Detection of Residual Donor Erythroid Progenitor Cells after Hematopoietic Stem Cell Transplantation for Patients with Hemoglobinopathies
Summary
Quantification of donor-derived cells is required to monitor engraftment after stem cell transplantation in patients with hemoglobinopathies. A combination of flow cytometry-based cell sorting, colony formation assay, and subsequent analysis of short tandem repeats may be used to assess the proliferation and differentiation of progenitors in the erythroid compartment.
Abstract
The presence of incomplete chimerism is noted in a large proportion of patients following bone marrow transplant for thalassemia major or sickle cell disease. This observation has tremendous implications, as subsequent therapeutic immunomodulation strategies can improve clinical outcome. Conventionally, polymerase chain reaction-based analysis of short tandem repeats is used to identify chimerism in donor-derived blood cells. However, this method is restricted to nucleated cells and cannot distinguish between dissociated single-cell lineages. We applied the analysis of short tandem repeats to flow cytometric-sorted hematopoietic progenitor cells and compared this with the analysis of short tandem repeats obtained from selected burst-forming unit – erythroid colonies, both collected from the bone marrow. With this method we are able to demonstrate the different proliferation and differentiation of donor cells in the erythroid compartment. This technique is eligible to complete current monitoring of chimerism in the stem cell transplant setting and thus may be applied in future clinical studies, stem cell research and design of gene therapy trials.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) is the only available curative approach for a variety of inborn genetic disorders of the hematopoietic system, achieving disease-free survival rates of more than 90% for otherwise highly compromised and life-limited patients1. The efficacy of this important therapeutic tool has been optimized by limiting the toxicity of pre-and post-transplant regimens2, but also by interventions aimed at sustaining stable graft function, which is quantified by close monitoring of donor-derived cells3,4,5.
In general, complete chimerism (CC) implies the total replacement of the lymphohematopoietic compartment by donor-derived cells, whereas the term mixed chimerism (MC) is used when donor- and recipient-derived cells are simultaneously present in various proportions. Split chimerism (SC) denotes the coexistence of mixed chimerism observed in single-cell lineages, such as in the erythroid compartment. Prompt determination of chimerism status following HSCT is critical, as it may help identify patients susceptible for disease relapse and initiate subsequent immunomodulatory strategies, such as donor lymphocyte infusions or reduction of immunosuppressive therapies6.
Several methods have been developed for monitoring engraftment after HSCT. Isotyping of immunoglobulins and analysis of cytogenetics have poor sensitivity and are limited in their ability to detect polymorphism7,8. The introduction of fluorescent in situ hybridization (FISH) can enhance sensitivity in chimerism monitoring after HSCT, but is restricted to sex-mismatched allografts9. Currently, polymerase chain reaction (PCR) is the most widespread method used to detect chimerism and is based on conventional agarose-acrylamide gel electrophoresis of variable number tandem repeats (VNTRs) or short tandem repeats (STRs). Routinely used quantitative PCR is able to detect an extremely small proportion of residual donor cells following HSCT. The major limitation of the studies so far is that MC detection is almost exclusively limited to the presence of nucleated cells, rather than mature erythrocytes, namely cells that are functionally crucial for patients affected by hemoglobinopathies. In patients with different blood groups, it is worth remembering that cytofluorometric analysis is able to identify chimerism in red blood cells by utilizing monoclonal antibodies directed towards the erythrocyte antigens ABO and C, c, D, E, and e10,11. A different, but very interesting means of assessing chimerism in the erythroid lineage is the combination of flow cytometric sorting of erythroid progenitors and selection of various erythroid progenitor types by culturing in clonogenic assays, followed by analysis of STR12. This approach is able to quantify relative proportions of donor-versus-recipient chimerism in the erythroid compartment and may be utilized in the strategy to sustain the bone marrow graft.
Protocol
1. Isolation of Hematopoietic Bone Marrow Cells by Multi-parameter Fluorescence-activated Cell Sorting
- Sample staining
- Before starting the process, the following items need to be collected and prepared:
- Gradient media for the preparation of mononuclear cells (GM), 50 mL conical tubes
- 12 x 75 mm flow tubes for staining cells
- Staining buffer: phosphate buffered saline (PBS) + 3% fetal calf serum (FCS)
- Pipettes
- FC block and staining antibodies (CD34 APC, CD36 FITC, CD45 V450, BD Biosciences). With this fluorochrome combination there is negligible spillover into other channels, but any other suitable fluorochrome combination is also possible.
