Deep Dermal Injection As a Model of Candida albicans Skin Infection for Histological Analyses
Summary
Here we describe a protocol that allows histological and molecular analysis of skin samples after Candida albicans intradermal injection. This protocol maintains the structural integrity of the skin and allows for the localization of tissue-resident or newly recruited immune cells as well as the pathogen distribution.
Abstract
The skin is an extremely extended organ of the body and, due to this large surface, it is continuously exposed to microorganisms. Skin damage can easily lead to infections in the dermis which can, in turn, result in the dissemination of pathogens into the bloodstream. Understanding how the immune system fights infections at the very early stage and how the host can eliminate the pathogens is an important step to set the base for future therapeutic interventions. Here we describe a model of Candida albicans infection that can visualize the processes that occur early during an infection, including when the pathogen has passed the epithelial barrier, as well as the immune response elicited by the C. albicans invasion. We used this infection model to perform histological analyses that show the immune cells that infiltrate the skin as well as the presence and localization of the pathogen. Samples collected after the infection can be processed for RNA extraction.
Introduction
The human body is covered with an extremely high number of microorganisms. The skin surface is the habitat of almost one million bacteria per square centimeter1. This number, however, does not reflect the full variety of microorganisms that colonizes the skin. In addition to bacteria, the human body is colonized by many fungal species, including C. albicans, which is able to survive both at the mucosal and the skin level2.
In recent years, the percentage of people diagnosed with fungal infections has enormously increased. This is mainly due to the higher number of immunocompromised persons, i.e., HIV-positive patients and patients that have gone through either chemotherapy or immunosuppressive drugs after transplantation3. In a surveillance study performed in the United States, Wisplinghoff et al. showed that 9.5% of the nosocomial bloodstream infections were caused by Candida species4. Due to the increased occurrence of fungal infections, and especially because of the elevated percentage of Candida species found during bloodstream infections, understanding how this pathogen escapes the control of the immune system is extremely important.
C. albicans is a dimorphic fungus that grows in different morphological forms such as yeast, blastospores, pseudohyphae, and hyphae depending on the environmental conditions5. In its hyphal form, C. albicans shows its highest invasiveness capacity and has the ability to penetrate the epithelium6.
C. albicans infections have been studied using several experimental approaches. The most common model of infection is the intravenous injection of C. albicans yeast7. However, this model does not take into consideration all the processes that happen before the fungus manages to spread into the bloodstream. Another model takes advantage of the ability of C. albicans to invade the epithelium. This method, also known as the sand paper model8, was developed by Gaspari et al. in 19989, and consists of using sand paper to abrade the skin, thus eliminating the stratum corneum before applying C. albicans. This procedure allows the fungus to penetrate the epithelium, thus enabling the analysis of the invasive abilities of this pathogen. Finally, other models of infections for the gastrointestinal10 and respiratory tracts11 have been used in different studies.
The formation of a wound (as in the sand paper model) causes the activation of several pathways, including immune cell recruitment and activation, in order to promote the healing process12. This can either alter or mask the immune response specifically elicited against the pathogen, thus leading to confounding results.
Here we describe a method of skin infection that avoids initial wound formation and the induction of a basal inflammatory environment. To maintain the intact epithelial structure, we directly inject C. albicans in its hyphal form in the deep derma. Even though a single injection can elicit mild inflammation, the amount of inflammation is limited and restricted compared to the formation of an open wound as in the sand paper model. The approach that we describe here allows the study of the immune response to fungal infection and spread while avoiding the excessive and pre-existing inflammatory environment caused by mechanical damage.
Protocol
All the procedures were approved under the Institutional Animal Care and Use Committee (IACUC) and operated under the supervision of the department of Animal Resources at Children's Hospital (ARCH) at Boston Children's Hospital or were approved by the Italian Ministry of Health and performed under the Institutional Animal Care Committee at the University of Milano-Bicocca.
