Visualization of Biofilm Formation in Candida albicans Using an Automated Microfluidic Device
Summary
This protocol describes the use of a customizable automated microfluidic device to visualize biofilm formation in Candida albicans under host physiological conditions.
Abstract
Candida albicans is the most common fungal pathogen of humans, causing about 15% of hospital-acquired sepsis cases. A major virulence attribute of C. albicans is its ability to form biofilms, structured communities of cells attached to biotic and abiotic surfaces. C. albicans biofilms can form on host tissues, such as mucosal layers, and on medical devices, such as catheters, pacemakers, dentures, and joint prostheses. Biofilms pose significant clinical challenges because they are highly resistant to physical and chemical perturbations, and can act as reservoirs to seed disseminated infections. Various in vitro assays have been utilized to study C. albicans biofilm formation, such as microtiter plate assays, dry weight measurements, cell viability assays, and confocal scanning laser microscopy. All of these assays are single end-point assays, where biofilm formation is assessed at a specific time point. Here, we describe a protocol to study biofilm formation in real-time using an automated microfluidic device under laminar flow conditions. This method allows for the observation of biofilm formation as the biofilm develops over time, using customizable conditions that mimic those of the host, such as those encountered in vascular catheters. This protocol can be used to assess the biofilm defects of genetic mutants as well as the inhibitory effects of antimicrobial agents on biofilm development in real-time.
Introduction
Candida albicans is a commensal member of the human microbiota, however it is also an opportunistic pathogen, capable of causing superficial and severe fungal infections1,2. A major virulence trait of C. albicans is its ability to form resilient and drug resistant biofilms, communities of cells adhered to a surface and enclosed in an extracellular matrix material1,3. C. albicans biofilms are highly structured, containing several layers of multiple cell types (round budding yeast-form cells, oval pseudohyphal cells, and tubular hyphal cells)4. C. albicans biofilm development begins with the adherence of round yeast-form cells to a surface (seeding the biofilm), followed by the proliferation of these cells on the surface, and then the maturation of the immature biofilm structure into a fully-formed biofilm that is surrounded by extracellular matrix material4. The mature biofilm is predominantly composed of elongated hyphal cells that form dense and interconnecting networks, providing the architectural stability to the biofilm4. Throughout the biofilm life cycle, round budding yeast cells disperse from the mature biofilm, and may travel to other regions of the body to cause disseminated infections or seed new biofilms at other sites4,5. C. albicans can form biofilms on biotic surfaces, such as mucosal surfaces and throughout host tissue, and on abiotic surfaces, such as catheters, pacemakers, dentures, and prosthetic joints. Due to the recalcitrant properties of biofilms, they are extremely difficult to eradicate, and in many cases the only effective treatment strategy is removal of the infected device4. It is thus crucial to investigate biofilm formation under conditions similar to those observed in clinical settings.
There are several critical in vivo animal models used to study C. albicans biofilm formation6,7,8; however, these studies can be costly, time consuming, and are limited by the number of strains and antimicrobial agents that can be tested at a given time. In vitro biofilm assays, on the other hand, allow for the rapid, high-throughput assessment of antifungal compounds and mutant strains, and are much more cost-effective and ethical than biofilm assays carried out in animal models9,10,11,12,13,14. Here we describe an in vitro assay that we developed and optimized to observe biofilm formation temporally under laminar flow using a customizable microfluidic device14,15. The assay allows for the visualization of each stage of biofilm formation, including the initial adherence step, cell proliferation, biofilm maturation, and cell dispersion. The assay is also useful to visualize cell morphology changes throughout the development of a biofilm.
Microtiter plates, which are typically utilized for in vitro biofilm assays, while high throughput, do not allow for controlled flow conditions. Traditional laminar flow cell systems allow for the continuous assessment of biofilm formation in controlled flow conditions, but these are often time consuming to set up and tend to have limited dynamic range control and throughput. The microfluidic device utilized here overcomes these limitations by combining high throughput plates (containing 48 wells) with a built-in laminar flow chamber and is highly reproducible, versatile, and customizable.
Here, we describe a protocol for the use of a commercially available automated microfluidic device to assess biofilm formation of a wild-type C. albicans strain, the effects of a known antifungal agent on the development of a biofilm, and biofilm formation in two mutant strains (bcr1Δ/Δ and efg1 Δ/Δ) that were previously reported to have biofilm defects in vitro and in vivo16,17,18. The described protocol can be used to test the efficacy of antimicrobial agents in inhibiting biofilm formation throughout the development of a biofilm, and to identify genes required for normal biofilm development by screening mutant libraries.
