Determination of the Optimal Chromosomal Location(s) for a DNA Element in Escherichia coli Using a Novel Transposon-mediated Approach
Summary
Here, the power of a transposon-mediated random insertion of a non-coding DNA element was used to resolve its optimal chromosomal position.
Abstract
The optimal chromosomal position(s) of a given DNA element was/were determined by transposon-mediated random insertion followed by fitness selection. In bacteria, the impact of the genetic context on the function of a genetic element can be difficult to assess. Several mechanisms, including topological effects, transcriptional interference from neighboring genes, and/or replication-associated gene dosage, may affect the function of a given genetic element. Here, we describe a method that permits the random integration of a DNA element into the chromosome of Escherichia coli and select the most favorable locations using a simple growth competition experiment. The method takes advantage of a well-described transposon-based system of random insertion, coupled with a selection of the fittest clone(s) by growth advantage, a procedure that is easily adjustable to experimental needs. The nature of the fittest clone(s) can be determined by whole-genome sequencing on a complex multi-clonal population or by easy gene walking for the rapid identification of selected clones. Here, the non-coding DNA region DARS2, which controls the initiation of chromosome replication in E. coli, was used as an example. The function of DARS2 is known to be affected by replication-associated gene dosage; the closer DARS2 gets to the origin of DNA replication, the more active it becomes. DARS2 was randomly inserted into the chromosome of a DARS2-deleted strain. The resultant clones containing individual insertions were pooled and competed against one another for hundreds of generations. Finally, the fittest clones were characterized and found to contain DARS2 inserted in close proximity to the original DARS2 location.
Introduction
The function of any genetic element can be affected by its location in the genome. In bacteria, this mainly results from interference by the transcription of neighboring genes, local DNA topology, and/or replication-associated gene dosage. In particular, the processes of DNA replication and segregation are controlled, at least in part, by non-coding chromosomal regions1, and the proper function of these regions depends on genomic location/context. In E.coli, examples are the dif site, required for sister chromosome resolution2; KOPS sequences, required for chromosome segregation3; and datA, DARS1, and DARS2 regions, required for proper chromosomal replication control (below; 4). We present a method allowing for the random relocation, selection, and determination of the optimal genetic context of any given genetic element, exemplified here by the study of the DARS2 non-coding region.
In E. coli, DnaA is the initiator protein responsible for DNA strand opening at the single replication origin, oriC, and for the recruitment of the helicase DnaB5,6,7. DnaA belongs to the AAA+ (i.e., ATPases associated with diverse activities) proteins and can bind both ATP and ADP with similar high affinities5. The level of DnaAATP peaks at initiation8, where DnaAATP forms a multimer on oriC that triggers DNA duplex opening9. After initiation, oriC is made temporarily unavailable for re-initiation due to sequestration by a mechanism involving the binding of the SeqA protein to hemimethylated oriC10,11. During sequestration, the level of DnaAATP is reduced by at least two mechanisms: the regulatory inactivation of DnaA (RIDA)12,13 and datA-dependent DnaAATP hydrolysis (DDAH)14,15. Both RIDA and DDAH promote the conversion of DnaAATP to DnaAADP. Prior to a new round of initiation, DnaAADP is re-activated to DnaAATP at specific DnaA-reactivating sequences (DARS): DARS1 and DARS216,17. The chromosomal datA, DARS1,and DARS2 regions are non-coding and act in a chaperone-like manner to modulate DnaAATP/DnaAADP interconversion. These regions, located outside the origin of replication, enable the assembly of a DnaA complex for either the inactivation (datA;14) or activation (DARS1 and DARS2;17) of DnaA. Deleting DARS2 in a cell does not alter mass doubling time but results in asynchronous replication initiation15,16,18. However, DARS2-deficient cells have a fitness cost compared to an otherwise isogenic wildtype during both continuous growth competition in rich medium or during the establishment of colonization in the mouse intestine18. This indicates that even minor changes in asynchrony/origin concentration have a negative effect on bacterial fitness. In E. coli, there is a selective pressure to maintain chromosome symmetry (i.e., two nearly equal length replication arms)19. The datA, DARS1, and DARS2 regions have the same relative distance to oriC in all E. coli strains sequenced18, despite large variations in chromosome size.
Here, we use the DARS2 region of E. coli as an example for the identification of the chromosomal position(s) optimal for its function. DARS2 was inserted into the NKBOR transposon, and the resultant NKBOR::DARS2 transposon subsequently inserted randomly into the genome of MG1655 ΔDARS2. We thus generated a collection of cells, each possessing DARS2 placed at a different location on the chromosome. An in vitro competition experiment, where all cells in the collection were pooled and competed against each other during continuous growth in LB for an estimated 700 generations, was performed. The outcome of the competition experiment was monitored/determined using Southern blot, easy gene walking, and whole-genome sequencing (WGS; Figure 1). End-point clones resolved by easy gene walking were characterized by flow cytometry to evaluate cell-cycle parameters. In a flow cytometric analysis, cell size, DNA content, and initiation synchrony can be measured for a large number of cells. During flow cytometry, a flow of single cells passes a light beam of the appropriate wavelength to excite the stained DNA, which is then simultaneously registered by photomultipliers that collect the emitted fluorescence, a measure of DNA content, provided the cells are stained for DNA. The forward-scattered light is a measure of cell mass20.
