected both by the motional constraints of the probe and by dipolar coupling. To determine the contribution of the two, L241(17) is co-labeled with a diamagnetic spin-label. The under-labeled and fully-labeled spectra are compared at pH 7.0 and pH 3.0 (Figure 4B). The extent of dipolar broadening at this position decreases when the channels are under labeled, which indicates an interaction between spin-labels among different subunits.
Figure 5 shows spectra for two positions on the M4 helix which is located at the periphery of the channel. ΔHo-1 is measured as the inverse of the central line width (shown in inset). As the nitroxide rotational motion is reduced, as witnessed during the formation of tertiary or quaternary contacts, the line width increases (and hence ΔHo-1 decreases) for any particular motional geometry. On the contrary, structural motions leading to an increase in the probe's freedom of movement is reflected as an increase in ΔHo-1. F312R1 is more mobile while R296R1 is immobile, consistent with their exposed and highly constrained environments, respectively.
Based on the accessibility parameters, a spin-label at a specific position on the protein can be classified as one of the following: buried within the protein core inaccessible to either water or lipids, on the surface exposed to an aqueous solution, or on the surface exposed to lipids (Figures 5 and 6). The accessibility parameters obtained for a series of consecutive sites can then be used to determine the secondary structure of a region, probe areas of protein-protein contact, measure the tilt of a protein within the membrane, estimate the immersion depth of interfacial loops, and identify water vestibules and lipid-bound pockets within the transmembrane helices.26 As an example, power-saturation curves in Figure 6 show that F312R1 has a relatively higher P1/2 for O2, in comparison to R296R1, suggesting the side-chains at position F312 are lipid-exposed. In addition, F312R1 is also more accessible to NiEDDA in comparison to R296R1 consistent with its location at the lipid-water interface. On the other hand, L180R1 in the loop C region of the extracellular domain is mobile and accessible to NiEDDA, consistent with its location in a water-exposed loop.
Probe dynamics and accessibility data at a given position provide local structural information about that specific position within the overall protein-fold. These parameters determined for a linear sequence of residues along the protein can be further analyzed using analytical approaches based on Fourier transform methods. These analyses are useful to evaluate periodic variations in EPR environmental parameters and to guide in predicting and orienting secondary structures. For a particular protein segment, a discrete Fourier transform power spectrum P(ω) is calculated as a function of the angle between adjacent positions (ω) in that segment. This plot is then used to determine the main angular frequency component of the spectrum and compared to the expected (ω) values for an α-helix (80 – 120º) or β-strands (150 – 180º).
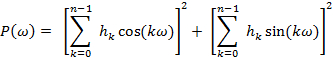
where P('ω') is the Fourier transform power spectrum as a function of angular frequency ω, n is the number of residues in the segment, is environmental parameter at the position . P('ω') gets evaluated for the value ω= 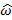 that maximizes P('ω').
that maximizes P('ω').
Further, a periodicity index parameter (gives the significance of the predicted secondary structure) is calculated as a ratio of the area under the power spectrum for the angle that represents a given structural element to that of the total area of the spectrum. A sliding window method is then used to identify the beginning and the end of a given secondary structural element. The relative orientation this segment with respect to the rest of the protein can be estimated from the direction of the P('ω') moment.
For α-helix:
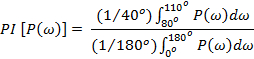
For β-strand:
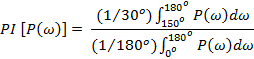
These methods have been successfully applied to several systems to determine three dimensional topologies and predict conformational changes underlying protein function 14,23,24,41,44-53.
Determining Distance Distribution from EPR
Inter-subunit nitroxide distances can be determined from electron-electron dipolar interactions. In CW-EPR, as shown in Figure 4, through-space dipolar interactions lead to spectral broadening and are a function of proximity between the labels. For distances up to ~15 – 20 Å, the extent of broadening (compared between fully labeled spectra and the under-labeled spectra) can be used to semi-quantitatively estimate distances using Fourier deconvolution methods.28,29
Inter-subunit distances (<50 Å) can be measured using Double Electron-Electron Resonance (DEER) methods.30-34 DEER data are collected in detergent samples or reconstituted in nanodiscs because of the limitations associated with these measurements in liposomes 54. For spin-labeled samples in detergent (~100 μM) in Buffer A with 0.5 mM DDM, 30% (weight/volume) glycerol is used for cryoprotection. The dipolar time evolution data are obtained at 83 K using a standard DEER four-pulse protocol (π/2)mw1-τ1-(π)mw1-τ1-(π)mw2-τ2-(π)mw1-τ2-echo55 on a pulsed EPR spectrometer operating at Q-band frequency (33.9 GHz). The pulse lengths for (π/2)mw1 and (π)mw1 are 10 and 20 nsec, respectively, and 40 nsec for (π)mw2. DEER signals are then background corrected assuming a 3D homogeneous background and analyzed by the Tikhonov regularization31,56 to determine average distances and distributions in distance.