Protocol
1. Fungal Cell Culture Preparation
NOTE: Conduct cell culture work (i.e. opening cryogenic stock tubes, cell culture tubes, and flasks) within a biosafety cabinet. Turn on the cabinet's ultraviolet (UV) germicidal lamp at least 1 h prior to work, and turn off the UV lamp while actively working in the cabinet. Wear gloves, safety glasses, and appropriate personal protective equipment, and decontaminate the surface of the bench and pipettes with 70% ethanol prior to the start of the experiment. Use of sterile filter tips and familiarity with basic aseptic microbiological techniques are recommended.
- Streak C. albicans strains (wild-type, bcr1 Δ/Δ, and efg1 Δ/Δ) on yeast extract peptone dextrose (YPD) medium (1% yeast extract, 2% peptone, 2% glucose; pH 6.8) agar plates. Incubate at 30 °C for 48 h, following standard microbiological practices19.
- Select a single isolated colony from the plate to inoculate into 4 mL of YPD liquid medium (1% yeast extract, 2% peptone, 2% glucose; pH 6.8) and incubate at 30 °C with shaking at 225 rpm in a standard shaking incubator for 12 – 15 h, following standard microbiological practices19.
- Determine the cell density of the culture by measuring optical density (OD) at 600 nm in a cuvette (1 mL, 1 cm path length), following standard microbiological practices19.
- Dilute the cell culture to a final OD600 of 0.5, equivalent to 2 x 107 cells/mL, in Roswell Park Memorial Institute (RPMI)-1640 medium (containing 165 mM 3-morpholinopropane-1-sulfonic acid (MOPS), L-Glutamine, and without sodium bicarbonate, pH 7.0) or Spider medium (1% nutrient broth, 1% mannitol, 0.4% K2HPO4, pH 7.2).
NOTE: Alternate media can be used to support the specific growth of strains of interest.
NOTE: Do not make cell dilutions too far in advance. It is recommended to begin dilutions after step 3.3 (described below) in order to prevent the initiation of hyphae formation prior to cells being seeded in the viewing channel.
2. Preparation of Microfluidic Channels of the Microfluidic Plate
NOTE: Refer to user manual of microfluidic system (see table of materials) for information on plates and instrument setup.
- Prior to running the microfluidic experiment, warm 20 mL of RPMI-1640 media or Spider media in an incubator at 37 °C. Additional media volume may be required based on calculations in step 2.8. Pre-warming media prevents the formation of air bubbles during the experiment.
- Turn on the microfluidics system which includes the controller, the two power supply units, the microscope, the camera, and computer attached to the system. Open the software that controls the system (see table of materials) from the program menu on the computer. Two windows will appear once the program is opened.
- Locate the bar code number of the plate in use and input the number when prompted by the software. Use two separate computer screens to view the software on the two windows. One window (control module) contains the controls for the plate and the second window (Montage, imaging module) contains the controls for the microscope and camera.
- Switch on the heater power button on the controller and set the microfluidics system temperature to 37 °C using the arrow buttons on the temperature controller.
- Clean the interface plate (Figure 1) using sterile water as follows. Spray the bottom of the plate with sterile water and use lens paper to remove dirt/dust. Rest the interface plate face down on clean lens paper to air dry. Clean the top of the interface plate using 70% isopropanol and lens paper.
NOTE: The interface plate connects the microfluidic system to the plate that will contain the cells and media. Ensure that no liquid remains and that all fibers and dirt are removed. Improper cleaning may result in low quality images and videos. - Remove condensation from the tubes on the interface plate.
- Leave the plate face down resting on the lens paper. In the control module window click on the manual mode for the plate. In the shear control menu section, click on the fluid selection for columns 1 – 4 and 5 – 8, and select 37 °C from the drop-down menu. In the shear control section, set the flow mode to constant and set the max shear to 2 dyne/cm2 (0.2 Pa) (Figure 2A).
- Click on the inlet wells to start the flow of sterile air through the interface plate tubes to remove condensation. After 5 min, click on the stop button in the well plate control section (Figure 2A). Observe the tubes in the interface plate to confirm that all condensation has been removed. If droplets are still visible, repeat the flow of sterile air, as mentioned above, for another 5 min.
NOTE: The inlet tubes are attached to 0.22 micron filters to ensure only sterile air is introduced to the microfluidics system. The flow can be monitored by the event log on the screen in the control module window.
- Place the 48-well 0-20 dyne microfluidic plate on the microscope stage.
3. Candida albicans Biofilm Formation in the Microfluidic System
- To record biofilm formation, add 600 µL of pre-warmed RPMI-1640 medium or spider medium to the inlet wells (see Figure 1 for plate layout). Have two replicates for each experimental condition.
- For testing biofilm formation of C. albicans wild-type and mutant strains, add 600 µL Spider medium to inlet wells of the experiment plate.
- For testing biofilm formation in the presence of antifungal drugs (or other compounds of interest), add 600 µL of RPMI-1640 medium to two wells (negative control) and 600 µL of RPMI-1640 medium supplemented with amphotericin B (16 µg/mL) (or drug of choice at desired concentration) to two other inlet wells.
