Large-Scale Production of Cardiomyocytes from Human Pluripotent Stem Cells Using a Highly Reproducible Small Molecule-Based Differentiation Protocol
Summary
Here, we present a robust, fast and scalable cardiomyocyte differentiation protocol for human pluripotent stem cells (hPSCs). Cardiomyocytes derived using this large-scale method can provide sufficient cell numbers for their effective use in human cardiovascular disease modeling, high-throughput drug screening, and potentially clinical applications.
Abstract
Maximizing the benefit of human pluripotent stem cells (hPSCs) for research, disease modeling, pharmaceutical and clinical applications requires robust methods for the large-scale production of functional cell types, including cardiomyocytes. Here we demonstrate that the temporal manipulation of WNT, TGF-β, and SHH signaling pathways leads to highly efficient cardiomyocyte differentiation of single-cell passaged hPSC lines in both static suspension and stirred suspension bioreactor systems. Employing this strategy resulted in ~ 100% beating spheroids, consistently containing > 80% cardiac troponin T-positive cells after 15 days of culture, validated in multiple hPSC lines. We also report on a variation of this protocol for use with cell lines not currently adapted to single-cell passaging, the success of which has been verified in 42 hPSC lines. Cardiomyocytes generated using these protocols express lineage-specific markers and show expected electrophysiological functionalities. Our protocol presents a simple, efficient and robust platform for the large-scale production of human cardiomyocytes.
Introduction
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), have the ability of self-renewal and the capacity to differentiate into cells of the three embryonic germ layers 1,2. Due to these characteristics, hPSCs provide a valuable and unlimited source for the generation and scalable production of disease-relevant cell types for modeling human disease 3-5, for high-throughput drug screening and toxicity assays 6,7 and potentially for clinical applications 8. Generation of cardiomyocytes from hPSCs provides the opportunity to specifically investigate the mechanisms of complex human cardiovascular diseases and their possible treatments, previously beyond the scope of our capabilities due to the lack of relevant animal models and/or the availability of affected primary tissues.
All of the aforementioned applications of hPSCs necessitate the production of massive numbers of highly enriched and functional cardiomyocytes. Thus, the availability of an efficient, reproducible and scalable in vitro cardiac differentiation protocol suitable for multiple hPSC lines is crucial. Conventional cardiomyocyte differentiation protocols have employed different strategies such as embryoid body formation 9, co-culture techniques 10, induction with cocktails of cytokines 11 and protein transduction methods 12. In spite of advances in these techniques, most still suffer from poor efficiency, require expensive growth factors, or offer limited universality when attempting to use multiple hPSC lines. To date, these challenges have set limits to the production of hPSC-derived cardiomyocytes for cell therapy studies in animal models, as well as in the pharmaceutical industry for drug discovery 13. Therefore, the development of robust and affordable techniques for large-scale production of functional hPSC-derived cardiomyocytes in scalable culture systems would largely facilitate their commercial and clinical applications.
In this manuscript, we report the development of a cost-effective and integrated cardiac differentiation system with high efficacy, reproducibility and applicability to hESCs and hiPSCs generated from a variety of sources and culture methods, including a method for the large-scale production of highly enriched populations of hPSC-derived cardiomyocytes using a bioreactor. Additionally, we have optimized this protocol for hPSC lines not adapted to feeder free and/or single cell culture, such as newly established hiPSCs or large cohorts of hPSC lines relevant to analysis of disease mechanism.
Protocol
1. Preparation of Culture Media, Coating of Cell Culture Plates and Maintenance of Undifferentiated hPSCs
- Media Preparation
Note: Sterilize media using a 0.22 µm filtration device and store at 4 °C protected from light for up to 4 weeks. Reagent names, suppliers and catalog numbers are listed in Materials Table.- For Mouse Embryonic Fibroblasts (MEF) Medium, combine 445 ml DMEM, 50 ml Fetal Bovine Serum (FBS) and 5 ml cell culture media (e.g., Glutamax).
- For hESC Medium, combine 390 ml Knockout-DMEM (KO-DMEM), 100 ml Knockout Serum Replacement (KO-SR), 5 ml cell culture media, 5 ml MEM minimum essential amino acids solution and 0.5 ml 55 mM β-mercaptoethanol.
CAUTION: β-Mercaptoethanol is toxic. Avoid inhalation, ingestion and skin contact. - For RPMI-B27 (RB) Medium, combine 475 ml RPMI 1640, 10 ml B27 minus insulin, 5 ml cell culture media, 5 ml MEM minimum essential amino acids solution, 5 ml penicillin/streptomycin and 0.5 ml 55 mM β-mercaptoethanol.
- For the Dissociation Solution (DS), combine 10 ml 0.05% trypsin, 4 ml KO-SR, 1 ml collagenase type IV (1 mg/ml), 5 ml KO-DMEM and 20 µl CaCl2(1 M).
- For Feeder-cell Conditioned Medium, add 15 ml hESC medium (without bFGF) to a T75 flask with confluent feeder cells (MEF or human foreskin fibroblasts) which have been previously inactivated by treatment with either mitomycin C or by irradiation. Harvest conditioned medium after 24 hr and replace with fresh hESC medium. Cells can be used for up to 2 weeks.
