A Fast Silver Staining Protocol Enabling Simple and Efficient Detection of SSR Markers using a Non-denaturing Polyacrylamide Gel
Summary
Here, we report a simple and low-cost silver staining protocol which requires only three reagents and 7 min of processing, and is suitable for fast generation of high-quality SSR data in the genetic analysis.
Abstract
Simple Sequence Repeat (SSR) is one of the most effective markers used in plant and animal genetic research and molecular breeding programs. Silver staining is a widely used method for the detection of SSR markers in a polyacrylamide gel. However, conventional protocols for silver staining are technically demanding and time-consuming. Like many other biological laboratory techniques, silver staining protocols have been steadily optimized to improve detection efficiency. Here, we report a simplified silver staining method that significantly reduces reagent costs and enhances the detection resolution and picture clarity. The new method requires two major steps (impregnation and development) and three reagents (silver nitrate, sodium hydroxide, and formaldehyde), and only 7 min of processing for a non-denaturing polyacrylamide gel. Compared to previously reported protocols, this new method is easier, quicker and uses fewer chemical reagents for SSR detection. Therefore, this simple, low-cost, and effective silver staining protocol will benefit genetic mapping and marker-assisted breeding by a quick generation of SSR marker data.
Introduction
The development of PCR-based markers has revolutionized the science of plant genetics and breeding1. Simple sequence repeat (SSR) markers are among the most commonly used and most versatile DNA markers. Their broad genome coverage, abundance, genome specificity, and repeatability are some of the merits of SSR markers in addition to their codominant inheritance for the detection of heterozygous genotypes2. Several studies have used SSR markers to investigate genetic diversity, track ancestry, construct genetic linkage maps, and map genes for economically important traits3,4.
PCR products of SSR markers are commonly separated using agarose or polyacrylamide gel electrophoresis and then visualized with silver staining or under UV light after staining with ethidium bromide. Silver staining of DNA fragments in polyacrylamide gels is more sensitive than the other staining methods5,6 and has been widely used to detect DNA fragments such as SSR markers7.
Like many biological laboratory techniques, silver staining of polyacrylamide gels has steadily improved since its first being reported as a fragment visualization technique in 19798. The technique was initially modified for detection of DNA fragments by Bassam et al.6 in 1991 and then improved by Sanguinetti and coworkers9 in 1994. The method has been further optimized in the last few decades6,7,9,10,11,12,13,14,15. However, most of these updated versions of the protocols still have some drawbacks such as high technical demand and long processing time for fixation and mounting6, that limit the application of these protocols7,11. An optimal protocol that combines low-cost with high efficiency of DNA fragment detection is urgently needed for routine application of silver staining in biological research.
In addition, polyacrylamide gel can be divided into denaturing and non-denaturing polyacrylamide gels, and both can be used for the detection of SSR markers using the silver staining method. The effect and resolution of which do not significantly differ, but non-denaturing polyacrylamide gels are easier to process and take less time16.
On the basis of the previous research15, the aim of the current study is to describe an optimized silver staining protocol in detail for quick, easy and low-cost detection of SSR markers in a non-denaturing polyacrylamide gel.
Protocol
1. Preparation of PCR Products of SSR Markers
- Prepare all chemicals and reagents for PCR reactions including template DNA (30 ng/µL), 2× PCR master mix (containing 2× PCR buffer, 0.4 mM of each dNTP, 3 mM of MgCl2, 0.1 U/µL of Taq DNA polymerase and dyes), 10 µM of each forward and reverse primers, and distilled or deionized water (dH2O).