The in vitro competition experiment we present here is used to address questions relating to the importance of the chromosomal position and genomic context of the genetic element. The method is unbiased and easy to use.
Protocol
1. Collection of the Transposon Library
NOTE: The chromosomal DARS2 locus was cloned into the mini Tn10-based transposon, NKBOR (on pNKBOR)21, resulting in NKBOR::DARS2 (pJFM1). pNKBOR can be obtained online22. pNKBOR is a R6K-based suicide vector that requires the initiator protein π for replication23. Plasmid pJFM1 is therefore able to replicate in an E. coli strain (e.g., Dh5α λ pir) containing a chromosomal copy of the pir gene. However, when pJFM1 is transformed into the Pir-deficient wildtype MG1655, pJFM1 cannot replicate, leading to the selection of kanamycin-resistant clones generated by the random insertions of NKBOR::DARS2 into the bacterial chromosome. For simplicity, these are referred to as DARS2 insertions. See Figure 1 for a schematic presentation of the methodology.
- Prepare electrocompetent MG1655 ΔDARS2 from actively growing cells in Lysogeny broth (LB) grown at 37 °C24.
- Electroporate pJFM1 into electrocompetent MG1655 ΔDARS2, according to Gonzales et al.24
- Add 1 µg of pJFM1 (in 1 µL of water) to 40 µL of electrocompetent E. coli MG1655 ΔDARS2 and transfer this mixture to a pre-chilled, sterile 0.2-cm gap cuvette. Insert the cuvette into the electroporation chamber. Electroporate at 18 kV, 500 Ω, and 25 µF; the time constant should be ~5.0 ms, and no arcing should occur.
- Quickly recover the bacterial suspension by resuspending it in 1 mL of pre-warmed LB broth and transfer to a 15 mL test tube.
- Let the cells recover by incubating under aerated growth conditions at 37 °C for 30 min, without antibiotic selection. Plate the bacteria onto LB agar plates supplemented with 50µg/mL kanamycin and incubate at 37 °C overnight.
- Count the colonies from the electroporation: one colony is considered equal to one chromosomal transposon insertion.
NOTE: Colonies can be counted by eye or with a colony counter. The number of colonies is inversely proportional to the average distance separating the transposons located in the chromosome of each clone. - Add 1 mL of LB broth to each plate and wash off all colonies; 1 mL of LB broth should be sufficient to wash off ~100,000 colonies. Re-use the same 1 mL of LB broth to increase the bacterial concentration. Pool all colonies in the same 50 mL tube.
- Vortex the tube and freeze the start material (t = 0). To freeze it, mix 1 mL of cell suspension with 1 mL of 50% glycerol on ice. Transfer the tubes to dry ice for 10 min. Once the cultures are frozen, transfer them to a -80 °C freezer.
NOTE: A cell concentration of a minimum of 50,000 CFU/mL is expected at this step.
2. Competition Experiment in LB
- Thaw the transposon library from -80 °C on ice. Mix by pipetting.
- Transfer 100 µL of the transposon library to 10 mL of LB in a 15 mL test tube.
- Grow the cells, aerated by continuous shaking (250 rpm), for 8 h at 37 °C to stationary phase (i.e., OD600 = ~4.0). Adjust these parameters to different growth conditions, as desired.
- Propagate the bacterial population by continuous transfers into fresh prewarmed medium every ~10 generations. Do this by transferring 10 µL of the previous stationary phase culture to 10 mL of fresh LB and growing for another 8 h to the stationary phase (i.e., OD600 = ~4.0).
NOTE: As the start and end optical density are the same, a 1,000-fold dilution corresponds to ~10 generations of growth (210 = 1,024). The bacterial population can either be propagated directly into fresh prewarmed medium or saved at 0-4 °C (on ice) and propagated the following day. LB was used here to ensure as many cellular doublings in as short a time as possible (a doubling time close to 22 min). - After each 100 generations of competition, save five 1-mL samples at -80 °C (as described in step 3.3).
- If the fitness differences between cells containing individual transposon insertions are expected to be small, keep the duration of competition long; here, 700 generations (t = 700) were used.
3. Southern Blot Analysis to Monitor the Competition Experiment Over Time
- Prepare the probe for the Southern blot (1-kb section of the NKBOR) by performing polymerase chain reaction (PCR) amplification using specific primers: NKBOR_Probe_FW: gatgttggacgagtcggaat and NKBOR_Probe_RV: cgttacatccctggcttgtt.
- Incubate at 98 °C for 30 s for initial denaturation. Then, perform 35 cycles at 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 45 s. For the final extension, use 72 °C for 10 min.
NOTE: The template for the PCR reaction was a purified pNKBOR plasmid21. - Label the resultant PCR fragment with [α-32P]dATP using the random primer system, according to Smith25.
- Incubate at 98 °C for 30 s for initial denaturation. Then, perform 35 cycles at 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 45 s. For the final extension, use 72 °C for 10 min.
- Prepare the total cellular DNA according to Løbner-Olesen and von Freiesleben26 for t = 0 and selected time points until t = 700 estimated generations of competition.
