Capturing the Interaction Kinetics of an Ion Channel Protein with Small Molecules by the Bio-layer Interferometry Assay
Summary
The protocol here describes the interactions of purified hEAG1 ion channel protein with the small molecule lipid ligand phosphatidylinositol 4, 5-bisphosphate (PIP2). The measurement demonstrates that BLI could be a potential method for novel small-molecule ion channel ligand screening.
Abstract
The bio-layer interferometry (BLI) assay is a valuable tool for measuring protein-protein and protein-small molecule interactions. Here, we first describe the application of this novel label-free technique to study the interaction of human EAG1 (hEAG1) channel proteins with the small molecule PIP2. hEAG1 channel has been recognized as potential therapeutic target because of its aberrant overexpression in cancers and a few gain-of-function mutations involved in some types of neurological diseases. We purified hEAG1 channel proteins from a mammalian stable expression system and measured the interaction with PIP2 by BLI. The successful measurement of the kinetics of binding between hEAG1 protein and PIP2 demonstrates that the BLI assay is a potential high-throughput approach used for novel small-molecule ligand screening in ion channel pharmacology.
Introduction
Targeting the cell surface-accessible ion channel proteins with small molecules offers a tremendous potential for the ligand screening and biological drug discovery1,2,3. Thus, an appropriate tool is needed for studying the interaction between ion channel and small molecules and their corresponding function. The patch-clamp recording has been demonstrated to be a unique and irreplaceable technique in ion channel functional assay. However, determining whether the small molecules directly target ion channels require other technologies. Traditionally, the radioactive ligand binding assay was used to observe the kinetics of binding between small molecule and its target ion channel protein. However, the usage of this technique is limited because of its requirement in radioactive labeling and detection. Moreover, the prerequisite step to label the small ligand in the study prevents its using in many types of ion channels without known specific ligand. Some label-free techniques such as NMR spectroscopy, X-ray diffraction, microscale thermophoresis (MST)4 and surface plasmon resonance (SPR) have been used to measure the protein-small molecule interactions. But these types of assays usually cannot provide sufficient information because of the difficulty to get the full-length protein, low resolution of dynamics, low throughput, and high cost5. In contrast with these techniques, bio-layer Interferometry (BLI) is emerging as a novel label-free methodology to overcome these drawbacks for detecting protein-small molecule interactions by immobilizing a tiny amounts of protein sample on the surfaces of biosensor and measuring the optical changing signals6,7. As a promising biosensor platform, BLI technique is already performed to observe the interaction of small molecules with natural water soluble proteins such as a human monoclonal antibody CR80208 and the detailed assay procedure has been reported in a previous article9. Although the key role of ion channel protein for new therapeutic targets discovery has been recognized, the ion channel protein-small molecule interaction assay based on BLI has not been described.
The human Ether à go-go channels (hEAG1) are expressed in various types of cancer cells and central nervous system which makes the channel a potential therapeutic target of many cancers and neuronal disorders10,11,12,13,14. The electrophysiological study in our lab has confirmed the inhibitory effect of phosphatidylinositol 4, 5-bisphosphate (PIP2) on hEAG1 channel15. Based on our results, testing PIP2 directly interaction with the hEAG1 by using BLI technique can be as a model for other types of ion channel protein-small molecule compound interaction especially for those channels lacking specific ligands. According to the instructions of BLI assay, we prepared biotinylated hEAG1 proteins and immobilized them on the surface of streptavidin (SA) biosensor tips followed by interaction them to PIP2 solutions to observe their direct binding between the protein and the lipid. After the attachment of PIP2 to the hEAG1 protein coated surface, the thickness of the layer on the surface increases, which directly correlates the spectral shift and can be measured in real-time16. The binding kinetics can be determined due to a positive shift in association step and a negative shift in dissociation step. According to this principle, we purified the functional hEAG1 ion channel protein from HEK-239T stable expression system by using affinity purification method to maintain the in vitro functional state, then measured the kinetics of binding of different concentration PIP2, and yielded a semblable kinetic data as observed in electrophysiological measurements15. The close correspondence between the results from the BLI and electrophysiological measurements demonstrate for the first time the suitability of BLI as an appropriate analytical tool for ion channel membrane protein-small molecule interaction.