Intramolecular distances for a representative position in M4 (F314R1) determined by DEER experiments at pH 7.0 are shown in Figure 7. Since there are five spin-labels within the GLIC pentamer, at least two distances are expected; one corresponding to the adjacent subunits and the other from nonadjacent subunits, although additional peaks may arise from alternate rotameric spin orientations. The DEER signal decays and the corresponding probability distributions are shown. The first short-distance peak (~42Å) represents distances between adjacent subunits, and the second peak (~53Å) corresponds to the non-adjacent distance. The peak for the diagonal distance appears to be at shorter distance, and this is likely to be underestimated because reliable measurements beyond 60 Å are not feasible under the experimental conditions due to technical difficulties with background correction.
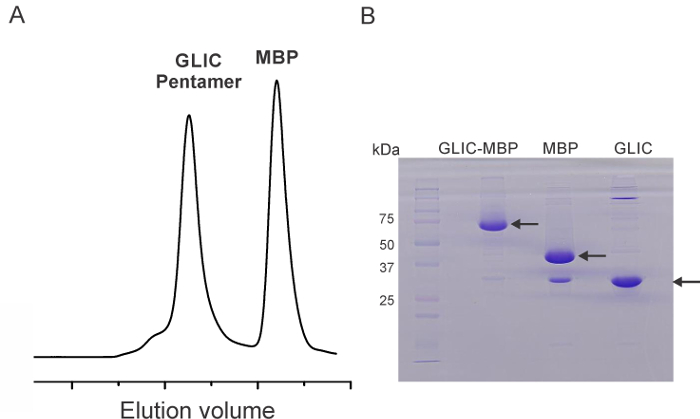
Figure 1. Biochemical Characterization Spin-labeled GLIC Mutants. (A) Representative gel filtration chromatogram of spin-labeled-GLIC after proteolytic cleavage of MBP tag. The peaks correspond to GLIC pentamer and free MBP. (B) SDS-PAGE showing uncut monomeric GLIC-MBP fusion protein (before gel filtration) and MBP and GLIC fractions pooled from gel filtration. Please click here to view a larger version of this figure.
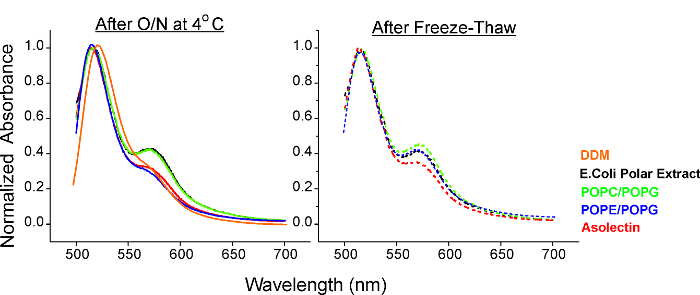
Figure 2. FRET Based Assay for Monitoring the Aggregation State of GLIC Reconstituted in Various Membrane Lipids. FRET measurements are shown for samples after O/N reconstitution (left) and after freezing/thawing the samples (right). (Modified from the original figure).43 Please click here to view a larger version of this figure.
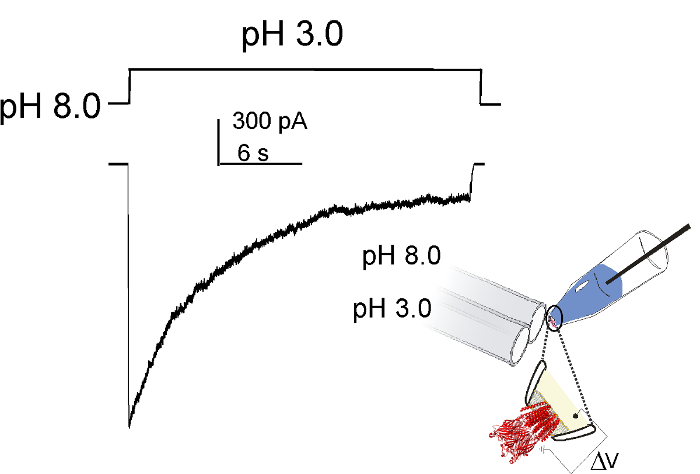
Figure 3. Macroscopic Behavior of GLIC in Proteoliposomes. In an inside-out patch excised from reconstituted asolectin vesicles, GLIC is activated by pH jumps using a rapid solution exchanger. The channels are seen to activate in ~10 msec and desensitize with a time constant of 1 – 3 sec. (Modified from the original figure).43 Please click here to view a larger version of this figure.