- Coating of Cell Culture Plates
- ECM Gel Coating:
- Upon purchase, thaw the ECM gel extract at 4 °C until it is in an evenly consistent liquid state, as per the manufacturer's instructions. Dilute at a ratio of 1:2 in cold KO-DMEM medium, aliquot and store at -20 °C.
Note: During the preparation of the ECM gel, keep all materials required for use in the aliquotting and coating procedure cold. The concentration of ECM gel varies with batch number. Ensure the concentration is noted on aliquots. Aliquots can be kept at -20 °C for up to 6 months. - Thaw an ECM gel aliquot at 4 °C. When thawed, add cold KO-DMEM medium for a final concentration of 0.34 mg/ml. Pipette well and add 0.75 ml per one well of a 6-well plate. Incubate for 1 hr at 37 °C.
- Upon purchase, thaw the ECM gel extract at 4 °C until it is in an evenly consistent liquid state, as per the manufacturer's instructions. Dilute at a ratio of 1:2 in cold KO-DMEM medium, aliquot and store at -20 °C.
- Mitotically Inactivated Mouse Embryonic Fibroblast Feeder Cell (MEF) Coating
Note: Preparation of MEF feeder cells has been described previously. 14- Coat a 6-well cell culture plate with Attachment Factor (AF) or 0.1% gelatin (see step 1.2.3) at RT for 5 min (0.75 ml per one well of a 6-well plate). Remove and leave plate in the biosafety cabinet (BSC) to dry.
- Thaw MEF by transferring a frozen vial to a 37 °C water bath for approximately 2 – 3 min.
- Add 7 ml MEF medium to a 15 ml tube. When the vial of cells is thawed, slowly transfer the contents to the 15 ml tube. Centrifuge at 300 x g for 4 min. Aspirate the supernatant and resuspend the cells in MEF medium.
- Count the cells and plate 1 × 106cells per 6 well plate (~ 1.7 x 105 cells/well). Transfer to a 37 °C / 5% CO2 incubator and allow cells to attach O/N before use.
- Gelatin Coating:
- Dissolve 1 g of gelatin powder in ultrapure water to make a 0.1% (w/v) solution. Incubate for 15 min at 37 °C then autoclave the solution at 121 °C for 15 min. When cooled, add to the required number of wells (0.75 ml per one well of a 6-well plate) and incubate for 15 min at 37 °C.
Note: Any remaining 0.1% gelatin solution can be kept at 4 °C for up to 1 month.
- Dissolve 1 g of gelatin powder in ultrapure water to make a 0.1% (w/v) solution. Incubate for 15 min at 37 °C then autoclave the solution at 121 °C for 15 min. When cooled, add to the required number of wells (0.75 ml per one well of a 6-well plate) and incubate for 15 min at 37 °C.
- Laminin Coating:
- Thaw laminin (1 mg/ml) at 4 °C until thawed. To 5 µl of laminin solution, add 1 ml of cold PBS. Pipette well then add to the required number of wells (0.75 ml per one well of a 6-well plate) and incubate for 1 hr at 37 °C.
- Silicon Coating of Spinner Flask Internal Surface:
- Thoroughly wash a 100 ml spinner flask (with 1 glass pendulum) with distilled water, using a cleaning brush to remove any dust and/or culture residue. Fill with 70% ethanol and after 30 min rinse with distilled water. Add 5 M NaOH and leave O/N.
- Remove NaOH solution and rinse the flask under running water for 5 min. Fill the flask with 1 M HCl and leave for 15 min. Wash the flask with running water for 5 min, then twice with double distilled water to completely remove any trace of HCl. Leave the flask to dry completely in the BSC.
- Add 1.5 ml of siliconizing solution to the flask and rotate horizontally to cover all surfaces. Repeat the same procedure for the glass pendulum.
- Heat the coated flask in a dry oven at 100 °C for 1 hr or allow to dry O/N at RT. If heated in an oven, leave to cool to RT for 30 – 60 min.
- Rinse the flask 3 times with deionized water for 15 min each, then sterilize by autoclaving at 121 ˚C for 20 min.
- ECM Gel Coating:
- Maintenance of Undifferentiated hPSCs
Note: For each of the protocols described, unless specifically stated, cells are grown and cultured in wells of 6-well culture plates and volumes will be given appropriate for this format.- hPSCs Cultured as Undifferentiated Spheroids in a Static Suspension System
Note: Cells used in these steps should already be adapted to single-cell culture. 15- Start experiment with hPSCs cultured on ECM gel in a 6-well culture plate at approximately 70 – 80% confluency. Remove any differentiated colonies using the stereomicroscope in the BSC. Add 10 µM ROCK inhibitor Y-27632 and incubate at 37 °C for 1 hr.
Note: For removing the differentiated colonies, use a pipette tip to detach and gently remove all the differentiated parts manually under the stereomicroscope. Be careful not to remove undifferentiated colonies. - Remove cells from incubator and aspirate medium. Wash the cells with 1 ml PBS, remove and add 0.5 ml cell dissociation enzyme. Incubate the cells for 4 – 5 min at 37 °C.
- Add 1 ml hESC medium and harvest the cells using a cell scraper. Dissociate the cells into single cells using a p1,000 pipette. Count the viable cells using Trypan blue and a hemocytometer.
- Resuspend cells to 2 × 105 viable cells/ml in feeder-cell conditioned medium supplemented with 100 ng/ml bFGF and 10 µM ROCK inhibitor Y-27632. Transfer cells to ultra-low attachment plates using a 5 ml pipette (3 ml per well of 6-well plate).