- Compensation beads (if necessary)
- Suspension buffer: Hank's Balanced Salt Solution (HBSS) + 25 mM HEPES + 3% FCS
- Trypan blue and hemocytometer
- Collection tubes: any rich medium with high serum tubes (containing 1 mL FCS + 25 mM HEPES) can be used for the collection of sorted cells.
- Bone marrow sample: informed consent for bone marrow biopsy from the iliac crest of the back of the hip bone is typically required. The bone marrow sample is than kept in heparinized syringes at RT until staining. The following protocol steps are performed in compliance with the guidelines of the institution's human research ethics committee for human welfare.
- Generate a single cell suspension of the bone marrow sample. Add 3 mL of GM to the centrifuge tube. Carefully layer the 4 mL of the single cell suspension onto the GM solution. Centrifuge at 550 g for 30 min at 20 °Celsius (C) (brake is turned off). Draw of the upper layer containing plasma and platelets using a sterile pipette, leaving the mononuclear cell layer undisturbed at the interface. Transfer the layer of mononuclear cells to a sterile centrifuge tube using a sterile pipette.
- After washing with 5 mL staining buffer, resuspend the cells in 2 mL suspension buffer and determine the cell concentration. Place 5 µL of cell suspension in a screw cap test tube. Add 95 µL of 0.2% Trypan blue stain. Mix thoroughly. Allow to stand for 5 min at room temperature. Fill a hemocytometer as for cell counting. Under a microscope, observe if non-viable are stained and count the viable cells.
- Centrifuge the cells at 250 g for 10 min at 20 °C, discard the supernatant and resuspend the cells in staining buffer at a concentration of up to 1 x 106 per 100 µL.
- Block FcR by the use of 2.5 µg Fc Block per 1 x 106 cells on ice for 10 – 15 min.
- Add the appropriate combination of 10 µg mAb to stain 1 x 106 cells and incubate for 20 min at 4° C in the dark, followed by a washing step with 3 mL staining buffer. For correct compensation, small aliquots of the cells (or preferably compensation beads) are stained with single antibodies each; one aliquot remaining unstained.
- Adjust the concentration to 20 x 106 cells/mL.
- Set up and optimize the cell sorter. The process of setting up a flow cytometer and software is standardized with a detailed instruction provided by the company and needs to be performed by appropriately trained personnel.
- Before starting the process, the following items need to be collected and prepared:
- Sorting on the cell sorter
- Prepare collection tubes from step 1.1.1.9.
- Set up a template that includes a bivariate plot to display forward scatter (FSC) and side scatter (SSC).
- Run the cells and adjust FSC/SSC to place the population of interest on scale
- Record the negative control tube.
- Run the single positive control tubes; record the data for each tube.
- Calculate compensation either manually or automatically with the function provided by the acquisition software.
- Acquire the experimental sample and gate them on a bivariate FSC/SSC dot plot (Figure 1A) to include both lymphocytic and myeloid cells. Exclude doublets and aggregates by comparing the different signals of FSC (i.e., height, width, and area) (Figure 1B). Visualize the singlet cells on a bivariate dot plot displaying CD45 and CD36 (Figure 1C). Events positive for CD45 are leucocytes (including their progenitors), those negative for CD45 and positive for CD36 are erythroid progenitors. Use gating tools to define CD36+ (if desired, these cells can be further divided in CD36 high and low expressing cells) and CD45+ cells. Visualize CD45+ events in another dot plot displaying CD34 and SSC parameters. Use gating tools to define CD34+ cells (leukocyte progenitors) and SSC high cells (myeloid cells at various stages of differentiation).
- Once gates have been determined, the gates can be selected for sorting into external collection tubes.
- Proceed with sorting at 4 °C until the required number of cells has been obtained
- Perform a post-sort analysis to determine the purity of the sorted cell populations (Figure 1E, 1F).
2. Clonogenic Assay
- Preparation of reagents
Before starting the process, the following items need to be collected and prepared:- Pipettes, 100 mm plates
- Trypan blue and hemocytometer
- Reconstitute human recombinant erythropoietin (EPO) at 500 U/mL in sterile PBS containing at least 0.1% human serum albumin. Divide this solution into small aliquots and keep at -20 °C to avoid repeated freeze-thaw.