1. C. albicans Preparation
- Culture C. albicans, strain CAF3-113, in tubes containing rich medium (yeast extract peptone dextrose (YEPD), 1% (w/v) yeast extract, 2% (w/v) of Bacto Peptone, 2% (w/v) glucose) supplemented with uridine (50 mg/L) at 25 °C. Under these conditions, C. albicans grows as a yeast.
NOTE: Other C. albicans strains can be used, such as the clinical isolate C. albicans strain SC531413. Since we have recently demonstrated that the ulcerative process is induced by the activation of the innate immune system14, theoretically every C. albicans strain that maintains both its cell wall components and structure, as well as the ability to originate hyphal projections, could be used. - Monitor the culture by counting the cellular concentration using a cell counter analyzer.
- Harvest the culture when it reaches a concentration of about 8 x 106 cells/mL. Alternatively, use a spectrophotometer to measure the OD600.
NOTE: Usually, a value of OD600 = 1 corresponds to a yeast concentration of 3 x 107 cells/mL, but titration is recommended. - Resuspend the culture in YEPD-uridine medium, using HEPES (50 mM, pH 7.5) as a buffer agent and incubate the culture at 37 °C to induce hyphae formation. Check hyphal formation under a microscope at 100X magnification. Hyphal formation is visible 5 h after culturing at 37 °C; however, use the cells 16 h after incubation at 37 °C when the hyphal percentage reaches almost 95% of the total culture.
- To further enrich the preparation in hyphal concentration, centrifuge aliquots of the culture at 3,300 x g for 5 min. Discard the supernatant and resuspend the pellet in sterile PBS at a concentration of 1 x 108 hyphae/mL.
2. Deep Dermal Injection
- 24 h prior the infection procedure, anesthetize the mice with an intraperitoneal injection of ketamine (80 – 100 mg/kg) and xylazine (10 mg/kg), and then shave the flank of the mice using an electric clipper.
NOTE: After any procedure involving anesthesia, place the mice under a heating lamp until they have efficiently recovered. - Rest the mice for 24 h to avoid any inflammation due to the shaving process.
- 24 h after the shaving procedure, anesthetize the mice with an intraperitoneal injection of ketamine (80 – 100 mg/kg) and xylazine (10 mg/kg).
- Check the paw reflex to ensure that the mice are deeply anesthetized. Assess the paw reflex by firmly pinching the paw. If the anesthesia is achieved, the animal does not feel pain and does not move.
- Inject C. albicans hyphae in the deep derma using a 0.3 mL insulin syringe with a 30 G x 8 mm needle in a final volume of 50 µL/injection, corresponding to 5 x 106 hyphae/injection.
- Pull the skin of the shaved flank taut using two fingers in order to inject the volume directly in the deep derma. Insert the needle with an angle of 10 – 15° with the bevel of the needle facing up.
- With this angle, the pathogen will be injected with a depth of approximately 300 – 500 µm. To ensure the injection is well performed, check that the injected volume forms a bump that is absorbed a few min after the injection.
NOTE: The bump formed after the injection will be of approximately 1 cm2. This is the area that will be removed at the designated time point for either molecular or histological analysis. - For time points of less than 24 h (recommended) mark the area of infection with a marker pen, since the bump formed with the injection persists for a few minutes, after which it is absorbed. For time points of 24 h or more, this step is not required, because either an ulcer or a cyst will be clearly visible.
3. Sample Collection and Preparation for Histological Staining
- Euthanize the mice according to institutional procedures.
- Using surgical forceps, pull the skin at the border of the injected site, previously marked with the marker pen. Using surgical scissors, excise the infected site by cutting the skin following the marked line.
NOTE: Be careful to separate the skin from the peritoneum using round tip scissors. - Immerse the skin in a disposable base mold filled with optimal cutting temperature (OCT) compound, with the internal side of the skin facing up. Freeze the sample in liquid nitrogen until the compound becomes white. Do not let the liquid nitrogen cover the sample before it is completely frozen as this would damage the sample.
NOTE: Caution! During step 3.3, wear a face shield for protection from liquid nitrogen. - If the samples are not used immediately for slide preparation, rapidly store them at -80 °C.