- After 2 days, carefully remove cells from incubator and aspirate half of the medium (approximately 1.5 ml). Replace with fresh conditioned medium supplemented with 100 ng/ml bFGF .
- Change half of the medium every day with conditioned medium supplemented with 100 ng/ml bFGF until day 5. Now, either further culture cells, passage for continued undifferentiated culture (step 1.3.1.2 – 5), or use for cardiac differentiation (Section 2). If continuing culture, passage cells every 4 – 8 days.
- Start experiment with hPSCs cultured on ECM gel in a 6-well culture plate at approximately 70 – 80% confluency. Remove any differentiated colonies using the stereomicroscope in the BSC. Add 10 µM ROCK inhibitor Y-27632 and incubate at 37 °C for 1 hr.
- hPSCs Cultured on Feeder Cells and Passaged Using Collagenase Type IV
- Before passaging hPSCs, remove any differentiated colonies using the stereomicroscope in the BSC. Aspirate the medium and wash cells with 1 ml PBS per well. Remove then add 0.75 ml collagenase type IV (1 mg/ml) and incubate at 37 °C for 5 – 15 min, as required to have the edges of the colonies starting to peel off.
- Aspirate collagenase and add 1 ml of hESC medium per well. Detach colonies to small clusters using a cell scraper, then transfer the cells using a p1,000 pipette into a 15 ml tube. Be careful not to break up the clusters into small pieces.
- Wash the well with 1 ml hESC medium to collect any remaining cells and add to 15 ml tube. Centrifuge at 100 x g for 2 min. Aspirate the supernatant and resuspend cells in hESC medium supplemented with 100 ng/ml bFGF.
- Remove a MEF coated plate from the incubator and aspirate the MEF medium. Transfer the cells at a ratio between 1:3 and 1:10, as appropriate. Change medium daily with fresh hESC medium supplemented with 100 ng/ml bFGF. Passage cells every 4 – 8 days.
- hPSCs Cultured as Undifferentiated Spheroids in a Static Suspension System
2. Differentiation of hPSCs as Spheroids in a Static Suspension System
- Start experiment using day 5 undifferentiated spheroids as prepared in Section 1.3.1.
Note: At this stage, the average size of spheroids should be 175 ± 25 µm. A minimum of 50 spheroids should be used when starting a differentiation experiment. - Transfer spheroids into a 15 ml tube using a 5 ml pipette. Wash the well with 1 ml RB medium to collect any remaining spheroids and add to the same 15 ml tube. Leave the cells for 5 min until spheroids have sedimented to form a loose pellet (DO NOT centrifuge). Aspirate the supernatant, being careful not to take any spheroids.
- Add 3 ml RB medium supplemented with 12 µM CHIR99021. Transfer the cells back into one well of a 6-well ultra-low attachment plate. After 24 hr, repeat step 2.2.
Note: RB medium is light sensitive. When working with RB medium, keep the BSC light switched off. - Add 3 ml RB medium to the cell pellet. Transfer the cells back into one well of a 6-well ultra-low attachment plate. After 24 hr, repeat step 2.2.
- Add 3 ml RB medium supplemented with IWP2, purmorphamine and SB431542 (5 µM each). Transfer the cells back into one well of a 6-well ultra-low attachment plate. After 2 days repeat step 2.2.
- Resuspend the cells in 3 ml of RB medium and transfer to a gelatin-coated plate to allow the cells to attach.
Note: At this stage, cells can also be kept in suspension. In order to continue with static suspension culture, transfer the cells back to an ultra-low attachment plate. - Change the medium with 3 ml RB medium after 2 days. The cultures will start to beat next day. Change the medium every 3 – 4 days with RB medium.
3. Differentiation of hPSCs as Spheroids in a Stirred Suspension Bioreactor
- Start experiment with undifferentiated spheroids cultured in static suspension for at least 3 passages (See Section 1.3.1). Add 10 µM ROCK inhibitor Y-27632 and incubate at 37 °C for 1 hr.
- Remove cells from incubator and transfer all spheroids into a 15 ml tube using a 5 ml pipette. Wash the well with 1 ml hESC medium to collect any remaining spheroids and add to the same 15 ml tube. Leave the cells for 5 min until spheroids have sedimented to form a loose pellet (DO NOT centrifuge). Aspirate the supernatant, being careful not to take any spheroids.
- Wash the cells by adding 1 ml PBS. Leave for 5 min until spheroids have sedimented to form a loose pellet (DO NOT centrifuge). Remove supernatant and add 0.5 ml 0.05% trypsin plus 0.53 mM EDTA. Incubate the cells for 4 – 5 min at 37 °C.
- Remove cells from the incubator and add 1 ml hESC medium. Using a p1,000 pipette, dissociate the cells into single cells, count the cells using Trypan blue and a hemocytometer. Resuspend to 2 × 105 viable cells/ml in conditioned medium supplemented with 100 ng/ml bFGF and 10 µM ROCK inhibitor Y-27632.
- Transfer 100 ml of cell suspension into the siliconized bioreactor flask with 1 pendulum (see Section 1.2.5). Start with 35 rpm agitation and after 24 hr increase to 40 rpm.