- Prepare complete Iscove's Modified Dulbecco's Medium (IMDM) with final concentrations of 5% FCS, 2 mM Glutamine, 100 units/mL Penicillin/Streptomycin (PS). Add human recombinant erythropoietin (EPO) at different concentrations in separate wells to the complete IMDM medium: 3 units EPO/well, 6 units EPO/well and 12 EPO units/well (1 x, 2 x, 4 x EPO respectively).
- Preparation of bone marrow cells
- 10 mL of the bone marrow sample diluted with 20 mL PBS are slowly layered on top of 15 mL density gradient medium in a 50 mL conical tube, maintaining clear interface between the two phases and under sterile conditions. Then centrifuge the 15 mL conical tubes at 550 x g for 30 min at room temperature (RT) with acceleration set as 9 and deceleration set as 0 (or Brake OFF).
- After centrifugation, remove the interface into a new 50 mL tube and add PBS to a final volume of 50 mL.
- After centrifugation with 250 g for 10 min at 10 °C remove the supernatant and resuspend the cells in complete IMDM medium at approximately 0.5 x 106 cells/mL. Take 10 µL of the cell suspension into a microtube, mix with the same volume of Trypan blue solution (0.4%) and count using a hemocytometer.
- Optional: CD34+ cells are enriched by magnetic cell sorting with Milteny CD34 beads according to the instructions given by the manufacture. The main advantage of this enrichment step is to detect the quality of hematopoietic stem cells, which are grown on a semi-solid matrix added with 50 ng/mL stem cell factor (SCF), 20 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF), 20 ng/mL interleukin-3 (IL-3), 20 ng/mL interleukin-6 (IL-6), 20 ng/mL granulocyte colony-stimulating factor (G-CSF), and 3 different concentrations of EPO as in 1.4.
- Dilute the cells to 0.1 – 0.15 x 106 mononuclear cells/mL or 2 x 103 CD34+ cells/mL by adding complete IMDM medium supplemented with EPO and transfer 300 µL to 3 mL Methocult H4434 medium. After agitation and a short incubation for 5 min plate three times 1,1 mL on a 35 mm dish. Two of these culture dishes together with a third – filled with water – are placed in 100 mm plates.
- Cells are cultured in a humidified 37 °C incubator with 5% CO2 for 14 days.
- Analysis of colonies
- Colonies are scored according to their morphology with an inverted microscope at 40X magnification in a culture dish marked with a scoring grid. For the purposes of our assay, the colony forming units (CFU) are classified into 4 categories: multipotential progenitor cells (CFU-GEMM), granulocyte-macrophage progenitor cells (CFU-GM), burst forming unit-erythroid (BFU-E) and colony forming unit-erythroid (CFU-E). See manufacturer's instructions for further colony subclassification.
- For further analysis, cells from the CFU assay plate are recovered separately according to the 4 categories by suspending in 4 mL room temperature PBS containing 2% FCS/IMDM. After centrifugation at 400 g for 10 min at 4 °C, the cells are resuspended in PBS, counted, and processed for DNA extraction.
3. Analysis of Chimerism
- DNA Isolation
- Pellet the cells by centrifugation at 400 g for 10 min.
- Isolate DNA using a blood DNA extraction kit (QIAmp DNA Blood extraction kit). Pre-warm the elution buffer (AE) at 37 °C to enhance the yield of eluted DNA.
- Measure the DNA concentration with Nanodrop ND-1000 spectra photometer. Samples with OD 260/280 between 1.8 – 2.0 are used for further analysis.
- Dilute DNA with DNAase free H2O to 0.1 ng/µL in a total volume of 50 µL.
- STR-Analysis
- Set up the PCR reaction by mixing Reaction Mix, AmplTaq Gold DNA Polymerase and Profiler Plus Primer Set with 20 µL genomic DNA (0.1 ng/µL) according to the manual (The primer kit contains the unlabeled primers in buffer to amplify the STR loci D3S1358, vWA, FGA, D8S1179, D21S11, D18S51, D5S818, D13S317, and D7S820, and the gender marker amelogenin). Include nuclease-free water as negative control and 9947A as positive control. Pre-transplant DNA from both donor and recipient are also included.
- Perform PCR reaction using Eppendorf mastercycler gradient with the following conditions: 95 °C for 11 min, 28 amplification cycles of 95 °C for 1 min, 59 °C for 1 min and 72 °C for 1 min, followed by 60° C for 45 min.