- Digest the total cellular DNA with PvuI, which cuts the NKBOR containing the region of interest once only in a region not covered by the probe; 1 unit of PvuI can be used to completely digest 1 µg of substrate DNA.
NOTE: The probe will recognize fragments containing part of the transposon, along with chromosomal DNA of various lengths, depending on the insertion site. - Perform a Southern blot using a 0.7% agarose gel according to Løbner-Olesen and von Freiesleben26.
4. Identification of the Fittest Clones
- Use easy gene walking, as described by Harrison et al.27, to identify DARS2 insertion sites from single clones (isolated on LB plates) after 700 estimated generations of competition; the more bands on the Southern blot, the more isolates are needed to cover all transposon insertions.
- Isolate genomic DNA for PCR template. Spread bacteria on an LB agar plate containing 50 µg/mL kanamycin and incubate at 37 °C overnight. Grow single colonies overnight in LB broth at 37 °C and subsequently transfer 20 µL into 200 µL of autoclaved distilled water (or use DNA purification, as in step 3.2).
- Vortex to mix. Heat the mix at 100 °C for 10 min and centrifuge at max speed for 5 min. Save the cell lysate at -20 °C for future use.
- Design three nested primers to anneal within the transposon of choice (see Figure 2 for a graphic representation of the PCR strategy).
NOTE: Designing Nested Primers should ensure that Nested Primer 3 lies closest to the known 5' end of the transposon DNA, followed by Nested Primers 2 and 1. The spacing between Nested Primer 3 and the 5' end of the known transposon DNA should be sufficient for a sequence read-through from the known DNA to the unknown DNA (to find the specific chromosomal transposon insertion site). For NKBOR the following nested primers were used: pNKBOR_Nested3_RV: gcagggctttattgattcca, pNKBOR_Nested2_RV: tcagcaacaccttcttcacg, and pNKBOR_Nested1_RV: actttctggctggatgatgg. Random primers containing the recognition sites of known restriction enzymes are also used. To ensure a result, one should use at least two different random primers. The restriction sites for the restriction enzymes (e.g.,Sau3AI and HindIII) should be located at the 3' end and preceded by ten random bases so that 5'-NNNNNNNNNNGATC-3' and 5'-NNNNNNNNNNAAGCTT-3' are the primers containing a Sau3AI site (GATC) and a HindIII site (AAGCTT), respectively, where N = A, T, G, or C. - Use 200 ng of genomic DNA in a 20 µL PCR reaction. Incubate at 98 °C for 30 s for the initial denaturation. Perform 25 cycles at 98 °C for 30 s, 50 °C for 15 s, and 72 °C for 4 min. For the final extension, use 72 °C for 10 min. Use primer pairs consisting of Nested Primer 1 and one of the random primers.
- Use primer pairs consisting of Nested Primer 2 and the same random primer from the second amplification, using 1 µL of its previous reaction as a template.
- Repeat step 4.1.4 with primer pairs consisting of Nested Primer 3 and the same random primer.
- Run the products from the final PCR reaction on a 1.5% agarose gel and stain with ethidium bromide. Cut out a band in the range 100-800 bp and isolate the DNA using any commercial DNA gel extracting kit according to the manufacturer's direction. Resuspend the DNA in 15 µL of water.
NOTE: Caution. Ethidium bromide is toxic and must be handled with care. - Sequence the band isolated in the previous step (step 4.1.6), with Nested Primer 3 as the sequencing primer, using Sanger sequencing at a commercial provider.
- Isolate genomic DNA for PCR template. Spread bacteria on an LB agar plate containing 50 µg/mL kanamycin and incubate at 37 °C overnight. Grow single colonies overnight in LB broth at 37 °C and subsequently transfer 20 µL into 200 µL of autoclaved distilled water (or use DNA purification, as in step 3.2).
- Perform WGS.
- Perform WGS on the total DNA extracted from selected samples collected during the competition experiment, as described by Frimodt-Møller et al4.
- Perform sequencing analysis.
- Align paired-end reads to 150 Ns contiguous to NKBOR, AF310136.1 1,904…2,204 using Bowtie228 with an interval between seed substrings (S,1,1.15) and a maximum number of ambiguous characters (L,0,0.9).
- Select the aligned reads and then re-align to both MG1655 ref|NC_000913.3 and NKBOR gb|AF310136 using blastN29
- Assign transposition insertion positions at junctions MG1655-NKBOR.
5. Flow Cytometry
- Collect samples for flow cytometry.
- Balance each culture by maintaining it in the exponential growth phase for at least 10 generations. Check the OD600 and ensure that it never exceeds 0.3.
- Collect two types of flow samples.
- For RIF-runout, transfer 1 mL of culture to a 15 mL tube containing 30 µL of RIF-CEF (300 µg/mL rifampicin and 36 µg/mL cephalexin dissolved in 12.5 mM NaOH). Leave it at 37 °C for a minimum of 4 h of shaking.
NOTE: Rifampicin indirectly blocks the initiation of chromosome replication while allowing for ongoing rounds of replication to continue to termination. Cephalexin prevents cell division, which results in replication runout (RIF-runout) and the accumulation of cells with fully replicated chromosomes. The number of fully replicated chromosomes is equal to the number of origins at the time of treatment with rifampicin and cephalexin20. - For the EXP-sample, transfer 1 mL of exponentially growing culture to a 1.5-mL tube on ice.