- After 48 hr, stop agitation for 5 – 10 min until all the spheroids have settled to the bottom of the flask. Replace half of the medium with conditioned medium supplemented with 100 ng/ml bFGF. Repeat the same process every 24 hr until day 5.
Note: At this stage, undifferentiated spheroids with an average size of 175 ± 25 µm should be formed. - Stop agitation for 5 – 10 min until all the spheroids have settled to the bottom of the flask. Transfer all spheroids to a 50 ml tube in a minimum amount of medium (less than 50 ml). Discard any remaining medium in the bioreactor flask, carefully ensuring no spheroids are remaining in the vessel.
- Leave for 5 – 10 min until all the spheroids have settled at the bottom of the 50 ml tube then carefully remove the supernatant without disrupting the loose cell pellet. Wash with 25 ml PBS with calcium and magnesium or RB medium, then leave again for 5 – 10 min until all the spheroids have settled at the bottom of the 50 ml tube to again form a loose pellet.
- Carefully remove the supernatant and add 40 ml RB medium supplemented with 12 µM CHIR99021, 10 µM Rock inhibitor and 0.1% poly vinyl alcohol (PVA). Resuspend the spheroids and transfer all cells back to the bioreactor flask. Add medium to make the total volume 100 ml. Restart agitation at 40 rpm for 24 hr.
Note: RB medium is light sensitive. When working with RB medium keep the BSC in light switched off mode. - Repeat Steps 3.7 – 3.8.
- Carefully remove the supernatant and add 40 ml RB medium supplemented with 0.1% PVA. Resuspend the spheroids and transfer all cells back to the bioreactor flask. Add medium to make the total volume 100 ml. Restart agitation at 40 rpm for 24 hr.
- Repeat Steps 3.7 – 3.8.
- Carefully remove the supernatant and add 40 ml RB medium supplemented with IWP2, purmorphamine and SB431542 (5 µM each). Resuspend the spheroids and transfer all cells back to the bioreactor flask. Add medium to make the total volume 100 ml. Restart agitation at 40 rpm for 2 days.
- Repeat Steps 3.7-3.8.
- Carefully remove the supernatant and add 40 ml RB medium only. Resuspend the spheroids and transfer all cells back to the bioreactor flask. Add medium to make the total volume 100 ml. Restart agitation at 40 rpm. Beating spheroids should be observed 48 – 72 hr later.
- Change half of the medium every 3 – 4 days with RB medium only by stopping the agitation for 5 – 10 min until all the spheroids have settled to the bottom of the vessel. Carefully remove 50 ml of medium without disturbing the spheroids and carefully replace with 50 ml of fresh RB medium.
4. Differentiation of hPSCs Using Cultures not Adapted to Single Cell Passaging
Note: This approach is specifically useful for the fast differentiation of a high number of hPSC lines without having to adapt to single-cell culturing techniques, an enormously labor-intensive and time consuming process. This technique is applicable to cell lines which are highly sensitive to enzymatic cell dissociation, such as newly established hPSC lines.
- Start experiment with hPSCs cultured on MEF (as in Section 1.3.2) which are approximately 70 – 80% confluent. Remove any differentiated colonies using the stereomicroscope in the BSC.
- Remove medium and add 0.5 ml Dissociation Solution (DS). Incubate for 0.5 – 1 min until MEF cells have rounded up and the edges of hPSCs colonies become clear. Quickly remove DS and add 0.75 ml collagenase type IV (1 mg/ml).
- Incubate the cells for 5 – 15 min at 37 °C. Check the cells under the microscope during the incubation period, and when whole colonies are starting to lift off and detach, remove collagenase and add 1.5 ml hESC medium.
Note: If after 15 min most of the colonies are still attached, put the cells back to incubator. Check the cells under the microscope every 5 min. Be careful not to leave the cells in collagenase for more than 30 min. If there are many floating colonies in the collagenase solution, do not remove the collagenase; just add 1 ml hESC medium. - Using a p1,000 pipette, gently pipette colonies to detach as whole colonies. Do not break the colonies into small pieces. Transfer the cells into a 15 ml tube using a 5 ml pipette. Wash the well with 1 ml hESC medium to collect any remaining cells to add to the same tube. Leave for 5 min until all colonies have sedimented (DO NOT centrifuge).
- Carefully remove the supernatant and add 2 ml hESC medium supplemented with 100 ng/mL bFGF. Transfer the cells into an ultra-low attachment plate using 5 ml pipette (1 ml into each well of a 24-well plate and 3 ml into each well of a 6-well plate). Incubate at 37 °C/5% CO2 for at least 6 hr (maximum of 12 hr).
- During this time, prepare the required number of laminin-coated wells/plates for step 4.7. Recommended cell transfer ratios are 1 confluent well of a 6-well plate to 2 wells of a 24-well plate (or equivalent amount per surface area).
- After 6 hr, carefully transfer the colony aggregates into a 15 ml tube using 5 ml pipette. Wash the plate with RB medium to collect any remaining aggregates and add to the same 15 ml tube (0.5 ml per well for 24-well and 1 ml per well for 6-well plates). Wait 5 min until the aggregates sediment, then carefully aspirate the supernatant. Be careful not to disturb the loose pellet.
- Add 2 ml RB medium supplemented with 12 µM CHIR99021 and carefully transfer with a 5 ml pipette to your pre-prepared laminin coated wells (as in Section 1.2.4) to allow the cells to attach.