NOTE: The SSR markers used in the present study were PT51333 (forward primer sequence: 5 '-GCACCTTTGGTTATCCGACA-3 ' and reverse primer sequence: 5 '-TGCTTTAAGTCATGTACCAAATTGA-3 ') and PT50903 (forward primer sequence: 5 '-AAATTTCTTTCGGTGTGATAACTG-3' and reverse primer sequence: 5'- AGAGACTGCCGTTTGATTTGA -3') for tobacco, and CX-43 (forward primer sequence: 5'-TGGGGATGTGAGCTTCTTCT-3' and reverse primer sequence:5'- AGGGTTCCTTTGGGGTGATA-3') and CX-157 (forward primer sequence: 5 '-TCGACGCTGACTTCACTGAC-3 ' and reverse primer sequence: 5 '-GGACAGCTTCACACATTTGC-3 ') for flowering Chinese cabbage. - Prepare 10 µL of PCR mix for amplification by adding 2 µL of template DNA, 5 µL of 2× PCR master mix, 0.2 µL of each forward primer and reverse primer and 2.6 µL of dH2O.
- Perform PCR amplification reaction in a thermocycler with an initial step of 94 °C for 5 min, follow by 38 cycles of 45 s at 94 °C for denaturation, 45 s at 55 °C for primer annealing and 1 min at 72 °C for extension, and a final extension step of 10 min at 72 °C; then store PCR products at 4 °C for later use.
2. Preparation of Solutions for Polyacrylamide Gels Casting
- Prepare 1 L of 5× TBE buffer by dissolving 54 g of Tris base and 27.5 g boric acid in dH2O, adding 20 mL of 0.5 M EDTA (pH 8.0) and adjusting the solution to a final volume of 1, 000 mL with dH2O.
NOTE: TBE buffer can be stored at room temperature for months. - Prepare 2 L of 0.5× TBE buffer by adding 200 mL of 5× TBE buffer to dH2O to a final volume of 2 L, and store at room temperature.
- Prepare a 6% non-denaturing polyacrylamide gel solution by dissolving 29 g of molecular biology grade acrylamide and 1 g of molecular biology grade N, N'-methylenebisacrylamide in 0.5× TBE buffer to a final volume of 500 mL. Cover the solution bottle with aluminum foil and store at 4 °C for later use.
Caution: Acrylamide is considered a potent neurotoxin and should be handled with care. Always wear gloves when weighing powder and handling solutions that contain it. - Prepare a 20% ammonium persulfate (APS) solution by dissolving 2 g of APS with dH2O to a final volume of 10 mL. It can be stored at -20 °C for months.
NOTE: For convenience, 10 mL of 20% APS solution can be aliquoted into 1.5 mL or 2 mL of centrifuge tubes for storage at -20 °C. Thaw and mix well before use. - Prepare a diluted bind-silane solution by dissolving 3 µL of bind-silane in 0.997 mL of 95% ethanol containing 0.5% of acetic acid. It can be stored at 4 °C for weeks.
Caution: Bind-silane is toxic, wear gloves when handling the solution and keep room ventilated.
3. Preparation of Polyacrylamide Gels for Electrophoresis
- Wash one set of glass plates and spacers with detergent in tap water, followed by a complete rinsing with dH2O. Air dry the plates by setting them in a plate drying rack.
- Disperse 1 mL of bind-silane solution onto the inner surface of a rectangular glass plate, and dry for 5 min.
NOTE: If the bind-silane solution is not spread uniformly on the surface of the plate, the gel may be detached during staining and result in an unsatisfactory staining effect. - Disperse 1 mL of repel-silane onto the inner surface of a notched glass plate with a swab, wipe off excess repel-silane with tissue paper, and air dry for 5 min.
NOTE: If the repel-silane does not coat the surface of the glass plate uniformly, it may damage the gel by partially stripping. - Assemble the glass plates with spacers with the notched plate on top, and clamp both sides of the assembly with casting clamps.
- Pour appropriate amount (40 mL) of 6% non-denaturing polyacrylamide gel solution to a beaker for the plate set of 33 cm width × 10 cm height and spacer thickness of 1.5 mm, add 20 µL of tetramethylethylenediamine (TEMED) and 200 µLfresh 20% APS and mix gently.