NOTE: Samples can be stored at -80 °C indefinitely.
4. Preparation of Slides
- Cut the sample in half to prepare the slices for histology starting from the center of the sample.
- Cut 5-µm thick slices with a cryostat.
NOTE: For skin samples, the temperature of the cryostat should not be higher than -25 °C. A higher temperature would cause a partial melting of the H-OCT and therefore the samples would not be maintained in the right position during the cutting procedure. - Use positively charged microscope slides to collect the slices.
- If the collected slices are not used soon after the preparation, store them at -20 °C. At this point, slices can be stored at -20 °C indefinitely.
5. Hematoxylin & Eosin Staining
- Stain the slides by immersion in Gill's hematoxylin for 4 min. Wash the slides in running tap water for 5 min.
- Stain the slides by immersion in Eosin Y solution for 1 min. Wash the slides in running tap water for 5 min. Rinse the slides using distilled water before continuing the protocol.
- Dehydrate by immersion in serially increasing concentrations of ethanol solutions (50%, 70%, 80%, 95%, 100%). Immerse the slides for 15 s in each ethanol solution.
- Clear the slides for at least 2 min by immersion in a histological clearing agent.
- Mount the slides using mounting medium and cover slips.
- Acquire images of the slides with a microscope slide scanner at a 40X magnification.
6. Periodic Acid-Schiff (PAS) Staining
- Fix the slides with ≥99.5% acetone at room temperature for 1 min. Wash the slides in running tap water for 1 min.
- Stain the slides by immersion in periodic acid solution (1 g/dL) for 5 min. Wash the slides in 3 changes of distilled water.
- Stain the slides by immersion in Schiff's reagent for 15 min. Wash the slides in running tap water for 5 min.
- Stain the slides by immersion in Gill's hematoxylin for 5 min. Wash the slides in running tap water for 30 s.
- Dehydrate the slides by immersion in 3 changes of 100% ethanol. Immerse the slides for 15 s each time.
- Clear the slides for at least 2 min by immersion in a histological clearing agent.
- Mount the slides using mounting medium and cover slips.
- Acquire images of the slides with a microscope slide scanner.
7. Sample Collection and Preparation for RNA Extraction
- Euthanize the mice according to institutional procedures. Remove the skin samples as in step 3.2.
- Immerse the samples in a 2 mL snap-cap tube with 1 mL of reagent for acid guanidinium thiocyanate-phenol-chloroform extraction.
NOTE: The protocol can be paused here and the samples can be stored at -80 °C. - Cut the sample into pieces of approximately 0.2 cm2 using surgical forceps.
NOTE: Caution! Most of the reagents for RNA isolation are toxic and mutagenic. Steps 7.3 and 7.4 must be done under a fume hood. - Smash the sample with a bead mill at a speed of 20 oscillations/s for 20 min. Alternatively, a mechanical potter can be used.
- Check the effective homogenization of the samples by looking for the presence of intact pieces of skin. Repeat this step if the samples are not completely homogenized.
- Centrifuge the samples at 16,000 x g for 1 min. Keep the supernatants for RNA extraction and discard the pellet. This step is performed to eliminate hair and debris that could block the extraction columns.
- Proceed with RNA extraction using RNA purification columns14.
- Use a spectrophotometer to assess RNA purification and concentration. If RNA extracts are not used immediately for cDNA preparation, store the samples at -80 °C.
Representative Results
Through the injection of the pathogen directly in the deep derma, the structural morphology of the tissue remains intact (Figure 1A).
The maintenance of skin structural integrity allows the detection of immune cells and their localization at the site of infection. The higher magnification shown in Figure 1B reveals that the abscess is mainly composed of polymorphonuclear leukocytes (PMCs) that contain the spread of the pathogen by its confinement to the site of infection. Recruitment of PMCs, such as neutrophils, allows the formation of the abscess, thus avoiding the spreading of the fungus within the tissue14. Higher magnifications (Figure 1C, D) show how the surrounding areas of the abscess are also enriched in PMCs.