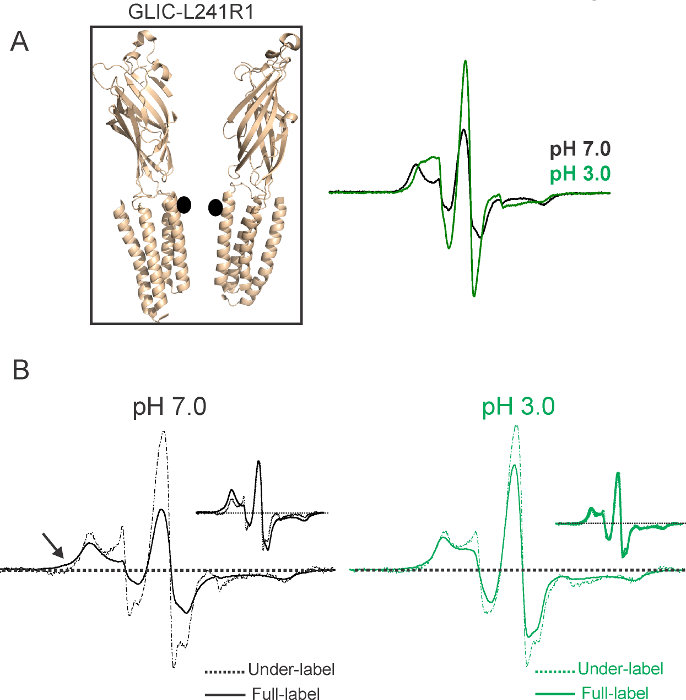
Figure 4. Structural Rearrangements During Channel Activation. (A) Representative CW-EPR spectra for a position in the pore lining M2 helix, displaying changes in amplitude and line shapes in response to pH changes. In each case, the spectra are normalized to the total number of spins. Spin-labeled position is shown as a black circle. Only two subunits are shown for clarity. (B) EPRspectra show dipolar broadening by spin-spin interactions. The spectra in dashed-line were from under-labeled channels (in the presence of diamagnetic labels) and in solid-line were from fully-labeled channels. The inset shows an overlay of amplitude-normalized spectra. Broadening under fully labeled conditions is highlighted by arrows. Scan width is 150 G. (Modified from the original figure).36 Please click here to view a larger version of this figure.
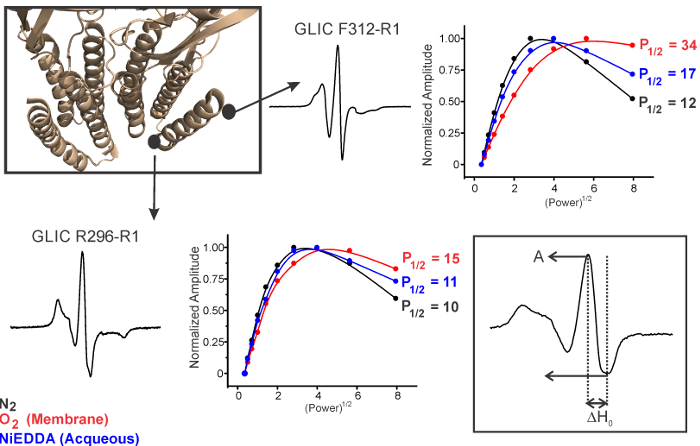
Figure 5. Residue Specific Environmental Parameters Measured by Power Saturation. Measurements of spectral width (ΔH0) and solvent accessibility (P1/2) for two contrasting positions in the M4 helix of the transmembrane domain. Please click here to view a larger version of this figure.