- For RIF-runout, transfer 1 mL of culture to a 15 mL tube containing 30 µL of RIF-CEF (300 µg/mL rifampicin and 36 µg/mL cephalexin dissolved in 12.5 mM NaOH). Leave it at 37 °C for a minimum of 4 h of shaking.
- Fix THE cells as follows. Harvest the cells by centrifugation at 15,000 x g for 5 min at 4 °C. Resuspend the pellet in 100 µL of ice-cold 10 mM Tris pH 7.5 and add 1 mL of ice-cold 77% ethanol. Store the samples at 4 °C until use.
- Stain the cells as follows. Harvest 100-300 µL of fixed cells by centrifugation at 15,000 x g for 15 min. Remove the supernatant and resuspend the pellet in 130 µL of "DNA Staining solution" (90 µg/mL mithramycin, 20 µg/mL ethidium bromide, 10 mM MgCl2, and 10 mM Tris pH 7.5). Put the samples on ice and in the dark; the samples are ready for flow cytometry analysis after 10 min.
NOTE: Other DNA stains than ethidium bromide and mithramycin can also be used30,31. - Determine the origin per cell, the relative cell mass, and the relative DNA content by flow cytometry analysis.
- Perform flow cytometry, as described previously32. Run RIF-runout and EXP-samples using the flow cytometer manufacturer's instructions. For mithramycin- and ethidium bromide-stained cells, use excitation wavelengths 395 and 440 nm and collect the fluorescence above 565 nm.
- To determine the relative cell mass and relative DNA content, run the EXP-samples to obtain single-parameter forward-light scatter versus cell number histograms and fluorescence intensity versus cell number histograms for a minimum of 30,000 events corresponding to the cell signal.
- Use forward-scattered light single-parameter distribution to measure the average relative cell mass.
NOTE: Forward-scattered light is a measure of the average cell mass20. - Use fluorescence intensity single-parameter distributions to measure the average DNA content20.
- Obtain the DNA concentration as the ratio of average DNA content to the average cell mass20.
- Determine the number of origins per cell.
- Run RIF-runout samples to obtain single-parameter fluorescence intensity versus cell number histograms for a minimum of 30,000 events corresponding to the cell signal.
- Use fluorescence intensity single-parameter distributions to determine the number of chromosomes in each cell20.
- Perform flow cytometry, as described previously32. Run RIF-runout and EXP-samples using the flow cytometer manufacturer's instructions. For mithramycin- and ethidium bromide-stained cells, use excitation wavelengths 395 and 440 nm and collect the fluorescence above 565 nm.
- Calculate the asynchrony index (Ai).
- Calculate the Ai,as described by Løbner-Olesen et al.32, using the fluorescence intensity single parameter distributions of the RIF-runout samples and the formula Ai = (f3 + f5 + f6 + f7) / (f2 + f4 + f8), where fx is the fraction cells with x number of fully replicated chromosomes. Consider initiations asynchronous when A >0.1.
Representative Results
A Southern blot was done to verify that DARS2 was distributed randomly throughout the chromosome in the transposon library (t = 0) and that the fittest clones would persist over time. The Southern blot was performed on DNA extracted from the initial transposon pool (at t = 0) and every estimated 100 out of 700 generations of competition (Figure 3). Here, the total cellular DNA from each time-point was digested with the PvuI restriction enzyme, known to cut transposon NKBOR::DARS2 once only in a region not covered by the probe. The Southern blot was probed with a radioactively labeled DNA fragment complementary to part of NKBOR. As seen from Figure 3, the initial DARS2 pool (t = 0) lacked distinct bands, which shows that DARS2 was inserted randomly throughout the chromosome. Over time, a pattern emerged where the initial large pool of DARS2 clones developed into only one or a few persisting DARS2 clones (Figure 3; t = 0 to t = 700).
In the example shown, DARS2 insertion sites from the competition experiment were identified using WGS and easy gene walking. Here, WGS was used to identify insertion sites from the start pool (t = 0) and after 300, 400, and 700 generations of competition. Note that the coverage in the present deep sequencing was insufficient for a complete mapping of insertion sites at t = 0; however, it gives a representative subset of the total number of insertions. WGS confirmed the Southern blot result (i.e., the selection of the fittest DARS2 clones), ending with approximately 98% of all DARS2 insertions close to the wildtype DARS2 chromosomal location (DARS2 Clone IR and Clone rppH), while the remaining 2% were elsewhere on the chromosome (Figure 4). This strongly suggests that the wildtype position is optimal for DARS2 function. At t = 400, an insertion was found on the opposite replication arm with an almost identical distance to oriC as the wildtype DARS2 position (Figure 4), but this insertion was not recovered after 700 generations. Thus, replication-associated gene dosage cannot be the single determinant for optimal position.