NOTE: For a 12 h biofilm formation experiment, add 600 µL of pre-warmed media to the inlet wells. For longer experiments add additional media (50 µL/h) to the inlet wells. Do not exceed the maximum volume of the well (1,500 µL).
- Slowly lower the cleaned interface plate on top of the 48 well microfluidic plate. Slide the interface plate slightly to the left until the tubes on the interphase plate are positioned on top of the inlet and outlet wells. Turn the lever on the interface plate from left to right to lock the interface plate in position, ensuring the experiment is airtight.
NOTE: Flow of sterile air from the tubes in the interface plate will push the liquid in the inlet or outlet wells (depending on the direction selected using the software on the computer), so that the liquid flows through the viewing channel between the inlet and outlet wells in the customized direction and speed dictated by the user. - In the shear control section, set the flow mode to constant and set the max shear to 1 dyne/cm2 (0.1 Pa). Click on inlet wells with media to begin the flow of media from inlet to outlet wells (Figure 2A) to prime the channel. After 5 min click on the stop button in the well plate control section. Remove the interface plate and observe the microfluidic plate wells. After priming the channels, only a small drop of media should be visible in the outlet wells. If the drop is not visible, prime the channels in 2 min increments until the small drop of media in the outlet wells is visible.
NOTE: Priming is required to remove air from the viewing channels and ensure that the channels are filled with the desired media. - Remove the interface plate from the microfluidic plate and set it aside on a sterile surface. Add 50 µL of cell culture (described in step 1.5) into each outlet well.
- For testing C. albicans wild-type and mutant strains, add 50 µL of wild-type (SC5314) cell culture in Spider media to two outlet wells, bcr1 Δ/Δ in Spider media to two outlet wells, and efg1 Δ/Δ in Spider media to two outlet wells. Place the interface plate back onto the microfluidic plate and lock the lid (see Figure 1 for plate layout).
NOTE: As described in step 3.1.1, all corresponding input wells contain Spider media. - For testing C. albicans biofilm formation in the presence of amphotericin B or other compound of interest, add 50 µL of wild-type (SC5314) cell culture in RPMI-1640 medium to four outlet wells. Place the interface plate back onto the microfluidic plate and lock the lid (see Figure 1 for plate layout).
NOTE: As described in step 3.1.2, all corresponding input wells contain RPMI-1640 media with and without amphotericin B or other compound of interest.
- For testing C. albicans wild-type and mutant strains, add 50 µL of wild-type (SC5314) cell culture in Spider media to two outlet wells, bcr1 Δ/Δ in Spider media to two outlet wells, and efg1 Δ/Δ in Spider media to two outlet wells. Place the interface plate back onto the microfluidic plate and lock the lid (see Figure 1 for plate layout).
- In the shear control section, set the flow mode to constant and set the max shear to 2 dyne/cm2 (0.2 Pa). Click on outlet wells with cells to begin flow of media from inlet to outlet wells (Figure 2A) to begin seeding of C. albicans. After 3 s click the stop button in the well plate control section to prevent cells from entering the input wells.
NOTE: The cells are moved from the outlet wells towards the inlet wells; it is essential that the flow is stopped before the cells reach the inlet wells. This ensures that the inlet wells do not get contaminated with cell culture and that the media in the inlet wells remains sterile for the duration of the experiment. The shear force and time needed is based on the viscosity of media used and the size of the cells. - Allow the cells to remain stationary with no flow (0 dyne/cm2) for 10 – 20 min in the viewing channel for initial cell adherence. During this 10 – 20 min adherence step, set up the computer to capture the camera stage positions and begin acquiring pre-flush images (described in detail in Sections 4 – 6).
4. Setting Up the Stage Positions for the Microfluidic Experiment
NOTE: The stage positions and plate calibration should be set just before starting the experiment. This setup allows the computer to store the positions of each viewing channel for capturing the images during the experiment. The microfluidic plate should not be disturbed after setting up the stage positions. If the plate is moved, the stage positions will have to be reset before the start of the experiment.
- Use the visualization analysis software (see table of materials) to set the stage positions; each viewing channel is a stage (1 – 24) (Figure 1D) and each stage has three sub-stages (1 – 3) for image capturing at different positions along the viewing channel (Figure 1C). In the imaging module window, open the 'Multi-dimensional Acquisition' (MDA) module by clicking on the MDA tab (Figure 2B).
- In the MDA module click on the 'Stage' tab in the menu on the left-hand side (Figure 2B). In the stage section, load the master stage list for the 48 well 0 – 20 dyne microfluidic plate. Each viewing channel should have three stages for image acquisition.