- Set up fragment analysis reaction by adding 12 µL Hi-Di Formamid and 0.5 µL GeneScan 500 ROX Size Standard (all Applied Biosystems) to 1 µL of PCR product. At first and last positon an Allelic Ladder (Genotyper AmpFlSTR Blue, Green II and Yellow, Applied Biosystems) is needed.
- Start electrophoresis and acquisition with the electrophoresis instrument.
- Analyze Loci, alleles and peak heights of samples and controls with the software13.
Representative Results
Separation of lymphohematopoietic progenitors by FACS cell sorting
We here demonstrate results from sorting the necessary cell populations for downstream STR analysis. Bone marrow cells were stained with V450-conjugated anti-CD45, FITC-conjugated anti-CD36 and APC-conjugated anti-CD34. The population of interest is the megakaryocyte erythroid progenitors (MEP), nucleated cells responsible for the development of erythrocytes. These cells express CD36, but are negative for the leukocyte common antigen CD45 (Figure 1C). If necessary, these cells can be further differentiated based on their CD36 expression level. Several additional populations can be sorted to serve as controls. We sorted CD45+CD34+ lymphoid/myeloid precursors as another progenitor cell population and CD45+ cells with high SSC signal as mature myeloid cells (Figure 1D). Sorting was performed with a BD FACSAria I instrument. After sorting, erythroid progenitor cell purity was >85% (Figure 1E) and myeloid cell purity was >95% (Figure 1F).
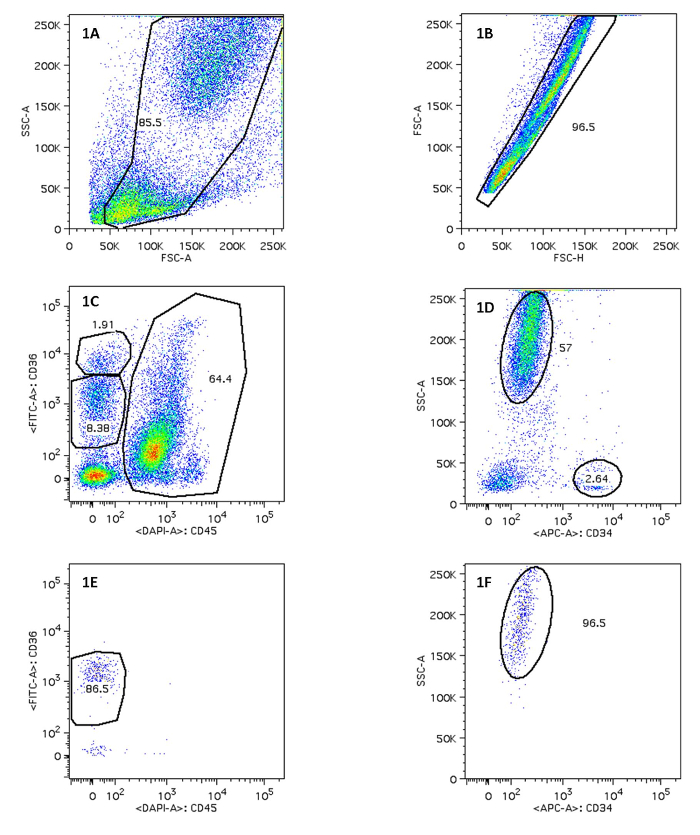
Figure 1. Sorting of Erythroid Progenitor Cells and Myeloid Progenitor Cells After FACS. Figure 1A/1B displays on FSC-A vs. SSC-A, FSC-H vs. FSC-A and the use of a polygon gate tool to select the population of interest. 1C indicates MEP on CD45 vs. CD36; 1D displays mature myeloid cells. The percentage of positive cells in 1E and 1F is shown. Please click here to view a larger version of this figure.
Generation of burst-forming units – erythroid colonies
Some cells derived from bone marrow and separated by CD34-specific magnetic beads appear to proliferate and differentiate. After a 14-day incubation period on the semi-solid medium colonies are formed. Stimulation with various concentrations of EPO greatly augmented the formation of prominent red (erythroid) colonies (Figure 2).