Easy gene walking was used to identify DARS2 insertions sites in single clones isolated after 700 estimated generations of competition. Here, the two DARS2 insertion sites mentioned above (DARS2 Clone IR and Clone rppH) were identified. Easy gene walking was only done on 20 clones, and this explains why all DARS2 insertions sites mapped in WGS were not identified. DARS2-deficient cells were previously shown to initiate replication in asynchrony and to have a decrease in origin concentration relative to wildtype cells4,16,17,18. We therefore used flow cytometry to resolve the synchrony in the initiation of DNA replication and cellular origin content for the two selected strains (DARS2 Clone IR and Clone rppH) which, in both cases, were restored to wildtype levels (Figure 5). A representative example of a strain possessing a single copy of DARS2 located in the terminus is shown (Figure 5E). Here, the presence of a DARS2 element in the terminus does not restore synchrony or the cellular origin content to wildtype levels, while the selected DARS2 clones IR and rppH do.
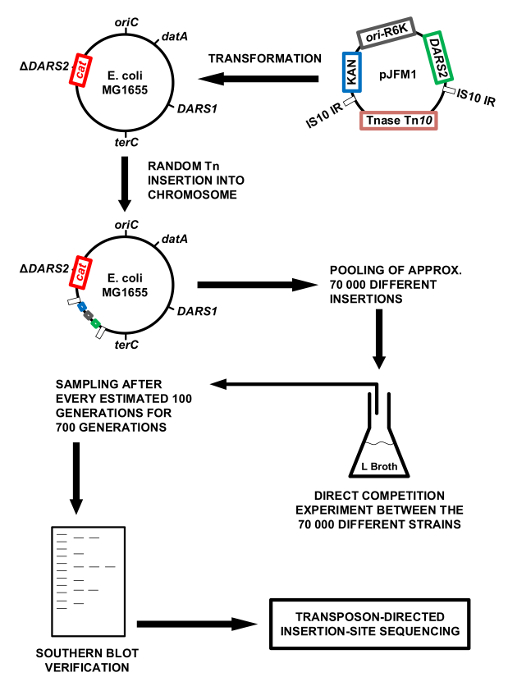
Figure 1: Methodology overview. Schematic presentation of the methodology. The chromosomal DARS2 region is cloned into the mini Tn10 on pNKBOR, creating pJFM1. pJFM1 is transformed into E. coli MG1655 ΔDARS2, which triggers a random insertion of DARS2 linked to Tn10 onto the chromosome of E. coli MG1655 ΔDARS2. Approximately 70,000 clones, each containing a different chromosomal DARS2 insertion, were pooled and competed directly against each other in LB broth at 37 °C. The direct competition experiment in LB broth was performed for an estimated 700 generations, where a sample was isolated for each 100 generations of direct competition. The total DNA was extracted from each isolated sample and used for Southern blotting and the identification of DARS2 insertions by WGS. Please click here to view a larger version of this figure.
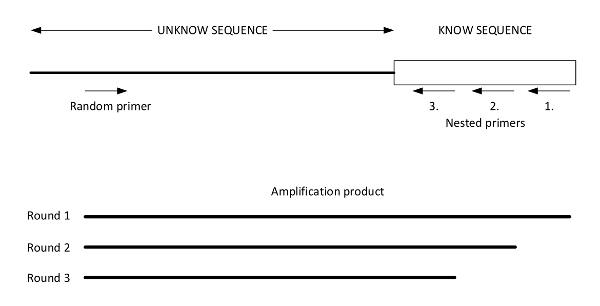
Figure 2: Graphic presentation of easy gene walking. This figure illustrates a genomic DNA template of an unknown DNA sequence adjacent to a known sequence with priming sites for the random primer and Nested Primers 1, 2, and 3. The results of the three successive amplifications performed using the three designed nested primers are illustrated below. The final product (from round 3) is sequenced using Nested Primer 3. This figure was adapted from Harrison et al.27. Please click here to view a larger version of this figure.
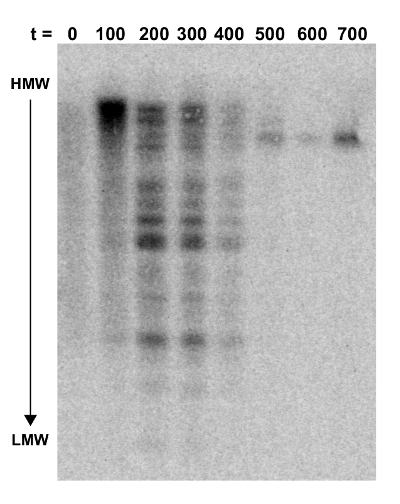
Figure 3: Southern blot probed for NKBOR. Southern blot analysis of DARS2 insertions into a DARS2-deficient strain. Genomic DNA extracted from every ~100 generations of direct competition, starting at t = 0 and ending at 700 generations, were digested with PvuI and gel-fractionated. The blot was hybridized with a NKBOR probe (see the Protocol). t indicates the number of generations of competition. This figure was adapted from Frimodt-Møller et al.4. HMW and LMW are high-molecular weight and low-molecular weight DNA, respectively. Please click here to view a larger version of this figure.