- After 24 hr, carefully remove the medium in each well and add 1 ml RB medium only.
Note: Some of the aggregates will have only attached very weakly to the culture plate; therefore it is important to not remove any large aggregates which may have come loose during the medium change procedure. It is important, however, to remove as much of the small cell debris as possible. - After 24 hr, carefully remove the medium and add RB medium supplemented with IWP2, purmorphamine and SB431542 (5 µM each).
- Change the medium after 2 days with RB medium only. Cells will start to beat next day.
- Change the medium every 3 – 4 days with RB medium only.
Representative Results
In order to establish a simple method for the large-scale differentiation of cardiomyocytes from hPSCs, we created a protocol in which cells were treated initially with a WNT/β-catenin activator (CHIR99021)16 and subsequently with inhibitors of the WNT/β-catenin and transforming growth factor-β (TGF-β) pathways (IWP216 and SB43154217, respectively) and finally an activator of the sonic hedgehog (SHH) pathway (purmorphamine)17 (Figure 1A). In our differentiation strategy, approximately 50% of the spheroids (175 ± 25 µm spheroid diameter size) started beating 7 days after initiation of differentiation, which then increased to 100% by day 10. Interestingly, using this protocol, differentiated spheroids have been observed to continue beating up to 60 days after differentiation initiation18. Population studies on dissociated spheroids examined by flow cytometry revealed that at day 15, more than 90% of the population contained cardiac troponin T positive (cTnT+) cells, while less than 12% of the cells were positive for smooth muscle and endothelial markers (3.1% von Willibrand Factor (vWF+), 8.4% alpha smooth muscle actin-positive (aSMA+))18. To date, the static suspension differentiation protocol has been tested on 5 hESC and 4 hiPSC lines, with outputs resulting in approximately 90% of beating spheroids from each line, showing the high reproducibility of this protocol among different hPSC lines (Figure 1B).
In order to develop an integrated platform for large-scale production of human cardiomyocytes, we applied our static suspension differentiation strategy to a stirred suspension bioreactor (Figure 2A). The differentiating spheroids showed similar behavior in the bioreactor environment as in the static system (Figure 2B) and approximately 100% of spheroids were observed to be beating at day 10 18. Immunostaining in sections of beating spheroids collected at day 30 using antibodies against cTnT and the cardiac transcription factor NKX2-5, showed cytoplasmic and nuclear expression of these proteins, respectively (Figure 2C). The total yield of cells in the dynamic suspension culture was approximately 90 – 100 million cells in 100 ml working volume after 15 days of differentiation culture. With this being the case, the cardiomyocyte yield can reach up to approximately 54-90 million cells (with the observed 60 – 90% differentiation efficacy) from 20 million starting hPSCs, inoculated into the hPSCs expansion phase as single cells. We additionally examined the electrophysiological properties of differentiated cardiomyocytes using the single-cell patch clamp method. Beating spheroids from the bioreactor cultures were dissociated into single cells at day 30 and action potentials recorded on representative cells using the whole cell patch clamp technique. Data showed the presence of the three main cardiac cell types (atrial-, nodal- and ventricular-like cells) in the population of single cells18.
For the two differentiation techniques mentioned above, hPSCs need to be adapted to feeder-free and/or single-cell suspension culture19-21. Some hPSC lines however, are highly sensitive to enzymatic cell dissociation and show significant cell viability loss after dissociation, such as can be observed in newly established hiPSC lines. Additionally, when working with large cohorts of lines, adapting all to feeder-free and single-cell culture would be enormously labor intensive and time consuming. To address these issues, undifferentiated spheroids were replaced with undifferentiated cell aggregates (Figure 3A). Our data show that using this modified technique, beating clusters appeared 7 days after differentiation initiation. The reproducibility of this modified version was confirmed using 42 hiPSC lines, where all tested lines generated on average more than 60% cTnT+ cells by day 15 for each experiment performed (Figure 3B). Immunostaining using antibodies against cTnT and NKX2-5 in dissociated beating clusters showed cytoplasmic and nuclear expression, respectively (Figure 3C).
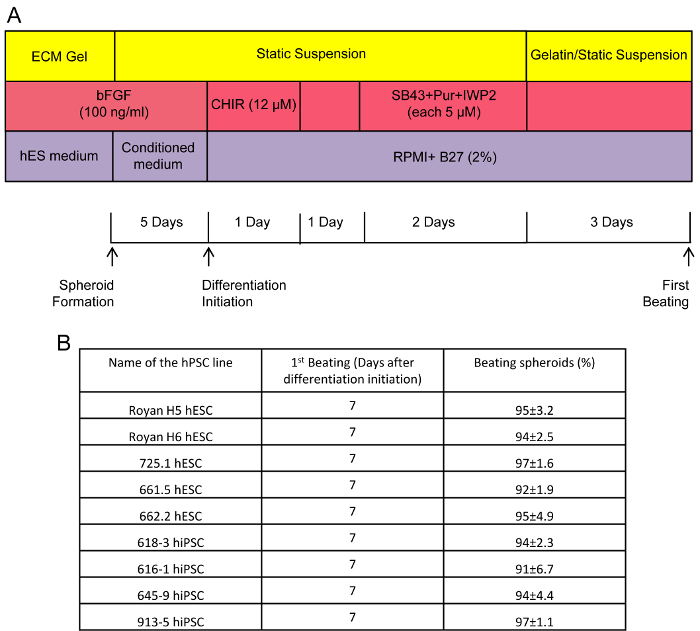
Figure 1. Schematic of Cardiomyocyte Differentiation from hPSCs in Static Suspension and Representative Data. (A) Experimental design of hPSCs differentiation to cardiomyocytes in static suspension system. (B) Evaluation of the efficiency of cardiac differentiation is measured by counting the number of beating spheroids (%). Shown here are the averages of at least 3 differentiation experiments from each of 5 hESC and 4 hiPSC lines18. Overall average of beating spheroids from all lines is approximately 90%. Error bars represent SD (n = 3). Please click here to view a larger version of this figure.