Protocol
NOTE: The HEK-293T cell line continuously expressing FLAG-tagged hEAG1 channel protein is constructed by transfecting a pCDH lentiviral plasmid containing the DNA sequence of hEAG1 with a FLAG at the distal C-terminus into HEK-293T cells followed by the puromycin-resistant selection as previously described15.
1. Affinity Purification of FLAG-tagged hEAG1 Channel Protein from HEK-293T Cells
- Thaw cells stably expressing hEAG1 channels from liquid nitrogen into the warm water (37 °C) quickly. Seed the cells (about 5 x 106 frozen cells) in a 10 cm dish. Grow the cell overnight in Dulbecco's Modified Eagle's (DMEM) medium supplemented with 8 mL 10% fetal bovine serum (FBS), 100 units/mL penicillin, 100 μg/mL streptomycin, 4 mM L-glutamine and changed the medium the next day.
- Check the GFP fluorescence of these HEK-293T cells by using a fluorescent microscope to make sure the high percentage of cells stably express hEAG1 channels in the culturing system. Exponentially trypsinize the growing cells with 1 mL of 0.25% trypsin for 1 min at room temperature and thenadd 2 mL of serum-containing culture fluid to terminate the digestion. Transfer 400 μL of cell suspension to a 15 cm dish containing 15 mL of complete DMEM medium. Prepare four 15 cm dishes in total.
- Harvest these cells after 2–3 days culture when the cells reach at about 90% confluency.
- Remove the growth medium from the cells and wash them twice with 4 mL of phosphate buffered saline (1x PBS, pH = 7.4).
- Discard PBS after washing.
- Scrape the cells into 2 mL 1x PBS for each dish by using cell scrape and transfer the scraped cells with 1 mL pipette into a 15 mL tube.
- Centrifuge the cell suspension for 5 min at 420 x g at 4 °C.
- Decant and discard the supernatant.
- Resuspend the cell pellet in total 4 mL of lysis buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM NaCl, 1% NP-40, pH 7.0, contained complete protease inhibitor) for 30 min on ice, and vortex thoroughly the lysate every 10 min.
- Centrifuge the cell lysate for 10 min at 12,000 x g at 4 °C.
- Transfer the supernatant to a 5 mL tube and keep it on ice for immediate use.
- Prepare the anti-FLAG M2 affinity resin.
- Thoroughly suspend the resin (supplied as a 50% suspension in store buffer) by gentle inversion to make sure the bottle of anti-FLAG M2 affinity gel is a uniform suspension of gel beads.
- Immediately transfer 400 μL of suspension to a chilled 1.5 mL tube.
- Centrifuge the suspension for 30 s at 8, 000 x g at 4 °C and discard the supernatant carefully to wash out the store buffer.
- Add 500 μL 1x PBS to the resin and suspend the pellet with 1 mL pipette. Then centrifuge the suspension for 30 s at 8, 000 x g at 4 °C and discard the supernatant PBS carefully. Repeat these wash step to clear the stored buffer.
- Add 500 μL protein extract supernatant prepared at step 1.3.8 to the resin pellet to suspend the pellet and transfer the suspension to a new chilled 5 mL tube. Repeat this step again to make sure no gel beads left. Add the left protein extract to the mixture.
- Incubate the mixture overnight on a shaker at 8 rpm at 4 °C to capture the FLAG fusion protein.
- Centrifuge the mixture after 12 h incubation for 10 min at 1, 000 x g at 4 °C.
- Discard the supernatant and wash the pellet three times with 500 μL of 1x PBS. Keep it on ice for immediate use.
- Elution the FLAG hEAG1 protein with 3x FLAG peptide.
- Prepare 3X FLAG elution solution. Dissolve 3X FLAG peptide in 500 μL stock solution (0.5 M Tris-HCl, 1 M NaCl, pH = 7.5) at a concentration of 8 μg/μL.
- Add 10 μL 3x FLAG elution solution to 390 μL of PBS to be a 200 ng/μL final concentration solution.
- Add 400 μL of 3x FLAG elution solution to the gel beads prepared at step 1.8.
- Incubate the sample at the shaker at 8 rpm for 2 h at 4 °C.
- Centrifuge the resin for 30 s at 8, 000 x g.
- Transfer the supernatant to a fresh 1.5 mL tube and store it at 4 °C for immediate use.