NOTE: The appropriate amount (40 mL) of gel solution was calculated from the inner volume of the two plates with a thickness of spacers (30 cm × 8.5 cm × 0.15 cm). As for the amount of TEMED and APS for gel polymerization, there is a standard ratio of gel solution to TEMED and APS based on the reference17. The gel solution described above is stored at 4°C. If gel solution is stored at room temperature, 20 µL of TEMED and 100 µL of 20% APS should be used. Gently stir the mixture of gel solution with a glass stick to avoid air bubbles in the solution. - Pour the solution immediately and carefully into the assembled glass plates along the edge of the notched plate, fill the space almost to the top, and insert the comb. Add a small volume of gel solution over the comb.
NOTE: Keep the glass plates horizontal to prevent the gel from leaking. Remove any bubbles with a bubble hook, if necessary. - Allow the gel to set for 30 min at room temperature.
NOTE: The time for polymerization may change with temperatures of gel solution and environment. - After the gel is fully polymerization, remove the casting clamps and position the plate set in the electrophoresis tank unit with the notched plate facing toward the upper buffer reservoir, and use large clamps to attach the plate set to the tank unit.
- Pour 1 L of 0.5× TBE buffer into each of the upper and the lower chambers. Remove the comb and flush all the wells thoroughly with buffer using a pipette or syringe.
NOTE: Ensure that the gel sandwich at the bottom of the plate is completely soaked in buffer without any bubbles.
4. Running Gels
- Load about 1 µL of PCR product into each well of the polyacrylamide gel. Load a DNA size ladder to both sides of the gel along with samples of PCR products. Fix the safety cover to the upper buffer chamber.
- Connect the leads to the power supply, matching the color-coded red to red and black to black. Run the gel at a constant voltage of 110 V until the dye reaches a defined position, normally for ~70 min.
NOTE: The voltage and running time can be adjusted according to the size of the PCR products.
5. Silver Staining for Detecting SSR Markers in a Non-Polyacrylamide Gel
- After electrophoresis, drain the buffer from the upper chamber into a large beaker via the valve. Detach the side clamps, remove the plates from the apparatus, carefully separate the notched glass plate along one side with a spatula, and make sure that the gel remains attached to the other glass plate coated with bind silane.
- Make 1 L of fresh impregnation solution by dissolving 1.5 g of AgNO3 in 1 L of dH2O. Prepare 1 L of fresh development solution by dissolving 10 g of NaOH in 900 mL of dH2O, add 1 mL of 37% formaldehyde, and adjust to a final volume of 1 L using dH2O.
NOTE: Wear gloves and handle the above chemicals and solutions with care. It is not necessary to keep the solutions and corresponding steps with the solutions in the dark. - Carefully rinse the gel and glass plate with ample dH2O to remove the electrophoresis buffer, then place the plate with the gel facing up onto a plastic tray and submerge the gel in 1 L of impregnation solution. Put the tray on a shaker.
- Shake the tray gently at 60 r/min for 3-4 min.
NOTE: Shaking time can be adjusted according to the gel thickness and concentration of silver nitrate in the impregnation solution. - Move the plate out of impregnation solution and put it onto another tray. Rinse the residual impregnation solution off from the surface of the plate and the gel using ample dH2O twice for 3-5 s each.
- Place the plate with the gel facing up onto another tray, submerge the plate in 1 L of development solution, then shake the tray gently at 50 r/min for ~3 min. Monitor the appearance of DNA fragments and stop development when the highest ratio of the signal of DNA fragments to the noise of background is observed (this step takes ~3 min).
- Rinse the plate and the gel with ample dH2O twice, and dry the gel plate with a tissue paper.
- Scan the gel using an appropriate scanner. A resolution of 300 dpi gives a clear visualization of DNA fragments. The gel can also be photographed using a camera.
- The banding pattern of SSR markers can be scored based on the DNA fragments in the image.
Representative Results
The PCR amplicons were produced using the corresponding SSR primer pairs in flowering Chinese cabbage and tobacco. After electrophoresis, the polyacrylamide gels were stained using the above silver staining protocol, which unambiguously detected the banding patterns of SSR markers (Figure 1).