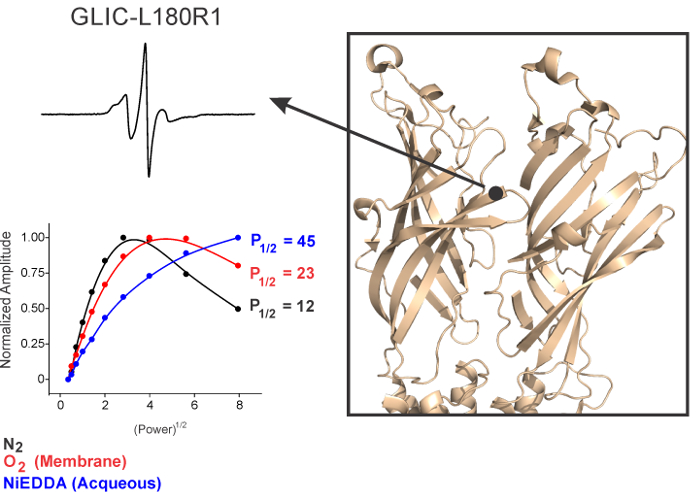
Figure 6. Environment of a Representative Loop C Residue in the Extracellular Domain of GLIC. Spin-normalized CW-spectra and the corresponding accessibility measurements for L180R1. Please click here to view a larger version of this figure.
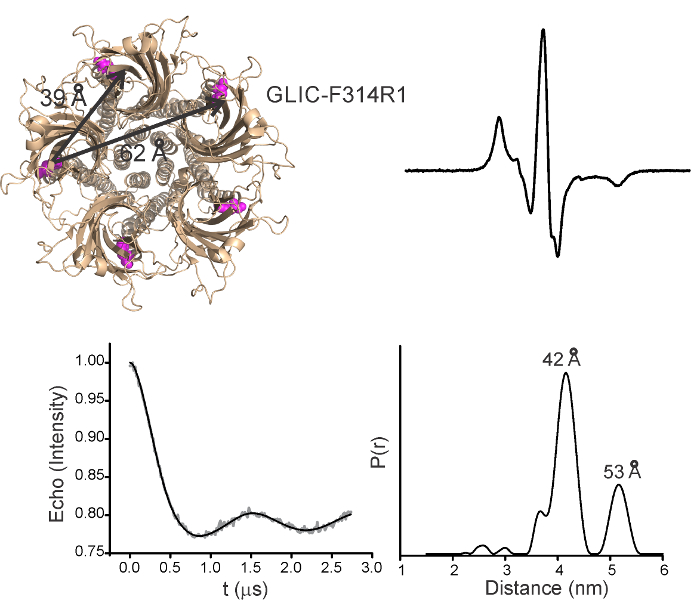
Figure 7. Distance Measurements by Pulsed-EPR Approaches (DEER). GLIC structure showing the location of Phe314 and corresponding cβ-cβ distances for the adjacent and non-adjacent subunits. CW-spectra for this position are shown on the right. Background subtracted DEER- echo intensity is plotted against evolution time and fit using model-free Tikhonov regularization for samples in detergent. The corresponding inter-spin distance distribution (right). Please click here to view a larger version of this figure.
Discussion
EPR spectroscopy has proven to be an unparalleled structural approach in quantifying conformational changes in membrane proteins in a near-native environment. This approach allows us a peek into the molecular details of protein dynamics that are obscured in high-resolution structures from X-ray crystallography and Cryo-electron microscopy. However, it is important to consider the technical limitations of this approach that may affect the general applicability to other systems and also to keep in mind the potential experimental roadblocks along the way. We discuss some of these aspects below along with recommended strategies for troubleshooting.
Among the more common issues that one encounters with a technique involving extensive mutational perturbations are the low yield and poor oligomeric stability of the protein. This aspect is further exacerbated when working with complex multimeric membrane proteins such as ion channels. It is therefore crucial to optimize these two biochemical aspects before embarking on a widespread mutagenesis. We found that expression of toxic GLIC mutants is better tolerated in C41 and C43 strains of BL21 cells.3,36 Further, lowering the post-induction temperature to 15 – 18 ºC, decreasing the IPTG concentration to 100 μM, and including 5% glycerol during induction seems to help with lowering protein aggregation presumably by slowing the rate of protein expression and reducing the extent of misfolding. Cysteine mutations at certain positions (particularly in the permeation pathway) have been shown to increase the constitutive activity of the channel and these mutants are likely to pose greater hurdles in expression. Use of divalent pore blockers (10 – 25 mM Ba2+) during induction alleviates toxicity and improves cell growth.57 These strategies have proven effective in many cases at enhancing yield, lowering protein aggregation and enhancing oligomeric stability.36,58
Another critical step in SDSL is the efficiency of the labeling process. While surface exposed cysteines should pose no problem in MTSL reactivity (>90% labeling yields), positions where the cysteine side-chains are buried and inaccessible may require higher spin-label concentrations and longer incubation times. Increasing the spin-label concentration to 30-fold molar excess and incubating O/N at 4 ºC helps with improving the labeling efficiency. In addition, it is also important to measure the spectra of the labeled Cys-less template to determine the background contribution. For sites that are poorly labeled (<10%), the background signal overwhelms the overall spectra. While determining the spin-labeling efficiency, note that the accuracy is affected by a number of factors that include sample volume, sample-capillary diameter, positioning of the capillary within the resonator, cavity temperature, and resonator Q value (which is the ratio of the energy stored in it over the energy dissipated out). Care must be taken to maintain these conditions identical between the samples and standard. Further, it should be noted that for immobile sites, where the spectra are intrinsically broad, the accuracy of the labeling measurement drops further. Alternatively, Liquid Chromatography Time of Flight-Electrospray Ionization Mass Spectrometry (LCT-ESI-MS) can be used to directly determine molecular mass and hence labeling efficiencies. However, the MS sensitivity is compromised for samples in detergents and for tightly folded membrane proteins.