2. Concentration Assay and Confirmation of Purified FLAG Fusion hEAG1 by BCA Protein Assay Kit and Western Blotting
- Determine the concentration of purified protein using the BCA protein assay kit according the manufacture's instruction.
- Use 30 μL sample for western analysis to confirm the interest protein had been purified by using an ANTI-FLAG antibody as previously described15.
3. Labeling the Purified Channel Protein with Biotin for the BLI Assay
- Prepare 5 mg/mL biotin stock solution in PBS. For each test (two biosensors), add a 3-fold molar excess of biotin to 20 μg purified protein to achieve a preferable N-terminal biotinylation of the purified protein in PBS.
- Incubate the sample in the dark on ice for at least 30 min.
- Prepare the sample dilution (SD) buffer: PBS with 0.02% polysorbate 20 and 0.1% bovine serum albumin (BSA, pH 7.4).
- Perform ultrafiltration to change the buffer of the purified channel protein to SD buffer. Remove the unbound biotin by using ultrafiltration device with molecular weight cutoff of 30 kDa, adding the SD buffer and centrifuging the sample at 12,000 x g, for 10 min at 4 °C.
- Remove the ultrafiltrate from the centrifuge tube of ultrafiltration device, add 200 μL SD buffer into the filter device and centrifuging the sample at 12,000 x g, for 10 min at 4°C. Repeat this operation at least three times.
- To collect the buffer exchanged sample, reversed insert the filter device into a 1.5 mL tube and centrifuging them at 2,000 x g, for 5 min at 4 °C. Keep the sample on ice for immediate use.
4. Preparation of PIP2 Solution for Assay
- Prepare the stock solution of PIP2 (1 mM) in deionized H2O by sonicating for 30 min on ice as previously described17. Store the solution in glass vials at -20 °C and dilute it to the final concentrations immediately before experiments by vigorous vortexing.
5. BLI Assay
- Turn on the equipment and check the "instrument status" window to confirm the machine is at "ready" state to prewarm the equipment at least for 30 min before the BLI study.
- Make sure the door of the instrument is closed before opening the Data Acquisition software and choose the "New Kinetics Experiment" in the Experiment Wizard.
- Define the wells to be used on the 96-well plate by right click to choose buffer, load, and sample. For sample wells, the unit of concentration of biotinylated FLAG fusion hEAG1 protein should be input as molar (10 μg, 0.09 μM).
- Define the assay steps including baseline, loading, association and dissociation. Choose an assay step and double click on the respective column. A duplicate of assay definition is set for control sensors. Set the rpm as 1,000. Choose 1 min for baseline step and 5–10 min for loading, association, and dissociation, respectively. Perform the test at room temperature (about 24 °C).
NOTE: There are two main procedures: loading the biotinylated channel protein to the sensors and assaying the interaction with small molecule compounds. They can be proceeded continuously (baseline, loading, baseline, association and dissociation). Alternatively, they can be proceeded separately to avoid wasting the test compounds when the first loading part unsuccessful. - Click the columns which contain the sensors and click the "Fill" to indicate the locations of sensors in the sensor tray.
- Review all planned steps to check for mistakes and go back to correct them.
- Set the location of data files and click "Go" to start the assay.
NOTE: The sensors need prewetted for at least 10 min in SD buffer, if this step has been done, then the "Delayed experiment start" setting should be skipped. If not, set a 600 s delay before prewetting the sensors. - Put a black 96-well plate in the bottom of the tray and insert the A1 corner of the plate into the notch on the tray to seat the plate. For the wells to load the sensors, 200 μL of assay buffer per well add into 2 wells in row A and 2 wells in row B of the 96-well plate.
- Prepare another black 96-well plate as the sample plate and fill the wells with 200 μL SD buffer in row B as control or biotinylated hEAG1 protein solution (10 μg) in row A as assigned during programming in step 5.3.
NOTE: Avoid introducing bubbles. - Open the door of the instrument and insert the sensor tray and sample plate into the left and right plate holder, respectively. Check that the sensor tray and sample plate are positioned correctly based on the shape of left plate holder and the "A1" marker on the top right corner of right plate holder. Close the door and start the assay.
6. Data Analysis
- Open the Data Analysis software and load the folder containing the assay data. Click "Processing" to get into the processing menu interface and we can see the colorful raw kinetic curves.