By maintaining the structural integrity, the localization of the pathogen during the infection can be determined. Taking advantage of the elevated content of polysaccharides in the cell wall of C. albicans, the identification of the pathogen was performed using PAS staining, which gives a purple-magenta color to the fungus. As shown in Figure 2A, PAS staining clearly shows that the pathogen is confined inside the abscess, formed by the recruitment of granulocytes14. C. albicans is clearly visible at the higher magnification (Figure 2B), and it appears to be surrounded by necrotic and immune cells.
Using the method described above, the infection process and the consequent inflammation can be followed over time. At the very beginning, immune cell recruitment leads to the formation of an abscess (Figure 1A and Figure 3A). When the analysis of the skin tissue was prolonged up to 48 – 72 h, the disruption of the skin structures and the formation of a scar was visualized (Figure 3B, C). The formation of this structure is due to the healing phase that follows the expulsion of the pathogen14.
The process that leads to the formation of an ulcer can be followed for several days after the infection. As shown in Figure 4A, C57BL/6 wild-type mice display an ulcerative process that increases over time. Around 70 – 80% of the mice show formation of an ulcer 72 h after infection. Due to experimental variability, at least 8 mice should be used in every experiment. For statistical analysis, it is recommended to use a minimum of 8 – 10 mice for each ulceration score if comparing different genotypes or treatments. In this case, even small differences should be visible. Illustrative pictures of the ulcerative process are shown in Figure 4B. Upon specific treatments or genetic deficiencies, the formation of the ulcer can be reduced and/or abrogated14. In some cases, instead of the ulcer, a cyst is formed, as illustrated in Figure 4C.
Other than Hematoxylin & Eosin and PAS staining, the slides can be stained for either specific cellular markers or with other immunohistochemical stainings to better visualize: collagen fibers; cytokine/chemokines location within the tissue; and immune cell identity and spatial distribution.
In addition to histological preparation, samples can be used also for molecular analyses. The entire injection site can be excised, as for histological preparations, and processed for RNA extraction. RNA extraction from the tissue allows the study and identification of those genes upregulated during the infection with the pathogen. The possibility to process the samples for molecular analyses is an important tool, together with histological analysis, in order to better characterize the type of immune response elicited against the pathogen and its regulation during the early and late stages after the challenge14.
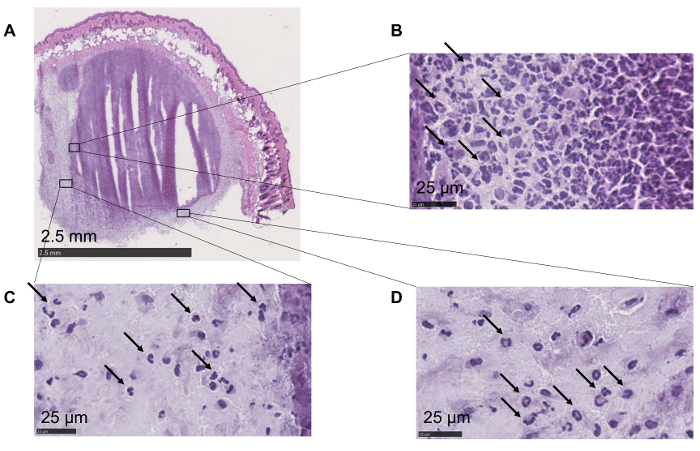
Figure 1: Maintenance of the skin morphology and cellular localization. (A) All the layers are preserved. Epithelium, adipocytes, and muscle cells are easily recognizable. (B) Polymorphonuclear leukocytes (highlighted with arrows) can be identified within the abscess. (C, D) Higher magnifications show the presence of polymorphonuclear leukocytes (arrows) in the area surrounding the abscess. Please click here to view a larger version of this figure.