- Open the 'Sample Reload Adjustment' (SRA) and 'Move Stage to Absolute Position' (MAP) modules by clicking on the SRA tab and the MAP tab (Figure 2B). In the SRA module, press the 'Live' button to see an image preview with crosshairs.
- Using the joystick, position the well plate such that the tip of the arrow (physically located on the plate) adjacent to viewing channel 4 aligns with the crosshairs on the software. In the MAP menu, press the 'Set Origin' button (Figure 2B). The current position should now read 0 in X, Y, and Z.
- In the SRA module, click on the 'Initial Reference Points' section in the left-hand menu (Figure 2B). Click on 'Load Settings' and load the file for the 48 well 0 – 20 dyne microfluidic plate.
- On the stage section of the MDA module, double-click on 'Stage Position 22,2'. This position indicates the middle (sub-stage 2) of the viewing channel number 22. This will bring the microscope viewing stage and the camera focus in close proximity to the arrow marker by viewing channel 22. Using the joystick, position the well plate such that the tip of the arrow (physically located on the plate) adjacent to viewing channel 22 aligns with the crosshairs.
- Copy the Y coordinate in the MDA module into the Y position of Point 2 in the 'Update Reference Points' tab of the SRA Module (Figure 2B).
- In the 'Update Stage Positions' tab of the SRA, click on 'Apply to MDA Stage List' (Figure 2B).
NOTE: This will update all viewing stage positions in the plate (1-24) to be calibrated to the specific microfluidic plate position on the microscope stage. The plate is now ready for image acquisition and will use the calibrated stage positions to move while capturing images from three positions in all 24 viewing channels.
5. Setting Up Acquisitions for Image Capture During the Microfluidic Experiment
- In the imaging module window, use the MDA module and click on the 'Save' tab in the left-hand menu (Figure 2B). Select the file name for the experiment and the folder to store the acquired images on the computer.
- In the MDA module, click on the 'Timelapse' tab in the left-hand menu and set it to acquire 145 total images; set the timelapse between images to 5 min. This will result in 1 image every 5 min for a 12 h biofilm development experiment.
NOTE: The time interval between images and the total number of images can be adjusted based on experimental requirements. If imaging for longer than 12 h, add additional media to the inlet wells at step 3.1 in order to ensure that the wells do not run dry. For longer experiments add additional media, as needed (50 µL/h) to the inlet wells. - In the MDA module, click on the 'wavelengths' tab in the left-hand menu (Figure 2B) and set the number of wavelength for the experiment as 1. Set the wavelength to capture at 50% Brightfield and 50% Camera with an exposure time of 12 – 20 ms, 0.6 gain and 20 MHz digitizer.
NOTE: Additional wavelengths can be used, including wavelengths for fluorescence imaging (for example, we have successfully visualized mCherry and GFP tagged C. albicans strains using this microfluidic device). The parameters of acquisition can also be altered based on experimental requirements. For instance, setting up a second wavelength and selecting 'Autofocus at each Timepoint' and a third wavelength and selecting 'Autoexposure at each Timepoint' is recommended for beginners to maximize image acquisition opportunities that are in focus. - In the MDA module, select the stage list tabs in the left-hand menu. Stages 1,1 – 24,3 will be displayed. One by one, click on each stage, and use the fine focus settings to manually focus on the cells in the viewing channel for each stage. Once the field of view is in focus, click the black arrow next to the stage list to update and save the settings for each stage. The computer will use these manually-defined position settings and focal settings to acquire images at every time point. Remove any unused stages from the list using the 'Remove' arrow.
NOTE: The MDA module and the program refer to viewing channels as stages interchangeably, and the same terminology is used by the software.
6. Running the Microfluidic Experiment
- In the imaging module window, using the MDA module, select 'Acquire' to begin capturing pre-flush images of adhered cells for all sub-stages (Figure 2B).
NOTE: The imaging module window and the camera controls will now show images that are being captured and the MAP, SRA, and MDA module will no longer be visible. The images captured will also show a timer on the side, to indicate the image number being captured and the time lapse before the next image capture cycle will start. Wait for one round of images to be captured before proceeding with the next step.
NOTE: Do not use the 'Pause' or 'Mark Event' buttons on the imaging module window screen. From this point on during the experiment, keep the mouse only on the control module window on the second computer screen (Figure 2A). This is important to avoid disturbing the imaging module window. - In the control module window stay on the manual mode for the plate. In the shear control section, set the flow mode to constant and set the max shear to 1 dyne/cm2 (0.1 Pa). Click on outlet wells O1, O7, O13 and O19 to begin the flow of media from inlet to outlet wells (Figure 2A) to remove non-adhered cells. After 5 min click on the stop button in the well plate control section. Allow for a second round of images to be acquired. Images can be visualized and the time lapse can be monitored on the second screen in the imaging module window.