- Under Step1: "Data Selection", click "Sensor Selection". On the "Sensor Tray #1", click the sensor wells only wetted with SD buffer and right click to "Change Sensor Type" to "Reference Sensor". On the 'Sample Plate Map", designate the all the non-specific binding wells and right click to "Change Well Type" to "Reference Well".
- Tick in the box before "Subtraction" of Step 2 and point "Double Reference".
- In Step 3: "Align Y Axis", select "Baseline" as the alignment step. For "Time Range", enter the last 10 s of that baseline (i.e. From: 0.1 To: 59.8).
- In Step 4: "Inter-step Correction", select "align to Baseline" to minimize signal shifts between the association and dissociation steps.
- In Step 5: "Process", select Savitzky-Golay filtering function in most cases and proceed "Process Data".
- Save Raw Data for further data analysis using other software in Step 7: "Save Results".
- Click "Analysis | Curve Fitting".
- For "Step to Analyze", choose "Association | Dissociation". For "Model", select 1:1.
NOTE: we choose 1:1 model because it fitted well on our original data and avoided the possibility of over fitting under 1:2 or 2:1 model due to their high freedom. But other options are available here and can be suitably chosen for other fitting analysis for different binding models of analyte. - For "Fitting", choose Global (Full). For "Group By", select "Color". Select "Rmax Unlinked By Sensor" to allow independent fitting of maximal signal response (Rmax).
- Click "Fit Curves!" to start the nonlinear regression analysis. Hill equation and single exponential function were used in our study.
- Click "Data Export |Save Report" to save fitting results. Or click "Data Export | Export Fitting Results" to save the raw data for further graphing and data analysis with other software.
- For "Step to Analyze", choose "Association | Dissociation". For "Model", select 1:1.
Representative Results
We purified the FLAG fusion hEAG1 channel protein from HEK-293T cells stably overexpressed hEAG1. The function of this fusion protein has been demonstrated by using the patch-clamp method and the quality and specificity of purified protein are confirmed by Western blot (Figure 1). The purified channel protein is biotinylated to perform an interaction assay with the lipids (PIP2) by using the real-time BLI assay. The BLI binding assay configuration is shown in Figure 2. A typical binding curve between hEAG1 and PIP2 is shown in Figure 3. In this case, 3 μM PIP2 is dissolved in PBS buffer (the configuration of PIP2 is shown in Supplementary Figure 1), and the signal is analyzed using a double reference subtraction protocol to subtract the non-specific binding (the binding between sensor and PIP2), background (the interaction between biotinylated hEAG1 protein and PBS), and signal drift (the binding between sensor and PBS) caused by sensor variability. And the binding trace is globally fit and shown a well-fitting overlay (Supplementary Figure 2). Also, we measure the kinetics of binding of PIP2 to the purified hEAG1 channel complex by incubating the proteins at different concentrations of PIP2. After analysis, we get a dissociation constant (Kd) value of 0.35 ±0.04 μM, which is similar to the IC50 value obtained from the electrophysiological measurements15. These results demonstrated that the BLI assay is appropriate for ion channel membrane protein and lipids interaction analysis.