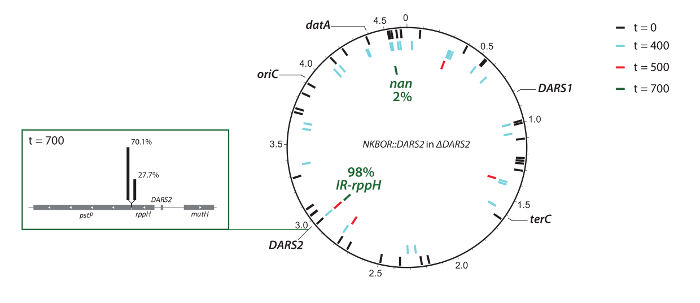
Figure 4: Graphic representation of resolved transposon insertions sites in ΔDARS2. The positions of oriC, datA, DARS1, DARS2, and terC are indicated. DARS2 insertion sites in ΔDARS2 at t = 0 (black bars), t = 400 (light blue bars), t = 500 (red bars), and t = 700 (green bars), resolved by full-genome sequencing. This figure was made using DNAPlotter33 and was adapted from Frimodt-Møller et al.4. Please click here to view a larger version of this figure.
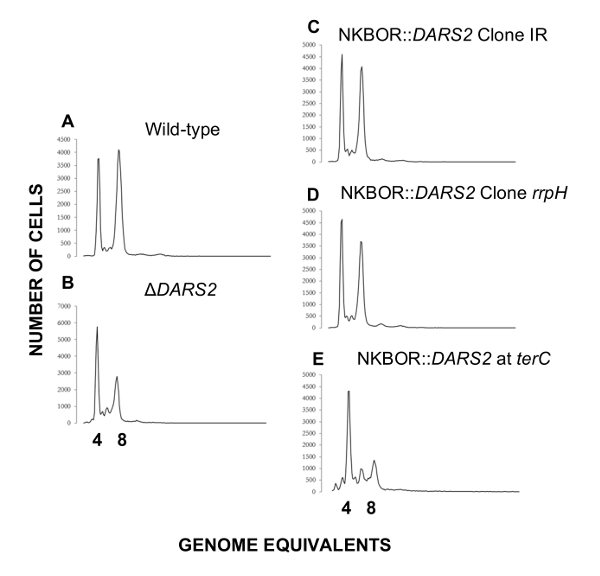
Figure 5: Representative flow cytometry histograms of transposon sites found by easy gene walking at t = 700. Cells were grown in AB minimal medium supplemented with 0.2% glucose, 10 µg/mL thiamine, and 0.5% casamino acids at 37 °C. Wildtype and ΔDARS2 are shown in A and B, respectively. Derivatives of the wildtype strain MG1655 devoid of DARS2 at the original locus and instead carrying a copy of DARS2 at the resolved transposon site rrpH, IR, and the terminus (terC) are shown in C, D, and E, respectively. This figure was adapted from Frimodt-Møller et al.4 to show a DARS2 location that results in a cell cycle anomaly (terC) or that restores the wildtype phenotype(rrpH, IR). Please click here to view a larger version of this figure.
Discussion
The methodology used here takes advantage of state-of-the-art techniques to answer a difficult question regarding the optimal genomic position of a genetic element. The random insertion of the genetic element (mediated by the transposon) enables the fast and easy collection of thousands of clones, which then can be made to compete against each other to select for the optimal position of the investigated genetic element (i.e., the fittest clone).
Here, DARS2 was inserted into the mini-Tn10-based transposon, NKBOR. The choice of transposon is important for the downstream analysis of insertions sites. Several different transposons have been used in transposon-directed insertion-site sequencing (TraDIS) experiments, such as mini-Tn5Km234 and the more popular choice, the Himar I Mariner transposon35,36 (for a recent review of this, see van Opijnen and Camilli36). We aimed for 70,000 initial random insertions of DARS2, which gives approximately a DARS2 insertion evert 65 bp in the MG1655 genome. This can easily be adjusted by decreasing or increasing the initial pool of colonies collected.
Here, we opted for continuous transfers in LB batch cultures, but the competition experiment can also be performed in a different medium or under different conditions, including using a chemostat37. Furthermore, the procedure can be modified to accommodate any conditions of choice, including various stresses, such as oxidative, osmotic, or antibiotic stress. To verify the presence of persisting clones over time, one can do at least three things: WGS; transposon sequencing (Tn-Seq); or, as here, a Southern Blot. Finally, easy gene walking can be done on a few clones, with the further advantage that transposon insertion sites are identified in single clones, such that the effect of the specific insertion site can be phenotypically analyzed.
The choice of phenotypic assay to evaluate the outcome of a competition experiment depends on the investigated region. We used flow cytometry, which is a powerful method to measure cell-cycle parameters of bacteria. Here, flow cytometry revealed that the selected chromosomal position(s) of DARS2, (i.e., the DARS2 insertions close to the wildtype DARS2 position) resulted in the correct regulation of replication initiation (i.e., synchrony and DNA concentration). The optimal chromosomal position(s) for DARS2 were selected in part due the proper replication-associated gene dosage and in part due to the favorable local genomic environment that is important for DARS2 function4.
Here, a non-coding DNA element was investigated, but this could be expanded to any coding region of choice. A recent study found that the level of gene expression varied ~300-fold, depending on its position on the chromosome, without clear correlation to replication-associated gene dosage38. The approach described here could give important insight into the relationship between the genomic location of a particular gene, its transcriptional activity, and the associated fitness advantage. This could in theory assist with the selection of genomic positions, leading to the stronger expression of a given gene, which in turn could be of interest to engineering new, improved strains for recombinant protein production.