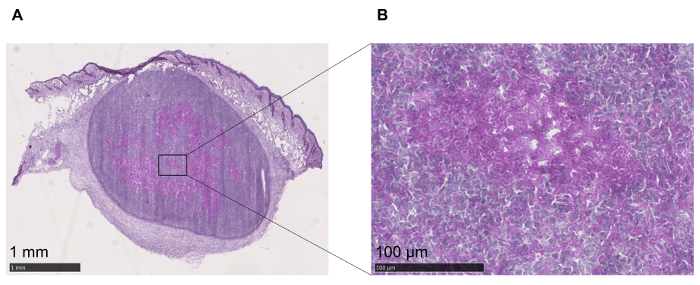
Figure 2: Identification of C. albicans. (A) C. albicans can be detected within the tissue with PAS staining. (B) Higher magnification to better visualize the fungus, stained in purple, confined inside the abscess. Please click here to view a larger version of this figure.
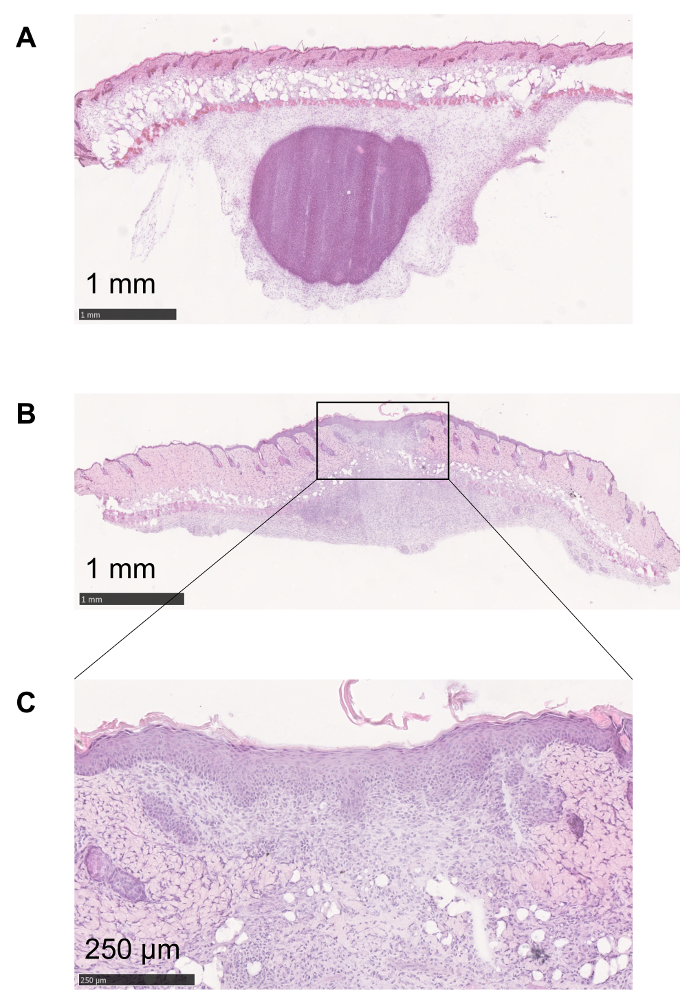
Figure 3: Follow up of the inflammatory process. (A) At the very beginning of the inflammatory process elicited by C. albicans injection, granulocytes are recruited in order to form an abscess to confine the pathogen. (B, C) The formation of an ulcer is necessary for the expulsion of the fungus. The healing tissue is characterized by an alteration in the structural integrity of the skin and can lead to the formation of a scar. Please click here to view a larger version of this figure.
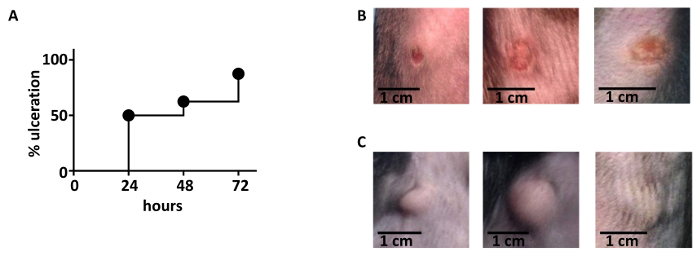
Figure 4: Kinetics of ulceration. (A) The ulcerative process follows a trend of increase during the first days after the infection. 72 h after the infection, 70 – 80% of the mice shows an ulcer at the site of infection. 16 C57BL/6 wild type animals were used. (B) Illustrative pictures of ulcers that developed 48 h after infection. (C) Illustrative pictures of cyst formation after infection. Please click here to view a larger version of this figure.