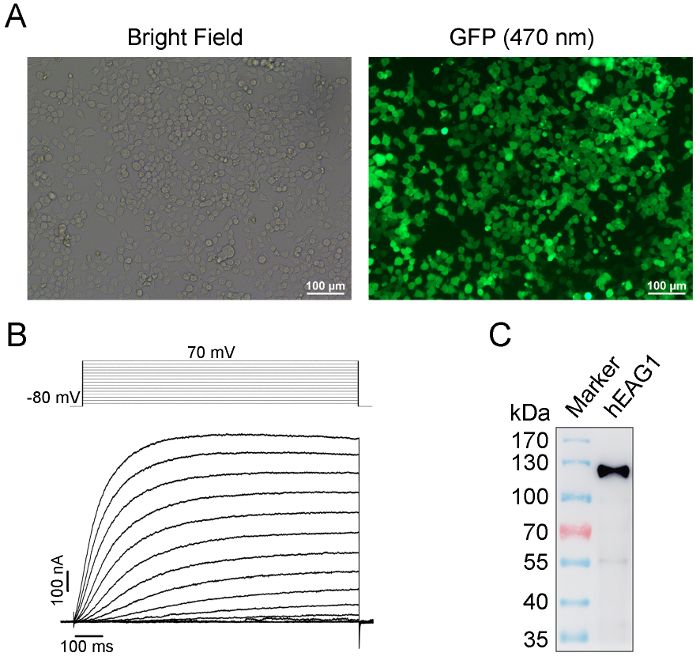
Figure 1: Identification of the expression, function, and specificity of recombinant hEAG1 protein from HEK-293T cells by GFP imaging, patch clamp, and western blot, respectively. (A) The stable expression hEAG1 HEK-293T system is successfully established by monoclonal puromycin-resistant selection after transfection with hEAG1-pCDH lentivirus system as evidenced by GFP expression in almost all cells. (B) Pulse protocol (top) and superimposed current traces from a representative whole-cell patch-clamp recording from hEAG1 channels in a stable cell in A. The current is elicited by depolarizing voltages from the holding voltage of -80 mV to 70 mV with the step of 10 mV followed by repolarization to -80 mV. The cells are incubated in the normal K+ channel recording solutions as described previously15. The voltage-dependent outward potassium currents suggest that functional hEAG1 channels are highly expressed in HEK293T cells. (C) Western blot of hEAG1 channel protein from purified protein samples. The anti-FLAG antibody recognizes a single protein band of ~110 kDa, demonstrating a full-length of FLAG-tagged hEAG1 channel expression. This Figure 1C has been modified from Han et al.15. Please click here to view a larger version of this figure.
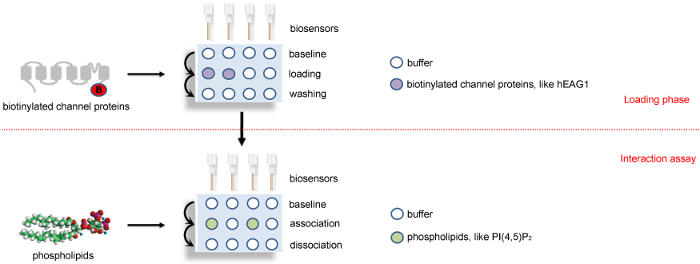
Figure 2: A schematic diagram showing the BLI binding assay protocol. Four sensors are used in parallel in biotinylated-channel proteins or their dissolved SD buffer to load the channel proteins and the references. After that, these four sensors are transferred to assay phase to detect the association and disassociation with phospholipids or its solution buffer. The positions of channel proteins, phospholipids, and buffer are colored as indicated. The horizontal red dotted line indicates the two major steps of BLI study: loading phase and interaction assay phase. This Figure 2 has been modified from Han et al.15. Please click here to view a larger version of this figure.
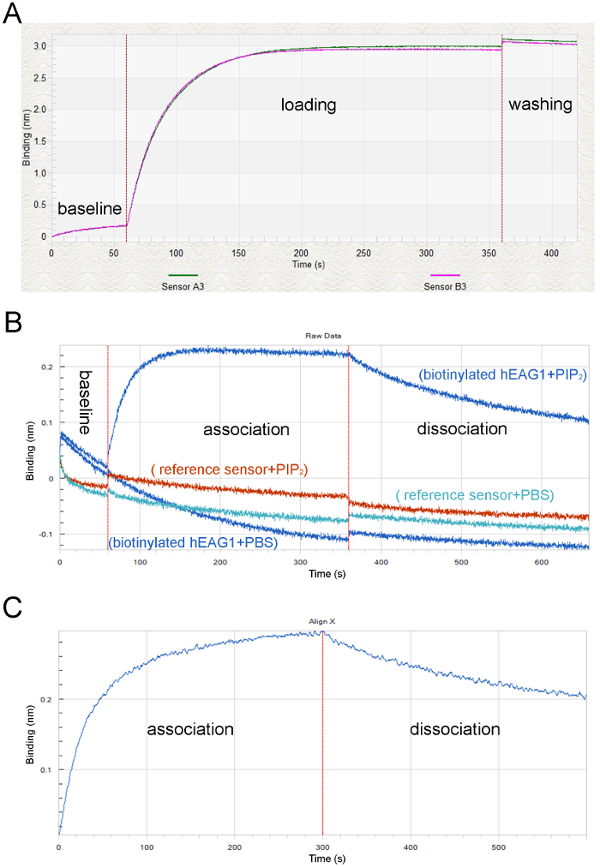
Figure 3: Screen captures showing the raw data and processed data in a typical BLI study. (A) Typical loading and equilibration curves showing the equilibration step (60 s) with SD-buffer (baseline), the loading step with hEAG1 proteins (loading) and the reference curve equilibrated and loaded with hEAG1 proteins (loading), simultaneous measurement of two individual sensor tips. (B) The vertical red lines indicate the transferring of sensors from the lipid solution to the buffer solution during assay operation. (C) The original optical signals at association and dissociation phases after processing the double reference subtraction to subtract the non-specific binding signals. Please click here to view a larger version of this figure.