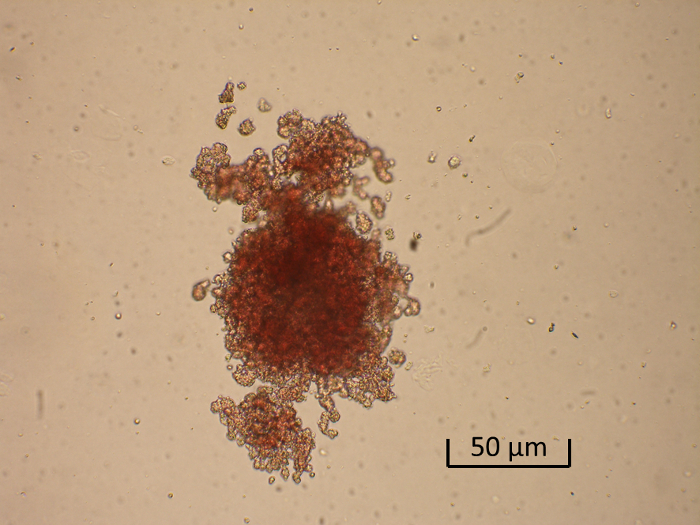
Figure 2. Generation of a Burst-forming Unit-erythroid (BFU-E). Shown is a representative low-power photomicrograph of a BFU-E colony. Please click here to view a larger version of this figure.
| MNC: | |||
| 1X EPO | 2X EPO | 4X EPO | |
| CFU-E | 2 | 2 | 2 |
| BFU-E | 9 | 10 | 8 |
| CFU-GM | 30 | 30 | 29 |
| CFU-GEMM | 1 | 1 | 2 |
| CD34+ cells: | |||
| 1X EPO | 2X EPO | 4X EPO | |
| CFU-E | 1 | 1 | 0 |
| BFU-E | 5 | 11 | 6 |
| CFU-GM | 25 | 26 | 32 |
| CFU-GEMM | 0 | 2 | 3 |
Table 1. Analysis of Colonies in Unsorted Bone Marrow Cells and CD34+ Selected Cells. Colonies are scored according to their morphology with an inverted microscope at 40X magnification in a culture dish marked with a scoring grid. For the purposes of our assay, the colonies are classified in four categories: colony-forming unit-erythroid (CFU-E), burst-forming unit-erythroid (BFU-E), granulocyte-macrophage progenitor cells (CFU-GM) and multipotential progenitor cells (CFU-GEMM).
Analysis of chimerism
Chimerism analysis is performed using the AmpFlSTR Profiler Plus Kit (Applied Biosystems, California) according to the manufacturer's protocol on ABI Prism 310 Genetic Analyzer (Applied Biosystems, California). Analysis is performed with GeneMapper Software (Applied Biosystems, California). Loci D21S11, D7S820, FGA and vWA were used to calculate results (percentage of recipient- and donor-specific DNA).
| Sample | Recipient DNA | Donor DNA |
| CD45 | 25% | 75% |
| CD36hi | 25% | 75% |
| CD36lo | 55% | 45% |
| CD34 | 25% | 75% |
Table 2. Percent Recipient and Donor DNA in Cells Obtained from Fluorescence-activated Cell Sorting of Bone Marrow Cells. Chimerism of donor cells in specific cellular lineages of the lymphohematopoietic compartment is given.
| Sample | Recipient DNA | Donor DNA |
| BM-MNC | 24.71% | 75.29% |
| BM-CD34 | 22.62% | 77.38% |
| BFU-E (BM-MNC) | 80.99% | 19.01% |
| CFU-GM (BM-MNC) | 0.00% | 100.00% |
| CFU-GEMM (BM-MNC) | 5.73% | 94.27% |
| BFU-E (BM CD34) | 30.13% | 69.87% |
| CFU-GM (BM CD34) | 0.00% | 100.00% |
| CFU-GEMM (BM CD34) | 9.87% | 90.13% |
Table 3. Percent Recipient and Donor DNA in Cells Obtained from the Clonogenic Assay. Chimerism of donor cells in specific cellular lineages of the lymphohematopoietic compartment is given.
Discussion
The objective of the current study is to provide the audience a combination of two approaches for analyzing donor/recipient chimerism in erythroid progenitors following HSCT in patients treated for hemoglobinopathies: 1.) fluorescence-activated cell sorting of hematopoietic progenitor cells in bone marrow samples followed by analysis of short tandem repeats and 2.) colony-forming unit growing of bone marrow cells, classification of colonies in various progenitor types followed by analysis of short tandem repeats. The novelty of this approach lies in combining the techniques in a protocol to evaluate donor/recipient chimerism in individual colonies.