Discussion
Here we described a method of C. albicans infection to study the inflammatory process that is initiated upon fungal entrance in the deep derma.
Even though skin abscess formation is a relatively rare event upon C. albicans infection15, injection of the fungus directly in the deep derma allows not only the study of fungal-driven abscess formation, but also the analysis of specific immune cells that participate to contain fungal spread. With the method described here, it is possible to study the interaction between immune cells, as well as the recruitment of the immune cells that are important to control the fungal spread, while avoiding any inflammatory response to mechanical damage or wound formation. Indeed, the possibility to keep the epithelial structure intact is important to avoid the development of an inflammatory environment caused by a mechanical stimulus, such as an abrasion. As theorized for the first time by Polly Matzinger in 199416, damage signals can, in fact, activate an immune response and, consequently, potently influence the immune response elicited by the pathogen.
Damage associated molecular patterns (DAMPs) can originate from dead cells but they can also be released under stress conditions. Since these signals can be produced in situations of tissue damage, the formation of a wound would cause a hyper-inflammatory environment even before the context of an infection. The method we have described limits the release of DAMPs and thus reduces the presence of confounding elements.
Moreover, the injection of a pathogen directly in the deep derma is a methodology that can be used not only for fungal infections but also for bacterial challenges. Another pathogen responsible for an elevated number of cases of both cutaneous and bloodstream infections is Staphylococcus aureus4. We have recently shown how this protocol is also an important tool to study S. aureus infection14. In particular, S. aureus is known in clinic for its ability to form abscesses in several districts of the body17. Injection of S. aureus in the deep derma leads to the formation of abscesses, thus enabling at the same time the study of the very early events driven by S. aureus encounter as well as the involvement of the immune responses that lead to the healing phase.
Finally, another important feature of the methodology described here is that the maintenance of an intact epithelial structure gives the possibility to histologically analyze the localization of immune cells and the activation of distinct pathways that can be spatially compartmentalized. Under these experimental conditions, in fact, the different anatomical structures of the skin are not altered and both immune cells and inflammatory mediators can be visualized in a precise location.
The use of this method that preserves the spatial distribution of immune cells is therefore ideal to better understand the roles of each cellular type during fungal and bacterial skin infections.
Disclosures
The authors have nothing to disclose.
Acknowledgements
FG is supported by Associazione Italiana per la Ricerca sul Cancro (IG 2016Id.18842), Cariplo Foundation (Grant 2014-0655), and Fondazione Regionale per la Ricerca Biomedica (FRRB).
IZ is supported by NIH grant 1R01AI121066-01A1, 1R01DK115217, HDDC P30 DK034854 grant, Harvard Medical School Milton Found, CCFA Senior Research Awards (412708), the Eleanor and Miles Shore 50th Anniversary Fellowship Program, and the Cariplo Foundation (2014-0859).