Once the mutants are purified as a homogenous population, it is important to ascertain the functionality of the preparation. Patch-clamp measurements of spin-labeled mutants in key regions of the channel will reveal the effect of perturbation on ligand affinity and channel gating. Alternatively, channel properties can be studied in Black Lipid Membranes (BLM)59 or by injecting proteoliposomes into oocytes and measuring currents by two-electrode voltage clamp.59-61 If it is found that the cysteine substitution and the spin-label at certain positions alter ligand-sensitivity or gating kinetics of the channels, experimental conditions (ligand concentration, modulators, lipids composition) can be varied appropriately to stabilize the conformational state under investigation. Further, it is to be noted that EPR measurements are made under steady-state conditions and therefore changes in rapid kinetics of the channel are less likely to impact the structural interpretation.
Clearly, a major advantage of this technique is the ability to study channel dynamics in membranes of defined composition and to directly probe how lipid-mediated effects on dynamics alters channel function. However, a limitation is that some lipid compositions may cause lateral aggregation of the channels on the membrane and therefore the suitability of lipids needs to be ascertained.45 For conditions that do cause aggregation, lowering protein-to-lipid ratio, carrying out reconstitution at lower temperature, and shortening the duration of reconstitution time may help in maintaining the channels monodisperse62 Alternatively, the channels can be reconstituted in nanodiscs, which are nanoscale lipid assemblies surrounded by an annulus of amphipathic membrane scaffold protein (apolipoprotein A1).63 By optimizing the molar ratio of the channel protein, lipid constituents, and membrane scaffolding protein, it is possible to achieve a homogenous and mono-disperse population of single channel units incorporated into individual nanodiscs. This system has several additional advantages in that the nanodiscs are optically clear and provide easy access to both the intracellular and extracellular sides of the membrane.
Finally, at the level of the EPR measurements, one might be faced with CW-EPR spectra that are not straightforward to interpret, especially for positions displaying multicomponent spectra. Such conformational heterogeneity may arise from slowly exchanging conformations of the protein or/and multiple orientations of the spin-label in a given protein conformation. Determining the relative contribution of the two scenarios is not trivial and may require the use of alternate spin-label derivatives, or changing experimental temperature,64 pressure,65 field strengths/microwave frequency,66 or by osmolyte perturbation.67
Furthermore, DEER measurements in liposomes may be limited by a number of factors, including lower sensitivity, higher background contribution resulting from enhanced intermolecular interactions, and a reduction in the measurable distance range (<50Å). In general, for longer distances (<50 Å), there is greater uncertainty in the widths of the distance distribution due to insufficient dipolar evolution times. However, the use of Q band (34 GHz) microwave frequency and labeling with a bifunctional spin label (with reduced intrinsic dynamics) have been shown to alleviate some of these limitations.54 Reconstituting in nanodiscs has also been shown to tremendously improve DEER sensitivity by decreasing background contributions that arises from intermolecular dipolar interactions.15
Despite these limitations, SDSL/EPR studies, particularly in systems with few native cysteines, have proven to be remarkably informative. Future technological advancements are geared toward adapting this powerful approach to study more complex human channels, where mutating native cysteines may not be feasible. In this regard, development of alternate labeling strategies such as those based on site-specific incorporation of unnatural amino acids,68 or synthesis of labels that are targeted towards other amino acids,69 appear to hold great promise.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are very grateful to the current and former members of the Chakrapani lab for critical reading and comments on the manuscript. This work was supported by the National Institutes of Health grant (1R01GM108921) and the American Heart Association (NCRP Scientist Development Grant 12SDG12070069) and to SC.