- In the shear control section, adjust the max shear flow to 0.5 dyne/cm2 (0.5 Pa) by clicking on the number in the max shear column and using the keyboard to enter 0.5. Press the enter key on the keyboard and confirm the change in flow rate in the event log in the control module window (Figure 2A). Leave the experiment undisturbed in the dark for 12 h.
NOTE: Exposure to light and movement/vibrations can severely reduce the quality of images acquired throughout the experiment. At the end of the microfluidic experiment, the imaging module window will not show any images and reverts back to showing the SRA, MAP, and MDA modules. The imaging module software can be used to view the images, assemble the videos, and quantify the biofilms formed (described below).
7. Analyzing the Results
- In the imaging module window, select the 'Analysis Tools' tab and click 'Review Multi-dimensional Data' (RMD) from the drop down menu. In the RMD module, click on 'Select Base File'. This will open the 'Multi-dimensional Data Set Utilities' window.
- Click on the 'Select Directory' button and choose the folder which was used to save the experiment in step 5.1. In the 'Data Sets' list, select the experiment file log (.nd extension). Once the log is loaded, click on the 'view' button.
- Select the wavelength by clicking on the options in the left-hand 'Wavelengths' menu and choose the stage position to view from a drop-down menu. Choose the image to load from the numbers on the top of the module by right-clicking the image number. Once selected, click on the 'load image(s)' button to load the images. You can view one image or all images at once.
- Select all images to make a time lapse video. The selected images will open as a video in a new window. Click on the 'file' tab in the imaging module window and choose 'save as' from the drop-down menu. Save the resulting video as the default file (TIFF/MetaSeries).
- Open and load the saved video file using ImageJ software. In ImageJ, click on the 'file' tab and click on 'save as'. Choose '.avi' from the drop-down menu and convert the file to an .avi file using the JPEG conversion set to 10 frames/s.
NOTE: The speed of the video can be adjusted by altering the frames/s. - To quantify biofilms, repeat steps 7.2, 7.3, and 7.4. Choose the last image (number 144) and click on 'load image(s)' button to load the image. The image will open in a new window. Click on the 'threshold image' button and select 'Inclusive' threshold. Move the cursor until the biofilm cells are colored red, and regions with no cells are not colored.
- Click on the 'Open Log' button to log measurements (the button will display 'Open Log' only initially; once a log is active the button will be renamed to 'F9: Log Data'). On the 'Open Data Log' window select 'Dynamic Data Exchange' and click 'ok.'
- In the imaging module window, select the 'Analysis Tools' tab and click 'Show Region Statistics'. This will open a new window displaying measurements from the image. Select 'Entire Image' in the 'Measure' options available and click 'F9: Log Data.' An excel file with all measurements will be displayed. Locate the values for 'Area' and 'Thresholded Area' and divide the latter by the former to obtain a numerical value for the area occupied by the biofilm.
NOTE: The 'Area' for each image obtained during the experiment is identical and thus the 'Thresholded Area' measurement can be used to evaluate differences in biofilm formation between wild-type strains and mutant strains or evaluate the effectiveness of the tested drug on biofilm formation.
Representative Results
We performed the microfluidic biofilm assay described here using a wild-type C. albicans strain under two media conditions (RPMI-1640 and Spider media), the wild-type strain in the presence of the known antifungal drug amphotericin B (16 µg/mL) in RPMI, and two mutant strains previously reported to have defects in biofilm formation (bcr1Δ/Δ and efg1 Δ/Δ) in Spider media.
Video 1 shows the development of a wild-type biofilm in RPMI-1640 medium and the effects of the antifungal drug, amphotericin B, on biofilm formation. Under wild-type conditions, several cells strongly adhere to the channel, the cells form hyphae as time progresses, and a thick biofilm can be observed with intercalating hyphae and yeast cells. Towards the end of the microfluidic experiment, the wild-type biofilm completely fills the viewing channel. The presence of amphotericin B, however, has a pronounced effect on reducing the biofilm. Specifically, the adherence of cells is diminished, and, unlike untreated conditions, in the presence of amphotericin B, several cells can be seen flowing away with the shear flow of the media. As time progresses, the cells fail to form hyphae and a biofilm is not formed. These differences are also shown in Figure 3, which depicts 4 time-points from the assay at 0 h, 2 h, 6 h, and 12 h.
Video 2 shows the development of a wild-type biofilm and two previously reported biofilm-defective mutants (bcr1 Δ/Δ and efg1 Δ/Δ) in Spider media. Both bcr1 Δ/Δ and efg1 Δ/Δ strains show severely reduced biofilm formation compared to the isogenic wild-type strain. For the efg1 Δ/Δ strain, we observed that the adhered cells do not form hyphae and the resulting biofilm is severely defective. For the bcr1 Δ/Δ strain, we observed that not only is the bcr1 Δ/Δ strain defective in biofilm formation, but it also displays a clear adherence defect, where cells can be seen drifting in the direction of the shear flow as they fail to adhere to the viewing channel. These differences are also shown in Figure 4, which depicts 4 time points from the assay at 0 h, 2 h, 6 h, and 12 h.