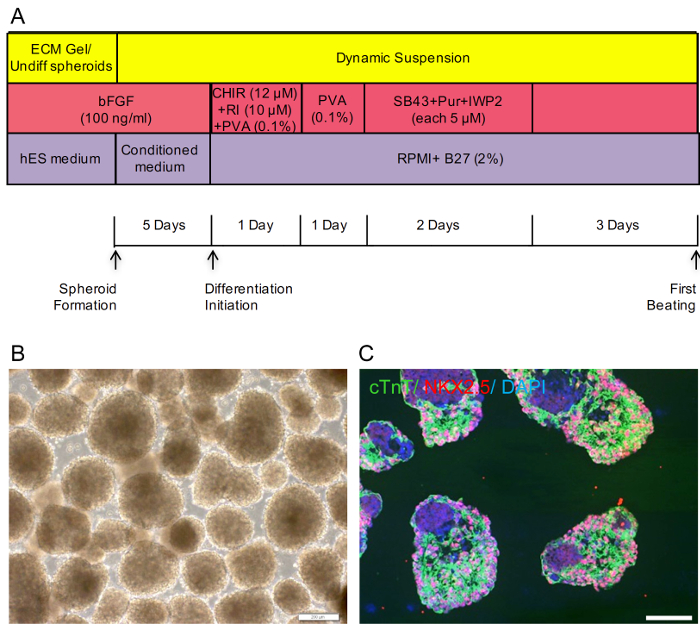
Figure 2. Overall Schematic of Cardiomyocytes Differentiation from hPSCs in Dynamic Suspension and Representative Data. (A) Experimental design of hPSCs differentiation to cardiomyocytes in dynamic suspension system. RI: Rock inhibitor (B) Phase contrast image of day 5 spheroids (Representative result from Royan H5 hESC line). Scale bar 200 µm (C) Immunostaining of hESC derived-beating spheroids sectioned at day 30 for NKX2.5 and cTnT. Scale bar 100 µm. Please click here to view a larger version of this figure.
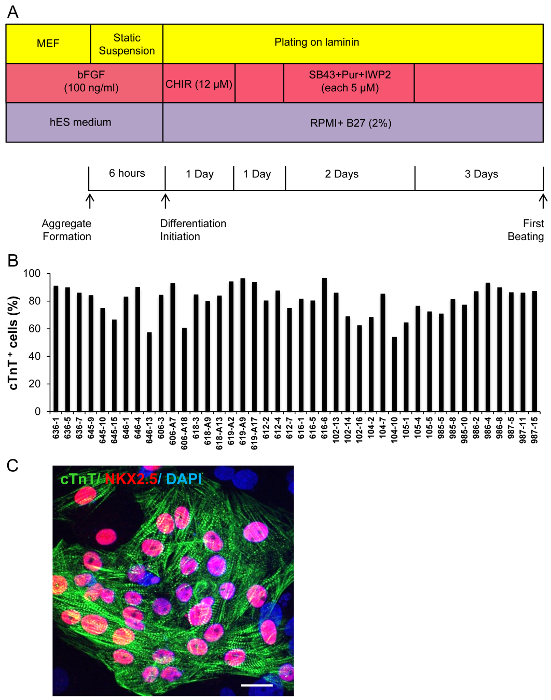
Figure 3. Overall Schematic of Cardiomyocytes Differentiation from hPSCs using Cultures not Adapted to Single Cell Passaging. (A) Experimental design of hPSCs differentiation to cardiomyocytes using cultures not adapted to single cell passaging. (B) Flow cytometry analysis of dissociated aggregates show on average more than 60% cTnT+ cells in 42 tested hiPSC lines. (C) Immunostaining of dissociated cardiomyocytes derived from hiPSCs for NKX2.5 and cTnT (Representative result from 646-4 hiPSC line). Scale bar 20 µm. Please click here to view a larger version of this figure.
Discussion
Cardiomyocytes derived from hPSCs are an extremely attractive source for use in human disease modeling, drug screening/toxicity testing and, perhaps in the future, regenerative therapies. One of the major hurdles to using these cells however, is the ability to provide enough high quality material for their effective use. Using our described protocol, we offer a method that overcomes this limitation.
Recently, synthetic small molecules targeting specific signaling pathways involved in cardiogenesis have been described to enhance cardiac differentiation 16,17,22,23. These are now being used as an alternative to recombinant cytokines and serum, containing many undefined factors and showing batch variation. The main advantage of a small molecule protocol, other than its specificity, is that it is inexpensive and component factors generally have an extended shelf-life in comparison to media containing cytokines or growth factors. As the small molecules used in our reported protocol are low molecular weight agents, they are structurally and functionally defined and can diffuse easily through the cell membrane 24,25.