Materials
| Site-Directed Mutagenesis and Cys mutations | |||
| 10x PfuUltra HF reaction buffer | Agilent Technologies | 600380-52 | |
| dNTPS | New England BioLabs Inc | N0447L | 10mM each dNTP |
| pfu Ultra DNA polymerase | Agilent Technologies | 600380-51 | 2.5 U/ul |
| DPNI | New England BioLabs Inc | R0176S | 20,000 U/ml |
| XL10 GOLD | Agilent Technologies | 200314 | |
| SOC media | New England BioLabs Inc | B9020S | |
| Kanamycin | Fisher Scientfic | BP905 | |
| LB media | Invitrogen | 127957084 | |
| Miniprep kit | QIAGEN | 27106 | |
| C43 competent cells | Lucigen | 60446 | |
| Expression and Purification | |||
| Glucose | Fisher Scientfic | D16 | |
| Tryptone | Fisher Bioreagents | BP1421-500 | |
| Yeast extract | Amresco | J850 | |
| Glycerol | Fisher Bioreagents | BP229 | |
| K2HPO4 | Amresco | 0705 | |
| KH2PO4 | Amresco | 0781 | |
| IPTG (isopropyl-thio-β-galactoside) | Gold Biotechnology | I2481C25 | |
| Trizma Base | Sigma Life Science | T1503 | |
| NaCl | Sigma-Aldrich | S7653 | |
| DNase I | Sigma Life Science | DN25 | |
| PMSF | Amresco | M145 | |
| Leupeptine | Amresco | J580 | |
| Pepstatin | Amresco | J583 | |
| DDM (n-Docecyl-β-D-Maltopyranoside) | Anatrace | D310S | |
| Amylose resin | New England BioLabs Inc | E8021L | |
| TCEP | Amresco | K831 | |
| EDTA | Fisher Scientfic | BP118 | |
| Maltose | Acros Organics | 329915000 | |
| Superdex 200GL | GE Healthcare | 17-5175-01 | |
| Empty polypropylene Chromatography column | BioRad | 731-1550 | |
| Site-Directed Spin Labeling | |||
| MTSL (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl) Methanethiosulfonate | Toronto Reaserch chemicals Inc | O873900 | |
| (1-acetoxy-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate | Toronto Reaserch chemicals Inc | A167900 | |
| DMSO | J.T. Baker | 9224-01 | |
| Reconstitution | |||
| Asolectin lipid | Avanti polar lipids Inc | 541602C | |
| Biobeads (Polystyrine beads) | Bio Rad | 152-3920 | |
| Methanol | Fisher chemicals | A413 | |
| FRET | |||
| Fluorescein-maleimide | ThermoFisher Scientific | F-150 | |
| Tetramethylrhodamine-maleimide | ThermoFisher Scientific | T-6027 | |
| POPC | Avanti polar lipids Inc | 850457C | |
| POPG | Avanti polar lipids Inc | 840457C | |
| E.Coli polar lipid extract | Avanti polar lipids Inc | 100600C | |
| HEPES | Sigma Life Science | H3375 | |
| EPR measurement | |||
| TPX plastic capillaries | Bruker | ER221 | |
| EDDA (Ethylenediamine-N, N'-diacetic acid) | Aldrich | 158186 | |
| Ni(OH)2 | Aldrich | 283622 |
References
- Tasneem, A., Iyer, L. M., Jakobsson, E., Aravind, L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 6, 4 (2005).
- Hilf, R. J., Dutzler, R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 452 (7185), 375-379 (2008).
- Bocquet, N., et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 457 (7225), 111-114 (2009).
- Hibbs, R. E., Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 474 (7349), 54-60 (2011).
- Miller, P. S., Aricescu, A. R. Crystal structure of a human GABAA receptor. Nature. 512 (7514), 270-275 (2014).
- Hassaine, G., et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 512 (7514), 276-281 (2014).
- Du, J., Lu, W., Wu, S., Cheng, Y., Gouaux, E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. , (2015).
- Sunshine, C., McNamee, M. G. Lipid modulation of nicotinic acetylcholine receptor function: the role of neutral and negatively charged lipids. Biochim Biophys Acta. 1108, 240-246 (1992).
- Fong, T. M., McNamee, M. G. Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry. 25 (4), 830-840 (1986).
- daCosta, C. J., Baenziger, J. E. A lipid-dependent uncoupled conformation of the acetylcholine receptor. J Biol Chem. 284 (26), 17819-17825 (2009).
- Criado, M., Eibl, H., Barrantes, F. J. Functional properties of the acetylcholine receptor incorporated in model lipid membranes. Differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J Biol Chem. 259 (14), 9188-9198 (1984).
- Hubbell, W. L., Lopez, C. J., Altenbach, C., Yang, Z. Technological advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 23 (5), 725-733 (2013).