Materials
| Reagents | |||
| PBS | Euroclone | ECB4053L | warm in 37 °C bath before use |
| H-OCT compound | histo-line laboratories | R0030 | |
| Gill's Hematoxilyn | histo-line laboratories | 09-178-2 | |
| Eosin Y solution, alcoholic | histo-line laboratories | 09-209-05 | |
| Ethanol absolute | scharlau | ET00232500 | |
| Citro-HISTOCLEAR | histo-line laboratories | R0050 | |
| Eukitt, mounting medium | bio-optica | ||
| Acetone | sigma-aldrich | 179124 | |
| PAS staining system | sigma-aldrich | 395B-1KT | |
| Bacto Peptone | BD | 211677 | |
| Bacto Yeast Extract | BD | 212750 | |
| D(+)-Glucose anhydrous for molecular biology | Applichem PanReac | 50-99-7 | |
| Uridine | Merck Millipore | 8451 | |
| HEPES | Applichem PanReac | A1070,0500 | |
| Safe-Lock tubes 2 mL | eppendorf | 30121597 | |
| TRIzol Reagent | Life Technologies | 15596018 | Toxic, corrosive and mutagen. Use all precaution needed |
| Rneasy Mini Kit | QIAGEN | 74104 | |
| liquid nitrogen | Wear eye protection | ||
| Instrument | |||
| Coulter Counter-Particle Count | Beckman Coulter | ||
| Centrifuge 5415 R | eppendorf | ||
| MC 3000 Microtome Cryostat | histo-line laboratories | ||
| TissueLiser | QIAGEN | ||
| Materials | |||
| 0.3 ml Insulin Syringe with a 30G x 8mm needle | BD | 324826 | |
| Surgical forceps | |||
| Surgical scissors | |||
| Base mould disposable | histo-line laboratories | 2781 | |
| Positively charged bio microscope slides | bio-optica | 09-2000 | |
| Cover slips 24 x 50 mm | thermo scientific | 11911998 |
References
- Belkaid, Y., Segre, J. A. Dialogue between skin microbiota and immunity. Science. 346 (6212), 954-959 (2014).
- Kashem, S. W., Kaplan, D. H. Skin Immunity to Candida albicans. Trends Immunol. 37 (7), 440-450 (2016).
- Havlickova, B., Czaika, V. A., Friedrich, M. Epidemiological trends in skin mycoses worldwide. Mycoses. 51, 2-15 (2008).
- Wisplinghoff, H., Bischoff, T., Tallent, S. M., Seifert, H., Wenzel, R. P., Edmond, M. B. Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases from a Prospective Nationwide Surveillance Study. Clin Infect Dis. 39 (3), 309-317 (2004).
- Romani, L. Immunity to fungal infections. Nat Rev Immunol. 4 (1), 11-24 (2004).
- Odds, F. C. Pathogenesis of Candida infections. J Am Acad Dermatol. 31 (3), S2-S5 (1994).
- MacCallum, D. M., Odds, F. C. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 48 (3), 151-161 (2005).
- Dai, T., Kharkwal, G. B., Tanaka, M., Huang, Y. -. Y., Bil de Arce, V. J., Hamblin, M. R. Animal models of external traumatic wound infections. Virulence. 2 (4), 296-315 (2018).
- Gaspari, A. A., Burns, R., Nasir, A., Ramirez, D., Barth, R. K., Haidaris, C. G. CD86 (B7-2), but not CD80 (B7-1), expression in the epidermis of transgenic mice enhances the immunogenicity of primary cutaneous Candida albicans infections. Infect Immun. 66 (9), 4440-4449 (1998).
- Koh, A. Y. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot Cell. 12 (11), 1416-1422 (2013).
- Mear, J. B., et al. Candida albicans Airway Exposure Primes the Lung Innate Immune Response against Pseudomonas aeruginosa Infection through Innate Lymphoid Cell Recruitment and Interleukin-22-Associated Mucosal Response. Infect Immun. 82 (1), 306-315 (2014).
- Leoni, G., Neumann, P. -. A., Sumagin, R., Denning, T. L., Nusrat, A. Wound repair: role of immune-epithelial interactions. Mucosal Immunol. 8 (5), 959-968 (2015).
- Fonzi, W. A., Irwin, M. Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 134 (3), 717-728 (1993).
- Santus, W., et al. Skin infections are eliminated by cooperation of the fibrinolytic and innate immune systems. Sci Immunol. 2 (15), (2017).
- Florescu, D. F., Brostrom, S. E., Dumitru, I., Kalil, A. C. Candida albicans Skin Abscess in a Heart Transplant Recipient. Infect Dis Clin Pract. 18 (4), 243-246 (2010).
- Pradeu, T., Cooper, E. L. The danger theory: 20 years later. Front Immunol. 3, 287 (2012).
- Cheng, A. G., DeDent, A. C., Schneewind, O., Missiakas, D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 19 (5), 225-232 (2011).