To compare the detection efficiency of different silver staining protocols, PCR products of SSR markers in tobacco and flowering Chinese cabbage were separated using polyacrylamide gel electrophoresis and visualized using five published silver staining protocols9,11,12,13,14 and the new protocol. All six protocols detected SSR banding patterns for tobacco and flowering Chinese cabbage genotypes (Figure 2), but the present protocol had the lowest background noise and highest contrast of DNA fragments, such that it produced the highest picture clarity for the unambiguous screening of SSR polymorphisms. The running time and number of the main steps and reagents used to complete the silver staining process are listed in Table 1 for each protocol, the new protocol takes the least time and requires fewer chemical reagents and process steps.
Figure 3 shows the sensitivity of the protocol as measured by 50-2,000 bp DNA marker on a non-denaturing polyacrylamide gel.
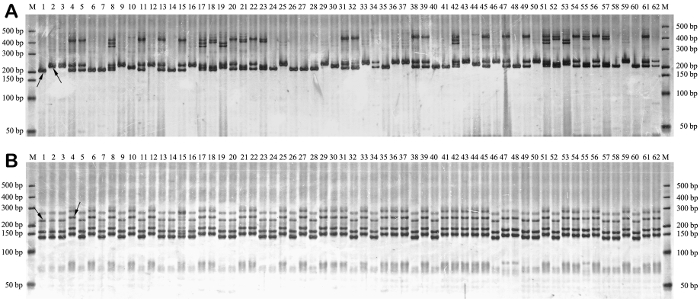
Figure 1: Detection of SSR banding patterns. Detection of SSR banding patterns for genotypes of flowering Chinese cabbage (A) and tobacco (B) using the new silver staining protocol. The PCR products were separated on 6% non-denaturing polyacrylamide gels and stained as per the above-reported silver staining protocol. Lane M is DNA size marker, lanes 1 to 62 represent the amplicons of 62 genotypes amplified by SSR primer pairs of "CX-157" (the sequences of forward and reverse primers are 5'-TCGACGCTGACTTCACTGAC-3' and 5'-GGACAGCTTCACACATTTGC-3', respectively) in flowering Chinese cabbage (Figure 1A) and "PT51333" (the sequences of forward and reverse primers are 5'-GCACCTTTGGTTATCCGACA-3' and 5'-TGCTTTAAGTCATGTACCAAATTGA-3', respectively) in tobacco (Figure 1B). The arrows point to the target SSR bands. Please click here to view a larger version of this figure.
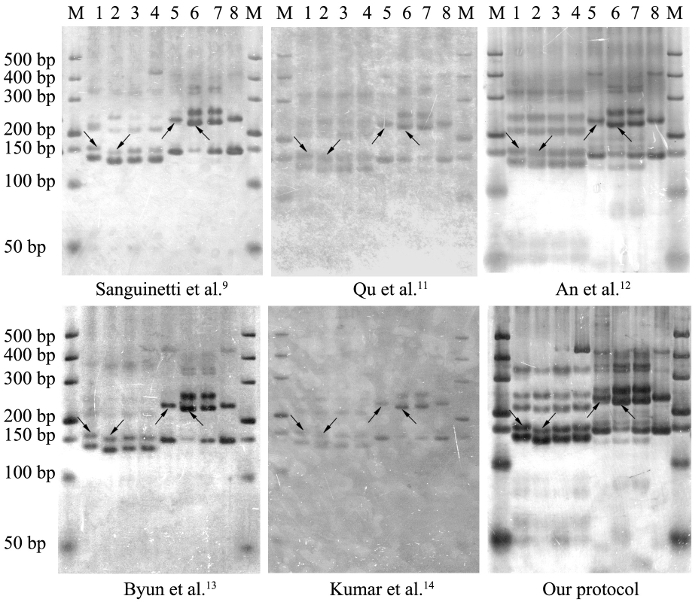
Figure 2: Comparison of the detection efficiency for SSR markers in tobacco and flowering Chinese cabbage genotypes using different silver staining protocols. The PCR products were separated in 6% of non-denaturing polyacrylamide gels and stained using five published silver staining protocols9,11,12,13,14 and the new protocol. Lane M is DNA size marker. Lanes 1 to 4 represent the amplicons of SSR primers of "PT50903" (the sequences of forward and reverse primers are 5'-AAATTTCTTTCGGTGTGATAACTG-3' and 5'-AGAGACTGCCGTTTGATTTGA-3', respectively) in four tobacco genotypes; lanes 5 to 8 indicate the amplicons of SSR primers of "CX-43" (the sequences of forward and reverse primers are 5'-TGGGGATGTGAGCTTCTTCT-3' and 5'-AGGGTTCCTTTGGGGTGATA-3', respectively) in four flowering Chinese cabbage genotypes. The target SSR bands are indicated with arrows. This figure has been modified from the Figure 2 of Liu et al.8 Please click here to view a larger version of this figure.