- Columbus, L., Hubbell, W. L. A new spin on protein dynamics. Trends Biochem Sci. 27 (6), 288-295 (2002).
- Hubbell, W. L., Cafiso, D. S., Altenbach, C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 7 (9), 735-739 (2000).
- McHaourab, H. S., Steed, P. R., Kazmier, K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 19 (11), 1549-1561 (2011).
- Bordignon, E. Site-directed spin labeling of membrane proteins. Top Curr Chem. 321, 121-157 (2012).
- Klare, J. P., Steinhoff, H. J. Spin labeling EPR. Photosynth Res. 102 (2-3), 377-390 (2009).
- Drescher, M. EPR in protein science : intrinsically disordered proteins. Top Curr Chem. 321, 91-119 (2012).
- Perozo, E., Cuello, L. G., Cortes, D. M., Liu, Y. S., Sompornpisut, P. EPR approaches to ion channel structure and function. Novartis Found Symp. 245, 146-158 (2002).
- Fanucci, G. E., Cafiso, D. S. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 16 (5), 644-653 (2006).
- Sahu, I. D., McCarrick, R. M., Lorigan, G. A. Use of electron paramagnetic resonance to solve biochemical problems. Biochemistry. 52 (35), 5967-5984 (2013).
- Hubbell, W. L., McHaourab, H. S., Altenbach, C., Lietzow, M. A. Watching proteins move using site-directed spin labeling. Structure. 4 (7), 779-783 (1996).
- Altenbach, C., Marti, T., Khorana, H. G., Hubbell, W. L. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 248 (4959), 1088-1092 (1990).
- McHaourab, H. S., Lietzow, M. A., Hideg, K., Hubbell, W. L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 35 (24), 7692-7704 (1996).
- Farahbakhsh, Z. T., Altenbach, C., Hubbell, W. L. Spin labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochem Photobiol. 56 (6), 1019-1033 (1992).
- Altenbach, C., Greenhalgh, D. A., Khorana, H. G., Hubbell, W. L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 91 (5), 1667-1671 (1994).
- Altenbach, C., Froncisz, W., Hemker, R., McHaourab, H., Hubbell, W. L. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys J. 89 (3), 2103-2112 (2005).
- Rabenstein, M. D., Shin, Y. K. Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci U S A. 92 (18), 8239-8243 (1995).
- Altenbach, C., Oh, K. J., Trabanino, R. J., Hideg, K., Hubbell, W. L. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry. 40 (51), 15471-15482 (2001).
- Borbat, P. P., McHaourab, H. S., Freed, J. H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J Am Chem Soc. 124 (19), 5304-5314 (2002).
- Chiang, Y. W., Borbat, P. P., Freed, J. H. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 172 (2), 279-295 (2005).
- Jeschke, G. DEER distance measurements on proteins. Annu Rev Phys Chem. 63, 419-446 (2012).
- Jeschke, G., Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 9 (16), 1895-1910 (2007).
- Jeschke, G., Wegener, C., Nietschke, M., Jung, H., Steinhoff, H. J. Interresidual distance determination by four-pulse double electron-electron resonance in an integral membrane protein: the Na+/proline transporter PutP of Escherichia coli. Biophys J. 86 (4), 2551-2557 (2004).
- Hilf, R. J., Dutzler, R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 457 (7225), 115-118 (2009).
- Velisetty, P., Chalamalasetti, S. V., Chakrapani, S. Conformational transitions underlying pore opening and desensitization in membrane-embedded Gloeobacter violaceus ligand-gated ion channel (GLIC). J Biol Chem. 287 (44), 36864-36872 (2012).
- Birnboim, H. C., Doly, J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7 (6), 1513-1523 (1979).
- Sanger, F., Nicklen, S., Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 74 (12), 5463-5467 (1977).
- Sanger, F., Coulson, A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 94 (3), 441-448 (1975).
- McHaourab, H. S., Kalai, T., Hideg, K., Hubbell, W. L. Motion of spin-labeled side chains in T4 lysozyme: effect of side chain structure. Biochemistry. 38 (10), 2947-2955 (1999).
- Gross, A., Columbus, L., Hideg, K., Altenbach, C., Hubbell, W. L. Structure of the KcsA potassium channel from Streptomyces lividans: a site-directed spin labeling study of the second transmembrane segment. Biochemistry. 38 (32), 10324-10335 (1999).
- Velisetty, P., Chakrapani, S. Desensitization mechanism in a Prokaryotic ligand-gated ion channel. J Biol Chem. 287 (22), (2012).