Because E. coli contains limited intergenic regions39, transposon insertions will, in most cases, disrupt gene(s). This may occasionally create false positives where the fittest clone(s) are not selected due to the optimal chromosomal position of the genetic element in question, but rather because a gene is disrupted – which, in other ways, provides a fitness advantage during the competition experiment4.
This methodology takes advantage of an easy-to-use transposon system, where any region of choice can be integrated at random locations. The designed competition experiment can be modified to include any number of selection forces other than pure growth, as seen here. The setup can also be modified to be purely based on Tn-Seq, which yields a greater sequencing resolution of insertions sites than WGS, used here. This unbiased approach should be used to elucidate exciting new features in thus-far uncharacterized organisms, which might show that common trends exist in the chromosomal organization of Eubacteria.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors were funded by grants from the Novo Nordisk Foundation, the Lundbeck Foundation, and the Danish National Research Foundation (DNRF120) through the Center for Bacterial Stress Response and Persistence (BASP).
Materials
| Autoclaved Mili-Q water | None | ||
| Electroporation Cuvettes, 0.1 cm | Thermo Fisher Scientific | P41050 | |
| Bio-Rad MicroPulser Electroporation System | Bio-Rad | 165-2100 | |
| LB Broth | Thermo Fisher Scientific | 12780029 | |
| LB Agar, powder (Lennox L agar) | Thermo Fisher Scientific | 22700025 | |
| Glycerol | Thermo Fisher Scientific | 17904 | |
| Fisherbrand Plastic Petri Dishes | Fisher Scientific | S33580A | |
| Falcon 50mL Conical Centrifuge Tubes | Fisher Scientific | 14-432-22 | |
| Falcon 15mL Conical Centrifuge Tubes | Fisher Scientific | 14-959-53A | |
| Nunc CryoTubes | Sigma-Aldrich | V7634 | |
| Phusion High-Fidelity DNA Polymerase (2 U/µL) | Thermo Fisher Scientific | F530S | |
| dATP, [α-32P]- 3000Ci/mmol 10mCi/ml, 250 µCi | PerkinElmer | BLU012H250UC | |
| DECAprime II DNA Labeling Kit | Thermo Fisher Scientific | AM1455 | |
| Spectrophotometer SF/MBV/03.32 | Pharmacia | ||
| Hermle Centrifuge SF/MBV/03.46 | Hermle | ||
| Ole Dich Centrifuge SF/MBV/03.29 | Ole Dich | ||
| Eppendorftubes 1.5 mL | Sigma-Aldrich | T9661 | |
| Eppendorftubes 2.0 mL | Sigma-Aldrich | T2795 | |
| Sodium Chloride | Merck | 6404 | |
| 96% Ethanol | Sigma-Aldrich | 16368 | |
| Trizma HCl | Sigma-Aldrich | T-3253 | |
| Phenol Ultra Pure | BRL | 5509UA | |
| Chloroform | Merck | 2445 | |
| Ribonuclease A type II A | Sigma-Aldrich | R5000 | |
| Sodiumdodecylsulphate (SDS) | Merck | 13760 | |
| Lysozyme | Sigma-Aldrich | L 6876 | |
| Isopropanol | Sigma-Aldrich | 405-7 | |
| 0.5M Na-EDTA pH 8.0 | BRL | 5575 UA | |
| Kanamycin sulfate | Sigma-Aldrich | 10106801001 | |
| PvuI (10 U/µL) | Thermo Fisher Scientific | ER0621 | |
| UltraPure Agarose | Thermo Fisher Scientific | 16500500 | |
| DNA Gel Loading Dye (6X) | Thermo Fisher Scientific | R0611 | |
| Tris-Borate-EDTA buffer | Sigma-Aldrich | T4415 | |
| Ethidium bromide | Sigma-Aldrich | E7637 | |
| Hydrochloric acid | Sigma-Aldrich | 433160 | |
| Sodium Hydroxide | Sigma-Aldrich | 71687 | |
| Whatman 3MM papers | Sigma-Aldrich | WHA3030931 | |
| SSC Buffer 20× Concentrate | Sigma-Aldrich | S6639 | |
| Amersham Hybond-N+ | GE Healthcare | RPN119B | |
| Ficoll 400 | Sigma-Aldrich | F8016 | |
| Polyvinylpyrrolidone | Sigma-Aldrich | PVP40 | |
| Bovine Serum Albumin – Fraction V | Sigma-Aldrich | 85040C | |
| Deoxyribonucleic acid sodium salt from salmon testes | Sigma-Aldrich | D1626 | |
| Carestream Kodak BioMax light film | Sigma-Aldrich | Z373494 | |
| GenElute Gel Extraction Kit | Sigma-Aldrich | NA1111 | |
| GenElute PCR Clean-Up Kit | Sigma-Aldrich | NA1020 | |
| T100 Thermal Cycler | Bio-Rad | ||
| SmartSpec Plus Spectrophotometer | Bio-Rad | ||
| Rifampicin | Serva | 34514.01 | |
| Cephalexin | Sigma-Aldrich | C4895 | |
| Mithramycin | Serva | 29803.02 | |
| Magnesium chloride hexahydrate | Sigma-Aldrich | 246964 | |
| Apogee A10 instrument | Apogee |
References
- Frimodt-Moller, J., Charbon, G., Lobner-Olesen, A. Control of bacterial chromosome replication by non-coding regions outside the origin. Curr Genet. , (2016).