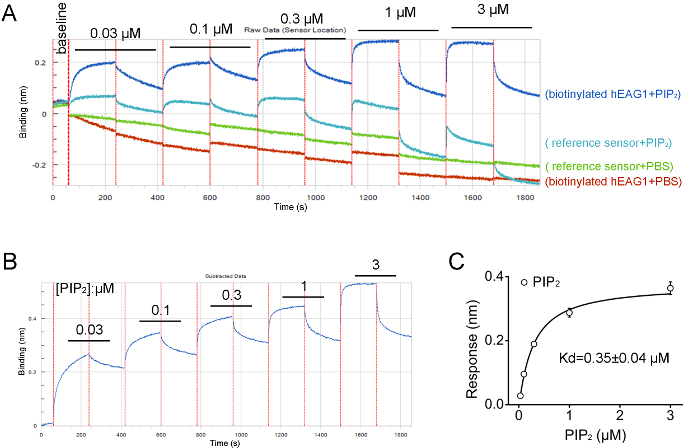
Figure 4: Results from BLI assay showing hEAG1 channel protein directly interacting with PIP2. (A) Screen capture showing the raw data of hEAG1 protein binding with PIP2 at concentration-dependence manner (0.03–3 μM). The accumulated concentrations of PIP2 denoted corresponding to the biosensors' data traces. (B) The raw data processing showing the changes in optical interference in different concentrations of PIP2 in a representative assay. (C) Curve fit with Hill equation obtained from the peak value of the optical interference signal measured at different PIP2 concentrations for determination of the equilibrium dissociation constants (Kd) of the interaction between the hEAG1 channel protein and PIP2 (n = 3).The Figure 4B and 4C have been modified from Han et al.15. Please click here to view a larger version of this figure.
Supplementary Figure 1: The configuration of long-chain phosphatidylinositol 4, 5-bisphosphate (PIP2). Please click here to download this figure.
Supplementary Figure 2: Screen capture of the fitted overlay from the BLI assay of Figure 3. The data is processed and fitted and only shown Association and Dissociation phases. The processed data curve is blue and the nonlinear fitting curve is red. Goodness of fit: R2 = 0.984789, X2 = 0.019109. The maximal binding parameter (Rmax) = 0.2875 nm (± 0.0006). Please click here to download this figure.
Discussion
Membrane ion channels have been verified as the primary therapeutic targets of over 13% of currently known drugs for the treatment of a variety of human diseases, including cardiovascular and neurological disorders18. Patch-clamp recording, the golden standard for measuring the functional of ion channels with small molecules, has been widely used for ion channel ligands screening. However, such electrophysiological approaches cannot demonstrate whether the small molecules binds to the channel directly or not19, because the small molecule could act on other proteins or intercellular pathways that interact with the channel. Compared to widely used radioactive ligand binding assay and other commonly used label-free biosensor methods, BLI has significant advantages in terms of relative simple arrangement, the unrestricted association phase, high throughout, assay design closer to the in-vivo system and a need of small amount of immobilized protein (a few μg), and it can provide detailed insights into kinetic data5,6,20.
In order to maximally simulate the in vivo situation, we purified the functional hEAG1 channel proteins from the mammalian stably expression system. An easy and efficient protocol for both purification and identification of the overexpressed ion channel protein from adherent mammalian cells is presented. Some important tips for successful purification of the membrane proteins are: 1) The purification process can be either scaled-down or scaled-up according the expression abundance of interest proteins in mammalian expression systems; 2) We fused a FLAG tag on the C terminus of hEAG1 channel to facilitate the purification of this protein with ANTI-FLAG affinity beads using the commercial kit and purification protocol. An electrophysiological measurement demonstrated that the fusion FLAG has no effect on the function of this channel15; 3) Incubation of the mixture of ANTI-FLAG beads and protein extract overnight at 4°C with gently shaking is helpful for capturing the FLAG fusion protein; 4) Using the affinity purification, we can get enough hEAG1 ion channel protein from four 15 cm dishes with above 90% cell confluency for once assay process.