- Velisetty, P., Chakrapani, S. Desensitization mechanism in prokaryotic ligand-gated ion channel. J Biol Chem. 287 (22), 18467-18477 (2012).
- Chakrapani, S., Cuello, L. G., Cortes, D. M., Perozo, E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 16 (3), 398-409 (2008).
- Cuello, L. G., Cortes, D. M., Perozo, E. Structural dynamics of the KvAP pore domain lacking the voltage sensing domain. Biophysical Journal. 88 (1), 19-20 (2005).
- Perozo, E., Cortes, D. M., Cuello, L. G. Structural rearrangements underlying K+-channel activation gating. Science. 285 (5424), 73-78 (1999).
- Chakrapani, S., Sompornpisut, P., Intharathep, P., Roux, B., Perozo, E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc Natl Acad Sci U S A. 107 (12), 5435-5440 (2010).
- Vasquez, V., Sotomayor, M., Cordero-Morales, J., Schulten, K., Perozo, E. A structural mechanism for MscS gating in lipid bilayers. Science. 321 (5893), 1210-1214 (2008).
- Perozo, E., Cortes, D. M., Sompornpisut, P., Kloda, A., Martinac, B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418 (6901), 942-948 (2002).
- Kim, M., Xu, Q., Murray, D., Cafiso, D. S. Solutes alter the conformation of the ligand binding loops in outer membrane transporters. Biochemistry. 47 (2), 670-679 (2008).
- Kazmier, K., Sharma, S., Islam, S. M., Roux, B., McHaourab, H. S. Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1. Proc Natl Acad Sci U S A. 111 (41), 14752-14757 (2014).
- Durr, K. L., et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 158 (4), 778-792 (2014).
- Borbat, P. P., et al. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5 (10), e271 (2007).
- Zou, P., McHaourab, H. S. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys J. 98 (6), 18-20 (2010).
- Pannier, M., Veit, S., Godt, A., Jeschke, G., Spiess, H. W. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 142 (2), 331-340 (2000).
- Jeschke, G., et al. DeerAnalysis2006-a comprehensive software package for analyzing pulsed ELDOR data. Applied Magnetic Resonance. 30 (3-4), 473-498 (2006).
- Ruta, V., Jiang, Y., Lee, A., Chen, J., MacKinnon, R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 422 (6928), 180-185 (2003).
- Velisetty, P., Chalamalasetti, S. V., Chakrapani, S. Structural basis for allosteric coupling at the membrane-protein interface in Gloeobacter violaceus ligand-gated ion channel (GLIC). J Biol Chem. 289 (5), 3013-3025 (2014).
- Dellisanti, C. D., et al. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLoS Biol. 11 (11), e1001714 (2013).
- Labriola, J. M., et al. Structural sensitivity of a prokaryotic pentameric ligand-gated ion channel to its membrane environment. J Biol Chem. 288 (16), 11294-11303 (2013).
- Kinde, M. N., et al. Conformational Changes Underlying Desensitization of the Pentameric Ligand-Gated Ion Channel ELIC. Structure. 23 (6), 995-1004 (2015).
- Vasquez, V., Cortes, D. M., Furukawa, H., Perozo, E. An optimized purification and reconstitution method for the MscS channel: strategies for spectroscopical analysis. Biochemistry. 46 (23), 6766-6773 (2007).
- Bayburt, T. H., Sligar, S. G. Membrane protein assembly into Nanodiscs. FEBS Lett. 584 (9), 1721-1727 (2010).
- Hanson, S. M., Francis, D. J., Vishnivetskiy, S. A., Klug, C. S., Gurevich, V. V. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 281 (14), 9765-9772 (2006).
- McCoy, J., Hubbell, W. L. High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc Natl Acad Sci U S A. 108 (4), 1331-1336 (2011).
- Mobius, K., et al. Combining high-field EPR with site-directed spin labeling reveals unique information on proteins in action. Magn Reson Chem. 43, 4-19 (2005).
- Lopez, C. J., Oga, S., Hubbell, W. L. Mapping Molecular Flexibility of Proteins with Site-Directed Spin Labeling: A Case Study of Myoglobin. Biochemistry. 51 (33), 6568-6583 (2012).
- Fleissner, M. R., et al. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Natl Acad Sci U S A. 106 (51), 21637-21642 (2009).
- Lorenzi, M., et al. Tyrosine-targeted spin labeling and EPR spectroscopy: an alternative strategy for studying structural transitions in proteins. Angew Chem Int Ed Engl. 50 (39), 9108-9111 (2011).

