Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ
Summary
Microglia are brain immune cells that survey and react to altered brain physiology through morphologic changes which may be evaluated quantitatively. This protocol outlines an ImageJ based analysis protocol to represent microglia morphology as continuous data according to metrics such as cell ramification, complexity, and shape.
Abstract
Microglia are brain phagocytes that participate in brain homeostasis and continuously survey their environment for dysfunction, injury, and disease. As the first responders, microglia have important functions to mitigate neuron and glia dysfunction, and in this process, they undergo a broad range of morphologic changes. Microglia morphologies can be categorized descriptively or, alternatively, can be quantified as a continuous variable for parameters such as cell ramification, complexity, and shape. While methods for quantifying microglia are applied to single cells, few techniques apply to multiple microglia in an entire photomicrograph. The purpose of this method is to quantify multiple and single cells using readily available ImageJ protocols. This protocol is a summary of the steps and ImageJ plugins recommended to convert fluorescence and bright-field photomicrographs into representative binary and skeletonized images and to analyze them using software plugins AnalyzeSkeleton (2D/3D) and FracLac for morphology data collection. The outputs of these plugins summarize cell morphology in terms of process endpoints, junctions, and length as well as complexity, cell shape, and size descriptors. The skeleton analysis protocol described herein is well suited for a regional analysis of multiple microglia within an entire photomicrograph or region of interest (ROI) whereas FracLac provides a complementary individual cell analysis. Combined, the protocol provides an objective, sensitive, and comprehensive assessment tool that can be used to stratify between diverse microglia morphologies present in the healthy and injured brain.
Introduction
Microglia have an immediate and diverse morphologic response to alterations in brain physiology1 along a continuum of possibilities that range from hyper-ramification and highly complex morphologies to de-ramified and amoeboid morphologies2. Microglia may also become polarized and rod-shaped3. Microglia cell ramification is commonly defined as a complex shape having multiple processes and is often reported as the number of endpoints per cell and the length of cell processes. Since microglia are finely tuned to neuronal and glial function through continuous cell-cell cross-talk and in vivo motility4,5, microglia morphologies may serve as indicators of diverse cell functions and dysfunctions in the brain. A quantitative approach is necessary to adequately describe the diversity of these morphologic changes and to distinguish the differences among ramified cells that occur with subtle physiologic perturbations (such as epilepsy5,6 and concussion7) in addition to gross injury (such as stroke8). An increased use of morphology quantification7,8,9,10,11,12,13,14 will reveal the full diversity of microglia morphologies during health and disease. The present study details the stepwise use of ImageJ plugins necessary to summarize microglia morphology from fluorescent or non-fluorescent photomicrographs of microglia acquired in fixed rodent tissue after immunohistochemistry (IHC).
Central to the analysis techniques described here are the ImageJ plugins AnalyzeSkeleton (2D/3D)15, developed in 2010 to quantify large mammary structures, and FracLac16, developed in 2014 to integrate ImageJ and fractal analysis to quantify individual microglia shapes. These plugins provide a rapid analysis of microglia ramification within entire photomicrographs or multiple microglia of a defined ROI within a photomicrograph. This analysis may be used alone or in complement with fractal analysis. The single-cell fractal analysis (FracLac) requires an investment of time but provides multiple morphology outputs regarding microglia complexity, shape, and size. The use of both tools is not redundant, as cell ramification is complementary to cell complexity, and the combination of multiple parameters may be used to distinguish between diverse microglia morphologies within datasets12,17.
Protocol
All experiments were approved by and performed in accordance with the guidelines established by the University of Arizona Institutional Animal Care and Use Committee and the NIH guidelines for the care and use of laboratory animals. Care was taken to minimize animal pain and discomfort. Euthanasia methods are according to an approved protocol and consist of cervical decapitation under isoflurane anesthesia.
1. Tissue Preparation
NOTE: Carry out microglia morphology analysis on fixed, cryoprotected tissue samples to preserve cell morphology. The following is a standard protocol to prepare and directly slice fixed tissue for fluorescence IHC.