The particular importance of this approach can be found in the context of mixed chimerism after HSCT for patients treated for hemoglobinopathies. Several studies have demonstrated that a significant proportion of patients express a low percentage of donor myeloid cells that correlates with that of erythroid precursor cells und results in long-lasting stable mixed hematopoietic chimerism3,11; in contrast, in the same patients a high percentage of donor-derived red blood cells (2- to 5-fold higher than that of mature leukocytes) is noted and suggests that the ineffective erythropoiesis takes place at a later stage of erythroid development. These observations have been attributed to the propensity for accelerated apoptosis in donor erythroblasts and the persistence of T and B residual lymphocytes responsible for allowing a mixed allograft14,15. In the context of immunomodulation following hematopoietic stem cell transplant we recently demonstrated that modification of immunosuppressive therapy after hematopoietic transplantation for ß-thalassemia results in a selective advantage for the genetically-corrected erythroid compartment, giving a 2- to 2.5-fold amplification of the residual donor stem cells12. This observation supports the utility of determining donor-versus-residual erythroid progenitors, as it provides an understanding of this phenomenon and supports future clinical trials studying gene therapy for the treatment of hemoglobinopathies. In fact, the proportion of gene-modified nucleated cells needed to achieve a therapeutic level of circulating red blood cells might be similar to that observed in patients treated with stem cell transplantation for hemoglobinopathies.
In order to determine the full picture of donor erythropoiesis we modified the protocol and provide a detailed, step-by-step experimental approach. The critical step in this protocol is the process of setting up a flow cytometer and software, which is standardized with detailed instructions and must be performed by appropriately trained personnel. Presentation of our protocol in the visualized format allows the audience to follow our protocol easily. We compared our PCR results using genomic DNA from erythroid progenitors obtained from colony-forming units in BM samples versus DNA obtained from FACS-sorted erythroid bone marrow progenitors. The donor engraftment data from the two approaches are basically consistent. However, usually less variation was found in samples obtained from colony-forming units. This is likely due to the improved survival of donor red blood cell precursors in the in vitro assay as compared to the untreated and sorted donor counterparts. In this light, this protocol can be expanded by artificially creating mixed chimeric combinations with known proportions of healthy progenitors and progenitors from a range of patients with various hemoglobinopathies. The clonogenic assay and assessment of chimerism should accurately reflect in-put proportions.
Although a precise understanding of the mechanisms involved in this particular setting is lacking, the method presented here is of paramount importance for providing relevant information for the routine monitoring of engraftment and prognostic information in clinical practice. Attempts to rescue graft function are limited by the risk of developing graft-versus-host disease and can be guided by serial engraftment monitoring. Finally, this method might also provide a useful basis for research areas committed to improving the care of patients with hemoglobinopathies, particularly those affected by acute graft-versus-host disease and autoimmune disease.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Kinderkrebshilfe Regenbogen Südtirol.
Materials
| Ficoll-Paque | GE Healthcare | GE17-1440-02 | Remove RBC |
| 50 mL conical tubes | Falcon | 14-432-22 | Sample preparation |
| 12 x 75 mm flow tubes | Falcon | 352002 | FACS sorting |
| Phosphate buffered saline | Gibco | 10010023 | PBS |
| Fetal calf serum | Invitrogen Inc. | 16000-044 | FCS (heat-inactivated) |
| CD34 APC | BD Bioscience | 561209 | FACS-Ab |
| CD36 FITC | BD Bioscience | 555454 | FACS-Ab |
| CD45 V450 | BD Bioscience | 642275 | FACS-Ab |
| Trypan blue | Gibco | 15250061 | |
| Hemocytometer | Invitrogen Inc. | C10227 | Automatic cell counting |