So far, different methods have been applied to hPSCs in order to establish robust, scalable differentiation techniques; however, most of these methods have been established as 2D and small-scale static cultures, which may offer poor scalability and homogeneity. Techniques such as forced aggregation, micro-printing technologies and micro-carrier cultures 26-29 are now coming into use, but in spite of some advances in this area, these methods are still far from providing the cell numbers required for their use in high-demand systems. Limited or unproven reproducibility and scalability, and the high costs due to the necessity for expensive media (mTeSR1 or StemPro-34) or micro-carriers for both expansion of hPSCs and their directed differentiation to cardiomyocytes 30,31, are the drawbacks of using these methods in high throughput hPSCs technologies.
In this study, we have reported a simple, robust and scalable protocol for the production of hPSC-derived cardiomyocytes. The reproducibility of this protocol has been validated with more than 40 different hPSC lines, the most extensive validation reported to date. Compared to previously reported suspension protocols, this method shows great advantages in terms of scalability, reproducibility, affordability, efficacy and functionality of the cardiomyocytes generated. The success of our differentiation protocol is thoroughly dependent on the high quality of the hPSCs used for study. Therefore it is crucial to check each line's karyotype and the high and sustained expression of pluripotency markers by immunostaining and PCR before starting the experiment. Particularly, when culturing cells in the bioreactor, it is crucial to use hPSCs that have been passaged at the single-cell level for at least three passages prior to the start of differentiation. In our protocols we have used spheroids with an average size of 175 ± 25 µm, which were spheroids generated after 5 days. The day which the spheroids meet this size restriction may vary depending on the growth rate of individual hPSC lines; therefore it is important that the size, more than the day of growth, is taken into consideration.
This protocol also can be manipulated to become suitable for sensitive cell lines and large cohorts of hPSC lines. Although this protocol results in highly enriched cardiomyocytes, purity may be improved by combining the differentiation protocol with a metabolic selection method such as lactate-enriched medium 32. Additionally, improvements in the maturation and functionality of the derived cardiomyocytes described in this protocol may be applied by the generation of three-dimensional, aligned cardiac tissues 33. One of the limitations of this protocol is that particular subtypes of cardiomyocytes are not specifically generated, simply a mixture of atrial, ventricular and nodal cells. Further investigation in this field is needed to develop differentiation protocols which can produce highly enriched subtype-specific cardiomyocytes in large-scale. Although some small-scale protocols have been developed to date, the methods would need to be rigorously tested to ensure scale-up is possible 34.
Development of such integrated platforms can be considered as an important step towards the commercialization of hPSC-derived cardiomyocytes technologies for clinical, pharmaceutical, tissue engineering, and in vitro organ/organoid development applications.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was funded by grants provided from Royan Institute, Iranian Council of Stem Cell Research and Technology, the Iran National Science Foundation (INSF), the National Health and Medical Research Council of Australia (NHMRC; 354400), the National Heart Foundation of Australia/Heart Kids Australia (G11S5629), and the New South Wales Cardiovascular Research Network. HF was supported by a University International Postgraduate Scholarship from the University of New South Wales, Australia. RPH was supported by a NHMRC Australia Fellowship. The authors express their gratitude to the human subjects who participated in this research.
Materials
| Knockout DMEM | Life Technologies | 10829018 | |
| Knockout Serum Replacement (KO-SR) | Life Technologies | 10828028 | |
| Glutamax | Life Technologies | 35050061 | |
| MEM Non-essential Amino Acids | Life Technologies | 11140-050 | |
| β-Mercaptoethanol | Life Technologies | 21985-023 | |
| Basic Fibroblast Growth Factor (bFGF) | Miltenyi Biotec | 130-093-843 | |
| RPMI1640 | Life Technologies | 11875093 | |
| DPBS, no calcium, no magnesium | Life Technologies | 14190144 | |
| DPBS | Life Technologies | 14287072 | |
| Attachment Factor (AF) | Life Technologies | S006100 | |
| ECM Gel | Sigma-Aldrich | E1270 | |
| Laminin | Invitrogen | 23017-015 | |
| DMEM | Life Technologies | 11965-092 | |
| Fatal Bovine Serum (FBS) | Life Technologies | 16140-071 | |
| B27 minus insulin | Gibco | A18956-01 | |
| Penicillin/Streptomycin | Life Technologies | 15070063 | |
| 0.05% Trypsin/EDTA | Life Technologies | 25300-054 | |
| Collagenase Type IV | Life Technologies | 17140-019 | |
| Calcium Chloride (CaCl2) | Sigma-Aldrich | C7902 | |
| Mitomycin C | Bioaustralis | BIA-M1183 | |
| CHIR99021 | Miltenyi Biotec | 130-104-172 | |
| IWP2 | Miltenyi Biotec | 130-105-335 | |
| SB431542 | Miltenyi Biotec | 130-095-561 | |
| Purmorphamine | Miltenyi Biotec | 130-104-465 | |
| ROCK inhibitor Y-27632 | Miltenyi Biotec | 130-104-169 | |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | E6758 | |
| Poly Vinyl Alcohol (PVA) | Sigma-Aldrich | 363073 | |
| Gelatin | Sigma-Aldrich | G1890 | |
| Trypan Blue | Bio-Rad | 145-0013 | |
| Accumax | Innovative Cell Technologies Inc. | AM105 | |
| Sigmacote | Sigma-Aldrich | SL2 | |
| CELLSPIN | Integra Biosciences | 183 001 | |
| Spinner flask with 1 pendulum, 100 ml | Integra Biosciences | 182 023 | |
| Mouse Embryonic Fibroblasts (MEF) | Prepared in-house (or commercially available) | ||
| Human pluripotent stem cell (hPSC) lines | Prepared in-house (or commercially available) |
References
- Thomson, J. A., et al. Embryonic stem cell lines derived from human blastocysts. Science. 282, 1145-1147 (1998).