- Remove mouse or rat brains from a euthanized animal after the desired experiment and according to a standard laboratory protocol. Place the brain into a 10 mL vial containing 5 mL of a 4% paraformaldehyde solution for 24 h at 4 °C. Then rinse and place into 5-10 mL of a 30% sucrose phosphate buffered solution (PBS, 0.01 M) for 72 h at 4 °C. Store whole brains at -80 °C until tissue sectioning with a cryostat, or at 4 °C if sectioning with a microtome.
NOTE: This protocol has not yet been tested using tissue sliced from paraffin-embedded tissue. - Section the brain tissue to the desired section thickness and orientation using either a cryostat or microtome and store free-floating sections in a cryoprotection solution (50 mM PBS, ethylene glycol, glycerol) at -20 °C.
NOTE: This protocol has been successfully carried out on coronal tissue sections ranging from 50 µm to 200 µm in thickness. Tissue sections less than 50 µm may not capture the full span of microglia processes, while in thick tissue sections, IHC staining may be imperfect due to antibody penetration into tissue. Tissue may be sectioned either in a coronal or sagittal orientation and this choice will depend on the experimental goal and brain region(s) to be studied.
2. Immunohistochemistry
NOTE: Skeleton and fractal analysis methods can be applied to either fluorescence or 3,3′-diaminobenzidine (DAB) IHC. The following is a standard fluorescence IHC protocol and may be substituted as needed. Fluorescence IHC yields superior visualization of cell processes when compared to DAB IHC.
- Place the tissue sections into a 4 mL glass vial (up to 15 mouse brain sections/vial) and incubate with 1 mL of solution containing up to 10% horse serum, PBS (0.01 M), 0.5% Triton, 0.04% NaN3 at room temperature (23 °C) for 1 h.
- Wash for 5 min with PBS (0.01 M) three times at room temperature.
- Incubate with the primary antibody (Iba1, 1:1,000) at room temperature for 72 h in 1 mL of solution containing PBS (0.01 M), 0.5% Triton, 0.04% NaN3, and covered (to preserve the NaN3 effectiveness).
- Wash for 5 min with PBS (0.01 M) three times at room temperature.
- Incubate with the secondary antibody (anti-rabbit 488, 1:250) covered at room temperature for 4 h in 1 mL of solution containing PBS (0.01 M), 0.5% Triton, 0.04% NaN3
- Wash for 5 min with PBS (0.01 M) three times at room temperature.
- Mount the brain tissue sections (number and orientation based on preference) to subbed slides, apply a soft-set mounting medium to the slide, and place the coverslip over the tissue. Store the slides at 4 °C.
NOTE: A 1.5 glass coverslip thickness is required for confocal imaging. Soft-set is a preferred mounting medium because the high viscosity does not compress the tissue and best retains the morphology when compared to hard-set mounting media.
3. Imaging
- Image Iba1 positive cells in the brain tissue section using a bright-field or confocal microscope that has a z-stack acquisition capability using a 20X objective or greater.
NOTE: Imaging parameters and software set-up should be constant for all photomicrograph acquisition in an experiment. Excellent process detail is obtained using a 40X or 63X objective. It is possible to apply the skeleton analysis protocol described herein to an entire photomicrograph or a multi-cell ROI within a larger photomicrograph.- Acquire 8-bit images by using the appropriate software settings specific to the microscope software.
NOTE: The conversion of files to 8-bit post-acquisition may distort the data collection. - Acquire at least a 30 µm z-stack with no more than a 2 µm interval between images using the appropriate software settings specific to the microscope software.
NOTE: The microscope and software should allow for image acquisition in an X, Y, and Z axis. A z-stack may be increased, and the interval may be decreased to provide additional microglia detail. In exchange, imaging time will increase. Use Nyquist sampling where possible for fluorescence microscopy.
- Acquire 8-bit images by using the appropriate software settings specific to the microscope software.