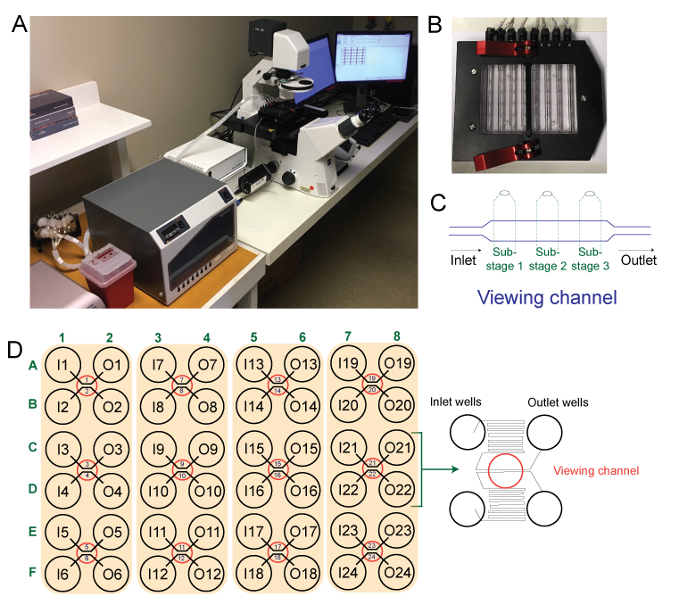
Figure 1: Microfluidic instrument used in this experiment. Panel A shows the microfluidic instrument used, panel B shows the interface plate, panel C shows a schematic outline of a single viewing channel (stage) and three sub-stages captured by the camera during the experiment, and panel D shows a schematic outline of the 48-well microfluidic plate used with a schematic of the outlet and inlet wells and viewing window. Please click here to view a larger version of this figure.
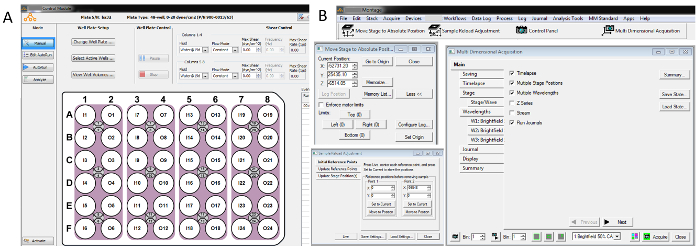
Figure 2: Select screenshots of imaging modulesoftware and microfluidic plate control system. Panel A shows the Control Module, that contains the controls for the microfluidic plate, and panel B shows the imaging module window, the 'Move Stage to Absolute Position' (MAP) module, the 'Multi-dimensional Acquisition' (MDA) module, and the 'Sample Reload Adjustment' (SRA) module. Please click here to view a larger version of this figure.
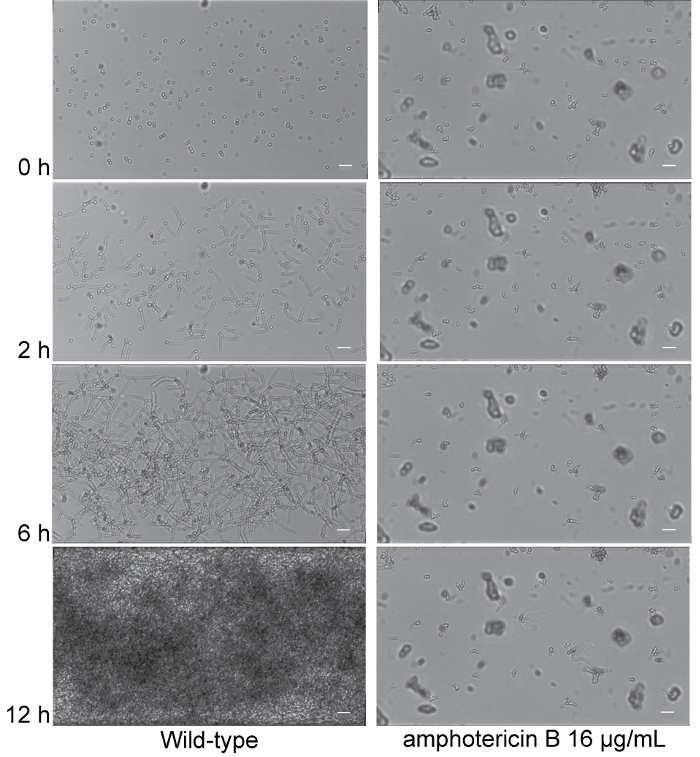
Figure 3: Exposure to amphotericin B results in biofilm formation defects. Time dependent visualization of biofilm formation in RPMI-1640 media (Column 1), and RPMI-1640 media supplemented with 16 µg/mL amphotericin B (Column 2) under dynamic flow (0.5 dyne/cm2) at 37 °C for 12 h post-adherence in the microfluidic system. Representative 0 h (post-adherence and initial wash), 2 h, 6 h, and 12 h images (top to bottom) are shown for the isogenic wild-type strain SN250 (Columns 1 and 2). Scale bars are 20 µm in each panel. Corresponding time lapse videos of biofilm formation are provided in Video 1. Please click here to view a larger version of this figure.