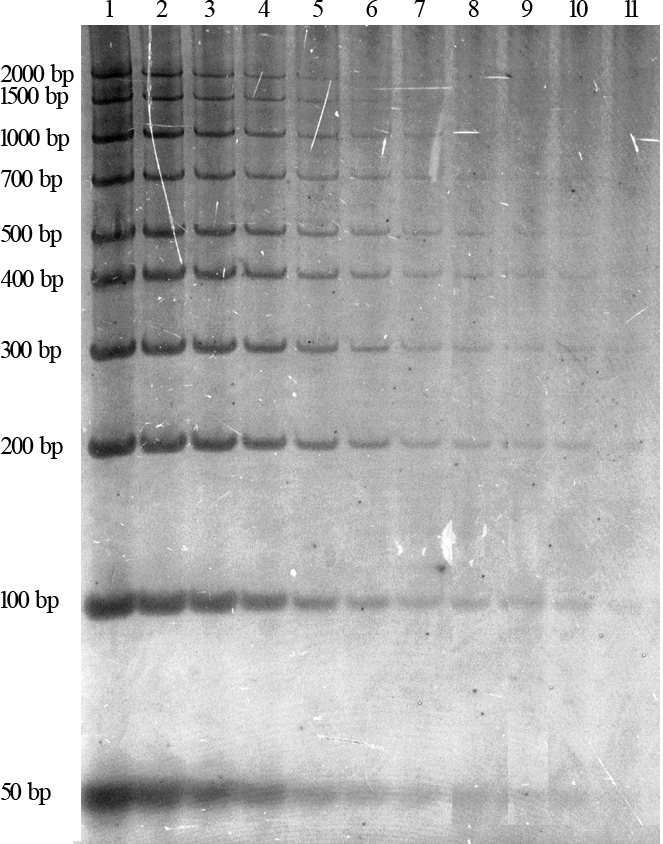
Figure 3: Sensitivity of the protocol as measured by 50-2000 bp DNA marker on a non-denaturing polyacrylamide gel. The first lane was loaded with 1µL of DNA marker with fragment sizes of 50, 100, 200, 300, 400, 500, 600, 700, 1000, 1500 and 2000 bp, each at 10 ng/µL. The remaining samples were loaded with a 1:2 serial dilution in preceding lanes. The minimum detectable amount for the protocol was 9.8 pg in the 11th lane (2.2 pg/mm2 in a 4.5 mm2 well). This figure has been modified from the Figure 1 of Liu et al.8. Please click here to view a larger version of this figure.
| Item | Sanguinetti et al.9 | Qu et al.11 | An et al.12 | Byun et al.13 | Kumar et al.14 | New protocol |
| Running time (min) | 30 | 12–25 | 8–9 | 9–31 | 42 | 6–7 |
| Number of steps | 5 | 4 | 3 | 3 | 3 | 2 |
| Number of reagents | 5 | 6 | 6 | 5 | 7 | 3 |
Table 1: Running time, number of key steps and reagents used in the different silver staining protocols.