- Save all files as Tif files or as required by the microscope software.
- Open the Tif files in ImageJ and use the toolbar to split channels by clicking Image | Color | Split Channels, and stack images by clicking Image | Stacks | X Project | Maximum Intensity Projection where appropriate. Save as .tif files.
4. Skeleton Analysis
- Download FIJI or ImageJ from <https://imagej.net/Fiji/Downloads>. For the individual plugins, download AnalyzeSkeleton(2D/3D) from <http://imagej.net/AnalyzeSkeleton>15 and download FracLac from <https://imagej.nih.gov/ij/plugins/fraclac/fraclac.html>16.
NOTE: The entire process of converting a photomicrograph to a binary skeletonized image takes less than 1 min. - If using a fluorescence photomicrograph, ensure the image is 8-bit and convert to grayscale to best visualize all positive staining. Use the toolbar and click Image | Lookup Tables | Greys. If using a DAB bright-field photograph, first use the FFT bandpass filter plugin (using the toolbar by clicking Process | FFT | bandpass filter) and then convert to grayscale.
NOTE: For the purposes of this protocol, ImageJ default settings for an FFT Bandpass Filter are sufficient (filter up to 3 pixels, down to 40, no stripe suppression). Applying an FFT Bandpass filter removes noise (small features) while preserving the overall larger aspects of the image. This is particularly useful in bright field images, where splits and cracks in the tissue can appear as background and thus complicate the skeleton analysis18. - Adjust the brightness and contrast if the image is too dim and the microglia process cannot be visualized. Use the toolbar and click Image | Adjust | Brightness/Contrast. Adjust the minimum or maximum sliders as needed, up to the edges of the histogram but no further.
NOTE: In ImageJ, brightness and contrast are changed by updating the image's lookup table (LUT), so pixel values are unchanged. Max and min sliders control the lower and upper limits of the display range, with pixel values above 255 appearing white and pixel values below 0 appearing black18. In the case of fluorescence photomicrographs, the maximum slider should be used, whereas the minimum slider will be used for photomicrographs of DAB stained microglia. - Run an Unsharp Mask filter to further increase contrast using the toolbar by clicking Process | Filters | Unsharp Mask. For the purposes of this protocol, ImageJ's default settings (a pixel radius of 3 and mask weight of 0.6) are used.
NOTE: Unsharp Mask sharpens and enhances the edge features of an image by subtracting a blurred version of the image (the Unsharp Mask) from the original, and then merging the resulting image with a high-contrast version of the original image. Therefore, the Unsharp Mask filter does not create details, but rather clarifies the existing detail in an image. The radius setting changes how blurry the Unsharp Mask will be (and thus how much blur will be removed), and the mask weight setting changes the degree of contrast that the Unsharp Mask will be merged with (and thus adjusts the contrast of the final image). - Perform a despeckle step to remove salt-and-pepper noise generated by the Unsharp Mask. Use the toolbar and click Process | Noise | Despeckle.
NOTE: Despeckle removes salt-and-pepper noise by replacing each pixel with the median value in its 3 × 3 neighborhood. The effect is that outliers in intensity are replaced by the median pixel without affecting the sharpness of edges18. - Convert the image to binary using the toolbar by clicking Image | Adjust | Threshold.
NOTE: Thresholding stratifies the grayscale images into features of interest versus background and converts the image into binary18. - Apply the despeckle, close-, and remove outliers functions: In the resulting binary image, there may be single-pixel background noise and gaps between processes.
- Apply the despeckle function using the toolbar by clicking Process | Noise | Despeckle.
NOTE: Applying despeckle to the binary image removes any remaining single-pixel noise. - Apply the close function using the toolbar by clicking Process | Binary | Close.
NOTE: This plugin connects two dark pixels if they are separated by up to 2 pixels. - Apply the remove outliers function using the toolbar by clicking Process | Noise | Remove Outliers.