| Hank’s Balanced Salt Solution | Gibco | 14025092 | Suspension buffer in FACS analysis |
| HEPES | Gibco | 15630080 | Component of suspension buffer |
| FcR | BD Bioscience | 564220 | Block FCR |
| FACS Aria I | BD Bioscience | 23-11539-00 | FACS Sorter |
| Recombinant human erythropoietin | Affimetrix eBioscience | 14-8992-80 | EPO |
| Isocove’s Modified Dulbecco’s Medium | Gibco | 12440053 | IMDM |
| L-Glutamine | Invitrogen | 25030-081 | Component of Culture Medium |
| CD34+ magnetic beads | Milteny Biotech | 130-046-702 | CD34+ purification |
| Recombinant human G-CSF | Gibco | PHC2031 | CFU-Assay |
| Recombinant human SCF | Gibco | CTP2113 | CFU-Assay |
| Recombinant human GM-CSF | Gibco | PHC2015 | CFU-Assay |
| Recombinant human IL-3 | BD Bioscience | 554604 | CFU-Assay |
| Recombinant human IL-6 | BD Bioscience | 550071 | CFU-Assay |
| Methocult H4434 Medium | Stemcell Technologies | 4444 | CFU-Assay |
| QiAmp DNA Blood extraction kit | Qiagen | 51306 | DNA Isolation |
| Nanodrop ND-1000 spectra photometer | Thermo Scientific | ND 1000 | DNA Quantification |
| DNAase free H2O | Thermo Scientific | FEREN0521 | DNA Preparation |
| AmplTaq Gold DNA Polymerase | Applied Bioscience | N8080240 | PCR |
| Eppendorf mastercycler gradient | Eppendorf | 6321000019 | PCR |
| Hi-Di Formamid | Applied Bioscience | 4311320 | PCR |
| GeneScan 500 ROX Size Standard | Applied Bioscience | 4310361 | PCR |
| 3130 Genetic Analyzer | Applied Bioscience | 313001R | PCR |
References
- Angelucci, E., et al. Hematopoietic stem cell transplantation in thalassemia major and sickle cell disease: indications and management recommendations from an international expert panel. Haematologica. 99, 811-820 (2014).
- Lucarelli, G., Isgro, A., Sodani, P., Gaziev, J. Hematopoietic stem cell transplantation in thalassemia and sickle cell anemia. Cold Spring Harb Perspect Med. 2, a011825 (2012).
- Andreani, M., Testi, M., Lucarelli, G. Mixed chimerism in haemoglobinopathies: from risk of graft rejection to immune tolerance. Tissue Antigens. 83, 137-146 (2014).
- Andreani, M., et al. Persistence of mixed chimerism in patients transplanted for the treatment of thalassemia. Blood. 87, 3494-3499 (1996).
- Andreani, M., et al. Long-term survival of ex-thalassemic patients with persistent mixed chimerism after bone marrow transplantation. Bone Marrow Transplant. 25, 401-404 (2000).
- Kumar, A. J., et al. Pilot study of prophylactic ex vivo costimulated donor leukocyte infusion after reduced-intensity conditioned allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 19, 1094-1101 (2013).
- Lawler, S. D., Harris, H., Millar, J., Barrett, A., Powles, R. L. Cytogenetic follow-up studies of recipients of T-cell depleted allogeneic bone marrow. Br J Haematol. 65, 143-150 (1987).
- McCann, S. R., Crampe, M., Molloy, K., Lawler, M. Hemopoietic chimerism following stem cell transplantation. Transfus Apher Sci. 32, 55-61 (2005).
- Dewald, G., et al. A multicenter investigation with interphase fluorescence in situ hybridization using X- and Y-chromosome probes. Am J Med Genet. 76, 318-326 (1998).
- Andreani, M., et al. Quantitatively different red cell/nucleated cell chimerism in patients with long-term, persistent hematopoietic mixed chimerism after bone marrow transplantation for thalassemia major or sickle cell disease. Haematologica. 96, 128-133 (2011).
- Andreani, M., Testi, M., Battarra, M., Lucarelli, G. Split chimerism between nucleated and red blood cells after bone marrow transplantation for haemoglobinopathies. Chimerism. 2, 21-22 (2011).
- Kropshofer, G., Sopper, S., Steurer, M., Schwinger, W., Crazzolara, R. Successful management of mixed chimerism after bone marrow transplant in beta-thalassemia major. Am J Hematol. 91, E357-E358 (2016).
- Kimpton, C. P., et al. Automated DNA profiling employing multiplex amplification of short tandem repeat loci. PCR Methods Appl. 3, 13-22 (1993).
- Centis, F., et al. The importance of erythroid expansion in determining the extent of apoptosis in erythroid precursors in patients with beta-thalassemia major. Blood. 96, 3624-3629 (2000).
- Miccio, A., et al. In vivo selection of genetically modified erythroblastic progenitors leads to long-term correction of beta-thalassemia. Proc Natl Acad Sci U.S.A. 105, 10547-10552 (2008).