Discussion
The washing of gel after impregnation is a critical step. Insufficient washing time and water volume may cause the incomplete removal of impregnation solution on the surface of the plate and the gel, and result in a dark background. The appropriate developing time is another key step, over-development can result in a dark-brown background with low contrast image of DNA fragments. In addition, the impregnation step significantly affects staining efficiency of DNA fragments. Although extending the impregnation time over 5 min or increasing AgNO3 amount in the impregnation solution do not significantly improve the staining quality, a reducing the impregnation time to less than 3 min or decreasing the amount of AgNO3 (<1 g) can result in grey or shallow black DNA fragments.
A quick and easy operation for efficient DNA detection is desirable for a silver staining method. The new protocol developed in this study avoids the fixing, stopping and several washing steps described in other protocols6,9,10,11,12,13,14 without affecting the staining quality (Figure 1 & Figure 2), and only requires two simple steps of impregnation and development that take 7 min. Therefore, the new protocol is faster than all the other staining protocols current available6,7,9,10,11,12,13,14. In addition, the new protocol only requires three chemicals, i.e. AgNO3, formaldehyde, and NaOH, at similar amounts as that used in other protocols6,7,9,10,11,12,13,14, therefore, the new protocol is more economical and generates less chemical hazards, which not only reduces the chemical cost but also minimizes the hazardous material handling process of extra chemicals in the laboratory.
The new protocol produced clear images with low background noise for the unambiguous detection of SSR banding patterns in tobacco and flowering Chinese cabbage genotypes (Figure 1). Compared with other protocols, the new protocol produced the best staining effect with the lowest background noise and highest picture contrast (Figure 2). At the range of 50-500 bp, the sensitivity of the new protocol for DNA detection was less than 19.5 pg/µL (4.3 pg/ mm2) with a minimum concentration of 9.8 pg/µL (2.2 pg/mm2) (Figure 3), which is higher than that reported in other protocols7,12,13.
Although fluorescence-labeling technologies can provide high throughput and resolution detection of DNA amplicons, they require specialized equipment and more expensive reagents that may not be accessible for many biological laboratories, especially those in developing countries. The new staining protocol is simple and fast that can screen ~800 DNA samples per person per day, thus, is a good option for these laboratories to perform routine research.
In conclusion, the new protocol developed in this study is quicker to process, easier to implement, cheaper, and only uses three reagents—without compromising detection effect or picture clarity—than all other existing silver staining techniques. The new silver staining protocol has been used successfully in tobacco and flowering Chinese cabbage research and can be a valuable tool for successful SSR genotyping in other species.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was funded by the Guangdong Natural Science Foundation of China (2015A030313500), the Provincial Key International Cooperative Research Platform and the Major Scientific Research Project of Guangdong Higher Education (2015KGJHZ015), the Science and Technology Plan of Guangdong Tobacco Monopoly Administration (201403, 201705), the Science and Technology Plan of Guangdong of China (2016B020201001), the National Innovation Training Project for Undergraduate Students (201711078001). Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Materials
| PCR master mix (Green Taq Mix) | Vazyme Biotech Co. Ltd, China | #P131-03 | |
| 50-2000 bp DNA Ladder | Bio-Rad, USA | #170-8200 | |
| DL500 DNA marker | Takara Bio Inc., Japan | #3590A | |
| Tris base | Sangon Biotech Shanghai, China | #77-86-1 | |