- Takahashi, K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131, 861-872 (2007).
- Carvajal-Vergara, X., et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 465, 808-812 (2010).
- Vitale, A. M., Wolvetang, E., Mackay-Sim, A. Induced pluripotent stem cells: a new technology to study human diseases. Int. J. Biochem. Cell Biol. 43, 843-846 (2011).
- Sharma, A., et al. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res. 115, 556-566 (2014).
- Zimmer, B., et al. Evaluation of developmental toxicants and signaling pathways in a functional test based on the migration of human neural crest cells. Environ Health Perspect. 120, 1116-1122 (2012).
- Diecke, S., Jung, S. M., Lee, J., Ju, J. H. Recent technological updates and clinical applications of induced pluripotent stem cells. Korean J Intern Med. 29, 547-557 (2014).
- Kimbrel, E. A., Lanza, R. Current status of pluripotent stem cells: moving the first therapies to the clinic. Nat. Rev. Drug Discov. 14, 681-692 (2015).
- Kehat, I., et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 108, 407-414 (2001).
- Mummery, C., et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 107, 2733-2740 (2003).
- Laflamme, M. A., et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 25, 1015-1024 (2007).
- Fonoudi, H., et al. ISL1 protein transduction promotes cardiomyocyte differentiation from human embryonic stem cells. PLoS One. 8, e55577 (2013).
- Zhu, W. Z., Hauch, K. D., Xu, C., Laflamme, M. A. Human embryonic stem cells and cardiac repair. Transplant Rev (Orlando). 23, 53-68 (2009).
- Jozefczuk, J., Drews, K., Adjaye, J. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J Vis Exp. (64), e3854 (2012).
- Stover, A. E., Schwartz, P. H. Adaptation of human pluripotent stem cells to feeder-free conditions in chemically defined medium with enzymatic single-cell passaging. Methods Mol Biol. 767, 137-146 (2011).
- Lian, X., et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci. 109, E1848-E1857 (2012).
- Gonzalez, R., Lee, J. W., Schultz, P. G. Stepwise chemically induced cardiomyocyte specification of human embryonic stem cells. Angew Chem Int Ed Engl. 50, 11181-11185 (2011).
- Fonoudi, H., et al. A Universal and Robust Integrated Platform for the Scalable Production of Human Cardiomyocytes From Pluripotent Stem Cells. Stem Cells Transl Med. , (2015).
- Abbasalizadeh, S., Larijani, M. R., Samadian, A., Baharvand, H. Bioprocess development for mass production of size-controlled human pluripotent stem cell aggregates in stirred suspension bioreactor. Tissue Eng Part C Methods. 18, 831-851 (2012).
- Larijani, M. R., et al. Long-term maintenance of undifferentiated human embryonic and induced pluripotent stem cells in suspension. Stem Cells Devt. 20, 1911-1923 (2011).
- Baharvand, H., Larijani, M. R., Yousefi, M. Protocol for expansion of undifferentiated human embryonic and pluripotent stem cells in suspension. Methods Mol Biol. 873, 217-226 (2012).
- Burridge, P. W., et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 11, 855-860 (2014).
- Minami, I., et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell reports. 2, 1448-1460 (2012).
- Buskirk, A. R., Liu, D. R. Creating small-molecule-dependent switches to modulate biological functions. Chem Bio. 12, 151-161 (2005).
- McKinsey, T. A., Kass, D. A. Small-molecule therapies for cardiac hypertrophy: moving beneath the cell surface. Nat Rev Drug Discov. 6, 617-635 (2007).
- Niebruegge, S., et al. Generation of human embryonic stem cell-derived mesoderm and cardiac cells using size-specified aggregates in an oxygen-controlled bioreactor. Biotechnol Bioeng. 102, 493-507 (2009).
- Bauwens, C. L., et al. Geometric control of cardiomyogenic induction in human pluripotent stem cells. Tissue Eng Part A. 17, 1901-1909 (2011).
- Hwang, Y. S., et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci U S A. 106, 16978-16983 (2009).
- Nguyen, D. C., et al. Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports. 3, 260-268 (2014).
- Kempf, H., et al. Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Reports. 3, 1132-1146 (2014).
- Hemmi, N., et al. A massive suspension culture system with metabolic purification for human pluripotent stem cell-derived cardiomyocytes. Stem Cells Transl Med. 3, 1473-1483 (2014).
- Tohyama, S., et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell stem cell. 12, 127-137 (2013).
- Nunes, S. S., et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat meth. 10, 781-787 (2013).
- Devalla, H. D., et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med. 7, 394-410 (2015).