The purified hEAG1 channel proteins are biotinylated by using excess biotin (3-10 fold) and exchanging the solution buffer with SD buffer for the further BLI assay. The excess biotin can maximally modify the membrane channel protein to increase the coated efficiency on the surface of biosensor tips, and the SD assay buffer containing low concentrations of BSA and Polysorbate 20 can minimize nonspecific binding9. In spite of proceeding these key operations, the non-ideal interactions still could exist especially when performing a binding study with high concentration of small molecule, which could non-specifically bound to the surface of SA sensors and result the false-positive interaction as the cyanine curve shown in Figure 4A. Thus, a double reference subtraction protocol is still necessary to get the reliable results by subtracting the non-specific bindings including the interactions between the sensor and small molecule, biotinylated hEAG1 protein with small molecule solution, and sensors with small molecule solution. This double reference subtraction operation needs four biosensors for once assay. Two biosensors will be coated with biotinylated membrane proteins and proceeding the interaction assay with small molecule and its buffer. The other two sensors will be wetted with SD buffer and interacting with small molecule and its buffer. Only the interaction of membrane protein immobilized biosensor and small molecule shows the positive signal and the rest of three interaction signals work as the controls (Figure 2). By using this procedure, as shown in Figure 3 and Figure 4, our results clearly demonstrate the strong interaction between hEAG1 channel proteins and PIP2 and show a concentration-dependence profile. It must be pointed out that there are several limitations in our measurement. For instance,there are some other membrane lipids which could bind to the purified channel protein. Also, it's still not clear that whether the anchored purified protein keep their original conformation and activity. It's very difficult to measure the real configurations of small molecule lipids when they interact with the protein. Although the detailed processes are unknown, our study show that the binding kinetics by BLI are consistent with those derived by electrophysiological recording, which makes the novel BLI application for ion channel protein-small molecule interaction in a supplementary manner.
In summary, our study confirms that it is a reliable strategy by purifying the functional membrane protein from mammalian expression system to detect the direct interaction with its ligands. The successful application of BLI for membrane protein-small molecule interaction will facilitate the small molecule screening and mechanism exploration in ion channel drug discovery.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by Bio-ID Center and SJTU Cross-Disciplinary Research Fund in Medicine and Engineering (YG2016QN66), National Natural Science Foundation of China (31271217), and National Basic Research Program of China (2014CB910304).
Materials
| DMEM/High Glucose Medium | HyClone | SH30243.01 | |
| Phosphate Buffered Saline (1x) | HyClone | SH30256.01 | |
| Fetal Bovine Serum | Gibco | 10270 | |
| Penicillin/Streptomycin | Gibco | 1697550 | |
| Cell Culture Dish | Corning | 430599 | 150 mm X 25 mm |
| Nonidet P-40 Substitute | Amresco | E109 | |
| Sodium chloride | BBI Life Sciences | A610476 | |
| Potassium chloride | BBI Life Sciences | A610440 | |
| Bovine Serum Albumin | BBI Life Sciences | A600332 | |
| Polyoxyethylene-20-Sorbitan Monolaurate | BBI Life Sciences | A600560 | |
| ANTI-FLAG M2 Affinity Gel | Sigma | A2220 | |
| 3x FLAG peptide | Sigma | F4799 | |
| Octet-RED96 | Pall/FortéBio | 30-5048 | |
| Data Acquisition software | Pall/FortéBio | Version 7.1 | |
| Data Analysis software | Pall/FortéBio | Version 7.1 | |
| Biosensor/Streptavidin | Pall/FortéBio | 18-5019 | |
| Microtiter plate | Greiner Bio-one | 655209 | |
| Sulfo-NHS-LC-LC-Biotin | ThermoFisher | 21338 | |
| Centrifugal Machine | ThermoFisher | 75004250 | |
| PageRuler Prestained Protein Ladder | ThermoScientific | 318120 | |
| Ultrafiltration device | MILLIPORE | UFC503008 | NMWL of 30 kDa |
| phosphatidylinositol 4, 5-bisphosphate (PIP2) | Sigma | P9763 | |
| Monoclonal ANTI-FLAG M2 antibody | Sigma | F1804 | 1:2000 dilution |
| goat anti-mouse HRP-conjugated secondary antibody | Santa Cruz Biotechnology | sc-2005 | 1:5000 dilution |
| Enhanced BCA Protein Assay Kit | Beyotime | P0010 | |
| Protease Inhibitor Cocktail Tablets | Roche | 04693159001 | |
| Amersham Imager 600 Imaging System | GE Healthcare Bio-Sciences | ||
| Western blot system | BIO-RAD |
References
- Peetla, C., Stine, A., Labhasetwar, V. Biophysical Interactions with Model Lipid Membranes: Applications in Drug Discovery and Drug Delivery. Mol. Pharm. 6 (5), 1264-1276 (2009).