- Sherratt, D. J., et al. Recombination and chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 359 (1441), 61-69 (2004).
- Bigot, S., et al. DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 24 (21), 3770-3780 (2005).
- Frimodt-Moller, J., Charbon, G., Krogfelt, K. A., Lobner-Olesen, A. DNA Replication Control Is Linked to Genomic Positioning of Control Regions in Escherichia coli. PLoS Genet. 12 (9), 1006286 (2016).
- Kornberg, A., Baker, T. A. . DNA Replication, Second Edition. , (1992).
- Kaguni, J. M. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol. 15 (5), 606-613 (2011).
- Leonard, A. C., Grimwade, J. E. Regulation of DnaA assembly and activity: taking directions from the genome. Annu Rev Microbiol. 65, 19-35 (2011).
- Kurokawa, K., Nishida, S., Emoto, A., Sekimizu, K., Katayama, T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18 (23), 6642-6652 (1999).
- Sekimizu, K., Bramhill, D., Kornberg, A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 50 (2), 259-265 (1987).
- Brendler, T., Austin, S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 18 (8), 2304-2310 (1999).
- Lu, M., Campbell, J. L., Boye, E., Kleckner, N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 77 (3), 413-426 (1994).
- Katayama, T., Kubota, T., Kurokawa, K., Crooke, E., Sekimizu, K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 94 (1), 61-71 (1998).
- Kato, J., Katayama, T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20 (15), 4253-4262 (2001).
- Kasho, K., Katayama, T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc Natl Acad Sci U S A. 110 (3), 936-941 (2013).
- Kitagawa, R., Ozaki, T., Moriya, S., Ogawa, T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12 (19), 3032-3043 (1998).
- Fujimitsu, K., Senriuchi, T., Katayama, T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23 (10), 1221-1233 (2009).
- Kasho, K., Fujimitsu, K., Matoba, T., Oshima, T., Katayama, T. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42 (21), 13134-13149 (2014).
- Frimodt-Moller, J., Charbon, G., Krogfelt, K. A., Lobner-Olesen, A. Control regions for chromosome replication are conserved with respect to sequence and location among Escherichia coli strains. Front Microbiol. 6, 1011 (2015).
- Bergthorsson, U., Ochman, H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 15 (1), 6-16 (1998).
- Boye, E., Steen, H. B., Skarstad, K. Flow cytometry of bacteria: a promising tool in experimental and clinical microbiology. J Gen Microbiol. 129 (4), 973-980 (1983).
- Rossignol, M., Basset, A., Espeli, O., Boccard, F. NKBOR, a mini-Tn10-based transposon for random insertion in the chromosome of Gram-negative bacteria and the rapid recovery of sequences flanking the insertion sites in Escherichia coli. Res Microbiol. 152 (5), 481-485 (2001).
- Shafferman, A., Helinski, D. R. Structural properties of the beta origin of replication of plasmid R6K. J Biol Chem. 258 (7), 4083-4090 (1983).
- Gonzales, M. F., Brooks, T., Pukatzki, S. U., Provenzano, D. Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J Vis Exp. (80), (2013).
- Smith, D. R. Random primed labeling of DNA. Methods Mol Biol. 18, 445-447 (1993).
- Lobner-Olesen, A., von Freiesleben, U. Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J. 15 (21), 5999-6008 (1996).
- Harrison, R. W., Miller, J. C., D’Souza, M. J., Kampo, G. Easy gene walking. Biotechniques. 22 (4), 650-653 (1997).
- Langmead, B., Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9 (4), 357-359 (2012).
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. Basic local alignment search tool. J Mol Biol. 215 (3), 403-410 (1990).
- Stepankiw, N., Kaidow, A., Boye, E., Bates, D. The right half of the Escherichia coli replication origin is not essential for viability, but facilitates multi-forked replication. Mol Microbiol. 74 (2), 467-479 (2009).
- Lobner-Olesen, A. Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J. 18 (6), 1712-1721 (1999).
- Lobner-Olesen, A., Skarstad, K., Hansen, F. G., von Meyenburg, K., Boye, E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 57 (5), 881-889 (1989).
- Carver, T., Thomson, N., Bleasby, A., Berriman, M., Parkhill, J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 25 (1), 119-120 (2009).
- Eckert, S. E., et al. Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J Bacteriol. 193 (7), 1771-1776 (2011).
- van Opijnen, T., Bodi, K. L., Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 6 (10), 767-772 (2009).
- van Opijnen, T., Camilli, A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 11 (7), 435-442 (2013).
- Jeong, H., Lee, S. J., Kim, P. Procedure for Adaptive Laboratory Evolution of Microorganisms Using a Chemostat. J Vis Exp. (115), (2016).
- Bryant, J. A., Sellars, L. E., Busby, S. J., Lee, D. J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 42 (18), 11383-11392 (2014).
- Blattner, F. R., et al. The complete genome sequence of Escherichia coli K-12. Science. 277 (5331), 1453-1462 (1997).