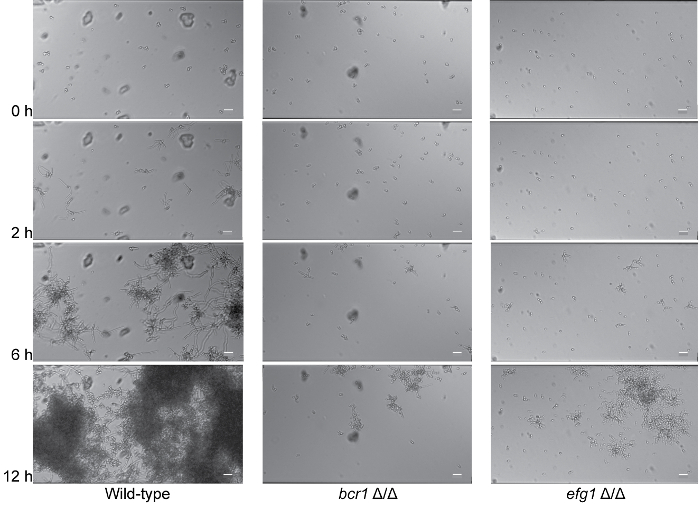
Figure 4: Deletion of BCR1 or EFG1 results in biofilm formation defects. Time dependent visualization of biofilm formation in Spider media (Columns 1-3) under dynamic flow (0.5 dyne/cm2) at 37 °C for 12 h post-adherence in the microfluidic system. Representative 0 h (post-adherence and initial wash), 2 h, 6 h, and 12 h images (top to bottom) are shown for the isogenic wild-type strain SN250 (Column 1), bcr1Δ/Δ strain (Column 2), and efg1 Δ/Δ strain (Column 3). Scale bars are 20 µm in each panel. Corresponding time lapse videos of biofilm formation are provided in Video 2. Please click here to view a larger version of this figure.
Discussion
The customizable microfluidic biofilm assay described here allows for the visualization of biofilm formation in real-time at a single cell level when exposed to a fixed rate laminar flow and constant temperature. It provides a powerful means to study the development of biofilms in wild-type and mutant strains, and the effects of antimicrobial agent treatments on biofilms under conditions that mimic physiological conditions observed in clinical settings. Unlike most in vitro biofilm assays, this method allows for the examination of a developing biofilm in real-time as it forms.
This method can be used in a high-throughput manner, where up to 24 samples can be run simultaneously to assess biofilm formation. The method can also be used to conduct genetic screens of mutant libraries to identify genes required for normal biofilm development and can be used to assess the effectivity against biofilms of antimicrobial compounds of interest in compound library screens. We anticipate that future applications of this microfluidic device will include such screens. We note, however, that certain drug treatment testing will be limited by the thickness of the biofilm formed using this microfluidic device as biofilms grown for >16 h tend to clog the viewing channel. Thus, drug testing experiments will need to be performed for <16 h.
Overall, this device is highly customizable and versatile, and can be adjusted to assess different microbial species across kingdoms, flow rates, temperatures, incubation times, and media. The method described provides information about a number of different aspects of biofilm formation, including initial cell adherence, biofilm maturation, and cell dispersal. Although we only report results for single-species biofilm formation here, this protocol could also be adapted to study dual- and mixed-species biofilm formation.
Two steps are critical to successfully perform this protocol. First, air bubbles in the system must be avoided by preheating the media at the experimental temperature. One common misstep is the dilution of cells in media that was not preheated. Cells should be diluted in the same media that is being used for the inlet wells. Second, the flow of cells from outlet to inlet wells must be carefully monitored; if the cells flow too far, the inlet media will become contaminated and the experiment will not yield interpretable results. On the other hand, if the cells are not allowed to move all the way into the viewing channel, then no cells will be visible in the channel during the experiment, leading to blank images. Additional key points to keep in mind are that the volume and movement between wells in the microfluidic plate are linked together for each column, and it is important to maintain as much media consistency as possible (with similar viscosity, cell density, and composition) and similarly sized cells (if possible), as different media and different cell sizes will have different flow rates in the column. During the experiment, the backflow of the cells is monitored using a microscope; however, only two channels can be viewed at one time (using a 10X objective). Thus, it is highly recommended to perform a mock experiment to measure the time required to flow from outlet to inlet wells using the cells and media planned to be used in the actual experiment.