| Boric acid | Sangon Biotech Shanghai, China | #10043-35-3 | |
| EDTA-Na2 | Guangzhou Chemical Reagent Factory, China | #6381-92-6 | |
| Acrylamide | Sangon Biotech Shanghai, China | #79-06-1 | |
| N,N'-methylene-bis-acrylamide | Sangon Biotech Shanghai, China | #110-26-9 | |
| N,N,N',N'-Tetramethylethylenediamine | Sangon Biotech Shanghai, China | #110-18-9 | |
| Ammonium persulfate | Guangzhou Chemical Reagent Factory, China | #7727-54-0 | |
| Bind-silane | Solarbio Beijing, China | #B8150 | |
| AgNO3 | Sinopharm Chemical Reagent Beijing Co.,Ltd, China | #7761-88-8 | |
| Formaldehyde | Tianjin DaMao Chemical Reagent Factory, China | #50-00-0 | |
| NaOH | Guangzhou Chemical Reagent Factory, China | #1310-73-2 | |
| Acetic acid | Guangzhou Chemical Reagent Factory, China | #64-19-7 | |
| Na2CO3 | Tianjin DaMao Chemical Reagent Factory, China | #497-19-8 | |
| Ethanol | Guangzhou Chemical Reagent Factory, China | #64-17-5 | |
| HNO3 | Guangzhou Chemical Reagent Factory, China | #7697-37-2 | |
| Na2S2O3.5H2O | Sinopharm Chemical Reagent Beijing Co.,Ltd, China | #10102-17-7 | |
| Eriochrome black T(EBT) | Tianjin DaMao Chemical Reagent Factory, China | #1787-61-7 | |
| Plastic tray | Shanghai Yi Chen Plastic Co., Ltd, China | – | |
| TS-1 Shaker | Qilinbeter JiangSu, China | – | |
| BenQ M800 Scanner | BenQ, China | – | |
| DYY-6C Power supply | Beijing Liuyi Instrument Factory, China | – | |
| High throughout vertical gel systems, JY-SCZF | Beijing Tunyi Electrophoresis Co., Ltd, China | – |
References
- Jiang, G. L., Anderson, S. B. Molecular markers and marker-assisted breeding in plants. Plant breeding from laboratories to fields. , 45-83 (2013).
- Powell, W., Machray, G. C., Provan, J. Polymorphism revealed by simple sequence repeats. Trend Plant Sci. 1, 215-222 (1996).
- Varshney, R. K., Graner, A., Sorrells, M. E. Genic microsatellite markers in plants: features and applications. Trend Biotechnol. 23, 48-55 (2005).
- Tuler, A. C., Carrijo, T. T., Nóia, L. R., Ferreira, A., Peixoto, A. L., da Silva Ferreira, M. F. SSR markers: a tool for species identification in Psidium (Myrtaceae). Mol. Biol. Rep. 42, 1501-1513 (2015).
- Rabilloud, T. Mechanisms of protein silver staining in polyacrylamide gels: a 10-year synthesis. Electrophoresis. 11, 785-794 (1990).
- Bassam, B. J., Caetano-Anollés, G., Gresshoff, P. M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196, 80-83 (1991).
- Liang, Q., et al. A rapid and effective method for silver staining of PCR products separated in polyacrylamide gels. Electrophoresis. 35, 2520-2523 (2014).
- Switzer, R. C., Merril, C. R., Shifrin, S. A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal. Biochem. 98, 231-237 (1979).
- Sanguinetti, C., Dias, N., Simpson, A. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques. 17, 915-919 (1994).
- Ji, Y., Qu, C., Cao, B. An optimal method of DNA silver staining in polyacrylamide gels. Electrophoresis. 28, 1173-1175 (2007).
- Qu, L., Li, X., Wu, G., Yang, N. Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis. 26, 99-101 (2005).
- An, Z., et al. A silver staining procedure for nucleic acids in polyacrylamide gels without fixation and pretreatment. Anal. Biochem. 391, 77-79 (2009).
- Byun, S. O., Fang, Q., Zhou, H., Hickford, J. G. H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 385, 174-175 (2009).
- Kumar, M., Kim, S. R., Sharma, P. C., Pareek, A. Simple and efficient way to detect small polymorphic bands in plants. Genome Data. 5, 218-222 (2015).
- Liu, W., et al. Development of a simple and effective silver staining protocol for detection of DNA fragments. Electrophoresis. 38, 1175-1178 (2017).
- Wang, D., Shi, J., Carlson, S. R., Cregan, P. B., Ward, R. W., Diers, B. W. A low-cost, high-throughput polyacrylamide gel electrophoresis system for genotyping with microsatellite DNA markers. Crop Sci. 43, 1828-1832 (2003).
- Echt, C. S., May-Marquardt, P., Hseih, M., Zahorchak, R. Characterization of microsatellite markers in eastern white pine. Genome. 39, 1102-1108 (1996).