NOTE: For the purposes of this protocol, bright outliers are targeted with a pixel radius of 2 and a threshold of 50. This plugin replaces a bright or dark outlier pixel by the median pixels in the surrounding area if it deviates by more than a certain value (the threshold)18.
- Apply the despeckle function using the toolbar by clicking Process | Noise | Despeckle.
- Save the image as a separate file for future use and/or fractal analysis and skeletonize the image using the toolbar by clicking Process | Binary | Skeletonize.
- Select the skeletonized image and run the plugin AnalyzeSkeleton(2D/3D) using the toolbar by clicking Plugins | Skeleton | Analyze Skeleton, and checking the Branch Information box.
NOTE: It is likely that the image processing will require optimization with the addition or deletion of the above suggested steps. In this process, skeletonized images are assessed for accuracy by creating an overlay of the skeleton and the original image. Somas should be single origin points with processes emanating from the center; circular somas confound the data and should be avoided through protocol adjustment. An example of a single origin point versus circular somas is illustrated in Figure 1.
Common problems resulting in non-representative skeletons and suggested solutions:
- Image too dim: convert to greyscale, adjust brightness/contrast sliders, and/or apply Unsharp Mask
- Too much background: adjust brightness/contrast sliders, apply Despeckle, and/or Remove outliers
- Circular somas in skeletonized image (particularly for fluorescence images): apply FFT Bandpass filter, and/or Unsharp Mask
- Cracks in tissue (particularly for bright-field images): apply FFT Bandpass filter, and/or Despeckle
- Copy all of the data from the Results and Branch information outputs and paste the data into an Excel spreadsheet.
- In Excel, trim the data to remove skeleton fragments that result from IHC and image acquisition.
- Duplicate the experiment workbook with the raw data output from skeleton analysis and add TRIM to the filename. All subsequent data trimming should occur in the duplicated workbook to preserve the raw data for future use and reference.
- Determine which length of fragments will be trimmed from the dataset by opening the skeletonized image in ImageJ and selecting the Line tool. Measure several fragments, taking note of the average length, and decide on a cutoff value.
NOTE: For the purposes of the data presented here, the cutoff length for undesired fragments is 0.5. This value should be consistent throughout a dataset. - Custom sort the Excel spreadsheet by clicking Sort & Filter | Custom sort. Sort by "endpoint voxels" from largest to smallest and, in a new level, by "Mx branch pt" from largest to smallest.
- Remove every row that contains 2 endpoints with a maximum branch length of less than the cutoff value (i.e., 0.5). Sum the data in the endpoints column to calculate the total number of endpoints collected from the image.
- Repeat for Branch information data: sort by 'branch length' from largest to smallest. Scroll through the data and remove every row that has a branch length of less than the cutoff value(i.e., 0.5). Sum the values in the branch length column to calculate the summed length of all branches collected from the image.
- Repeat steps 4.11.3-4.11.5 for every image/sheet until all data have been trimmed and summed.
- Divide the data from each image (summed number of endpoints and summed branch length) by the number of microglia somas in the corresponding image. Enter the final data (endpoints/cell & branch length/cell) into statistical software.
NOTE: The summed branch length/cell data may require conversion from length in pixels to microns.
5. Fractal Analysis
NOTE: FracLac is able to run a number of different shape analyses that are not covered in this protocol. For a more detailed explanation of FracLac's various functions, see the FracLac manual at <https://imagej.nih.gov/ij/plugins/fraclac/FLHelp/Introduction.htm> and associated references 2,16,19. Fractal analysis utilizes the protocol steps 4.1-4.7 described above.
- Determine the size of the ROI that will be used for all fractal analysis. Use the rectangle tool to draw the ROI. Ensure that the box is large enough to capture the entire cell and can remain consistent throughout the dataset.