In general, we highly recommend use of this microfluidic biofilm assay as an in vitro assay to be utilized before in vivo animal testing. The assay, however, is still an in vitro assay, and results will need to be validated in relevant animal models. In our experiences, the results of this microfluidic biofilm assay have had the best predictive value for in vivo biofilm assays compared with other in vitro assays we have assessed14.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank all members of the Nobile lab for helpful discussions on biofilm assays. This study was supported by National Institutes of Health (NIH) grant R21 AI125801 (to C.J.N.). D.L.R. was supported by a doctoral fellowship from The University of California Institute for Mexico and the United States (UC-MEXUS) and Consejo Nacional de Ciencia y Technologia (CONACYT).
Materials
| BioFlux 1000z | Fluxion | Automated microfluidic device for live cell analysis | |
| 48-well plate 0-20 dyne | Fluxion | 910-0047 | Microfluidic plate |
| Montage Software | Fluxion | Version 7.8.4.0 | Visualization analysis software |
| ImageJ Software | NIH | https://imagej.nih.gov/ij/ | |
| Yeast Extract | Criterion | C7341 | |
| Bacto Peptone | BD Biosciences | 211677 | |
| Dextrose (D-Glucose) | Fisher Scientific | D163 | |
| Potassium Phosphate Monobasic | Fisher Scientific | P285-500 | |
| RPMI-1640 | Sigma-Aldrich | R6504 | |
| MOPS | Sigma-Aldrich | M3183 | |
| Nutrient Broth | Criterion | C6471 | |
| Difco D-Mannitol | BD Biosciences | 217020 | |
| Agar | Criterion | C5001 | |
| Amphotericin B | Corning | 30-003-CF | |
| Sterile Inoculating Loops | VWR | 30002-094 | |
| Petri Dishes with Clear Lid | Fisher Scientific | FB0875712 | |
| Disposable Cuvettes | Fisher Scientific | 14-955-127 | |
| Lens Paper | VWR | 52846-001 | |
| Microplate and Cuvette Spectrophotometer | BioTek | EPOCH2TC | |
| Shaking Incubator | Eppendorf | M12820004 |
References
- Nobile, C. J., Johnson, A. D. Candida albicans Biofilms and Human Disease. Annu Rev Microbiol. 69, 71-92 (2015).
- Kojic, E. M., Darouiche, R. O. Candida infections of medical devices. Clin Microbiol Rev. 17 (2), 255-267 (2004).
- Fox, E. P., Nobile, C. J., Dietrich, L. A., Friedmann, T. S. . Candida albicans: Symptoms, Causes and Treatment Options. , 1-24 (2013).
- Gulati, M., Nobile, C. J. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 18 (5), 310-321 (2016).
- Uppuluri, P., et al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6 (3), e1000828 (2010).
- Andes, D., et al. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 72 (10), 6023-6031 (2004).
- Nett, J. E., Marchillo, K., Spiegel, C. A., Andes, D. R. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 78 (9), 3650-3659 (2010).
- Nett, J. E., et al. Rat indwelling urinary catheter model of Candida albicans biofilm infection. Infect Immun. 82 (12), 4931-4940 (2014).
- Krom, B. P., Willems, H. M. In Vitro Models for Candida Biofilm Development. Methods Mol Biol. 1356, 95-105 (2016).
- Hawser, S. P., Douglas, L. J. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 62 (3), 915-921 (1994).
- Ramage, G., Vande Walle, K., Wickes, B. L., Lopez-Ribot, J. L. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 45 (9), 2475-2479 (2001).
- Nett, J. E., Cain, M. T., Crawford, K., Andes, D. R. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J Clin Microbiol. 49 (4), 1426-1433 (2011).
- Krom, B. P., Cohen, J. B., McElhaney Feser, G. E., Cihlar, R. L. Optimized candidal biofilm microtiter assay. J Microbiol Methods. 68 (2), 421-423 (2007).
- Lohse, M. B., et al. Assessment and Optimizations of Candida albicans In Vitro Biofilm Assays. Antimicrob Agents Chemother. 61 (5), (2017).
- Winter, M. B., et al. Global Identification of Biofilm-Specific Proteolysis in Candida albicans. mBio. 7 (5), (2016).
- Nobile, C. J., et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 148 (1-2), 126-138 (2012).
- Fox, E. P., et al. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 96 (6), 1226-1239 (2015).
- Nobile, C. J., Mitchell, A. P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 15 (12), 1150-1155 (2005).
- Baker, K. . At the bench: A laboratory navigator. 27, (2005).