- van de Waterbeemd, H., Smith, D. A., Beaumont, K., Walker, D. K. Property-based design: Optimization of drug absorption and pharmacokinetics. J. Med. Chem. 44 (9), 1313-1333 (2001).
- Papo, N., Shai, Y. Exploring peptide membrane interaction using surface plasmon resonance: Differentiation between pore formation versus membrane disruption by lytic peptides. Biochemistry. 42 (2), 458-466 (2003).
- Wienken, C. J., Baaske, P., Rothbauer, U., Braun, D., Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100-106 (2010).
- Fechner, P., et al. Size does matter! Label-free detection of small molecule-protein interaction. Anal. Bioanal. Chem. 406 (17), 4033-4051 (2014).
- Wallner, J., Lhota, G., Jeschek, D., Mader, A., Vorauer-Uhl, K. Application of Bio-Layer Interferometry for the analysis of protein/liposome interactions. J. Pharm. Biomed. Anal. 72, 150-154 (2013).
- Wartchow, C. A., et al. Biosensor-based small molecule fragment screening with biolayer interferometry. J. Comput. Aided Mol. Des. 25 (7), 669-676 (2011).
- Ekiert, D. C., et al. A Highly Conserved Neutralizing Epitope on Group 2 Influenza A Viruses. Science. 333 (6044), 843-850 (2011).
- Shah, N. B., Duncan, T. M. Bio-layer Interferometry for Measuring Kinetics of Protein-protein Interactions and Allosteric Ligand Effects. J. Vis. Exp. (84), e51383 (2014).
- Occhiodoro, T., et al. Cloning of a human ether-a-go-go potassium channel expressed in myoblasts at the onset of fusion. Febs Lett. 434 (1-2), 177-182 (1988).
- Pardo, L. A., Stuhmer, W. Eag1: An emerging oncological target. Cancer. Res. 68 (6), 1611-1613 (2008).
- Simons, C., et al. Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat. Genet. 47 (1), 73-77 (2015).
- Kortum, F., et al. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 47 (6), 661-667 (2015).
- Han, B., Tokay, T., Zhang, G. M., Sun, P., Hou, S. Eag1 K+ Channel: Endogenous Regulation and Functions in Nervous System. Oxid. Med. Cell. Longev. 2017, (2017).
- Han, B., et al. Human EAG channels are directly modulated by PIP2 as revealed by electrophysiological and optical interference investigations. Sci. Rep. 6, (2016).
- Do, T., et al. A rapid method for determining dynamic binding capacity of resins for the purification of proteins. PREP. 60 (2), 147-150 (2008).
- Rohacs, T., Chen, J., Prestwich, G. D., Logothetis, D. E. Distinct specificities of inwardly rectifying K+ channels for phosphoinositides. J. Biol. Chem. 274 (51), 36065-36072 (1999).
- Overington, J. P., Al-Lazikani, B., Hopkins, A. L. How many drug targets are there?. Nat. Rev. Drug Discov. 5 (12), 993-996 (2006).
- Suh, B. C., Hille, B. PIP2 is a necessary cofactor for ion channel function: How and why?. Annu. Rev. Biophys. 37, 175-195 (2008).
- Yang, D., Singh, A., Wu, H., Kroe-Barrett, R. Determination of High-affinity Antibody-antigen Binding Kinetics Using Four Biosensor Platforms. J. Vis. Exp. (122), e55659 (2017).