NOTE: Use the rectangle selection rather than the freehand selection to ensure that all ROIs are the same sized rectangle, and therefore, the cells have the same scale. This would not be the case if the freehand selection were used because ImageJ will auto-scale a square ROI to fit different sized rectangular windows resulting in cells with different scales. While fractal shapes are scale-independent, the fractal analysis process using FracLac for ImageJ is dependent on scale16. Thus, there is the need for a consistently sized ROI throughout data collection. - Randomly select microglia for fractal analysis in each photomicrograph and corresponding binary image. In the ROI manager window, select Update to lock the ROI onto the cell's position. Use Ctrl + Shift + D to duplicate the area within the ROI as a new window, and then save the cropped cell. On the corresponding binary image (from the skeleton analysis), duplicate the area with the same cell using the ROI manager. Repeat until a sufficient number of cells have been randomly selected for fractal analysis and save all files.
NOTE: A random number generator and a numbered grid can be used to randomly select cells. - Open the binary image with the individual cell. Double-click the paintbrush tool, set the color to black, and adjust the brush width as needed. Using the matching photomicrograph as a reference, use the paintbrush to remove adjacent cell processes, connect fragmented processes, and isolate the cell of interest. Once the binary cell has been isolated, save the binary file.
NOTE: Holding 'alt' will switch the paintbrush from the foreground color (black) to the background color (white). - Convert the binary cell to an outline using the toolbar via Process | Binary | Outline.
NOTE: FracLac for Image J can be used on either solid shapes or shape outlines, however, current convention is to use shape outlines16. - In the toolbar, open FracLac using the toolbar by clicking Plugins | Fractal Analysis | FracLac and select BC (box counting). In the Grid Design settings, set Num G to 4. Under Graphics options, check the Metrics box to analyze the convex hull and bounding circle of the cell. When finished, select OK.
NOTE: Num G is the number of box counting grid orientations used during the scan and the recommended range for Num G is 4-12. The Num G setting and the suggested range is thoroughly reviewed in the FracLac manual16. Increasing Num G can significantly slow calculation times. FracLac settings will only need to be set up once per session. Once the settings have been entered, the Scan button will become available. - Select the Scan button to run a box-counting scan on the selected image.
NOTE: The scan will generate three windows with data outputs: Hull and Circle Results, Box Count Summary File, and Scan Types. The Scan Types window contains a log of the settings used, as well as standard deviations for certain measures. For the purposes of this protocol, the Scan Types window is not used and may be closed or saved for future reference. - In the Hull and Circle Results window, copy all desired data (i.e., density, span ratio, and circularity) results. In the Box Count Summary window, copy the desired data (i.e., fractal dimension and lacunarity) results. Transfer the copied data to an Excel file or statistical software.
NOTE: A full listing of the FracLac output data are provided and thoroughly explained in the FracLac for ImageJ manual16.
Representative Results
The microglia morphology analysis protocols described herein summarize steps helpful in processing fluorescent and DAB photomicrographs for morphometric analysis. These steps are visually summarized in Figure 2 and Figure 3. The goal of these steps is to create a representative binary and skeletonized image that appropriately models the original photomicrograph such that the accumulated data are valid. After protocol application, the AnalyzeSkeleton plugin results in a tagged skeleton image from which the number of endpoints and branch (i.e., process) lengthcan be summarized from resulting output files. Endpoints and process length data are then used to estimate the extent of microglia ramification in the photomicrograph or ROI. Figure 4 summarizes the resulting data (endpoints/cell and process length/cell) collected with and without the protocol application. While similar trends exist, the data summarized in Figure 4F are less variable than those in Figure 4E. In addition, these data illustrate increased sensitivity to detect differences between groups when the protocol is applied. Lastly, care must be taken concerning inter-user variability in the application of the protocol. Such differences are summarized by Figure 5 where the same data set was analyzed by two independent users applying an identical protocol as summarized above.
Additional morphology data are collected from single cells isolated from the binary images created during the protocol application. The protocol steps to analyze microglia morphology prior to and using the FracLac plugin are summarized in Figure 6. We illustrate this analysis in both uninjured (Figure 6A) and injured (Figure 6B) tissue. Representative images of binary, outlined, convex hull/encapsulating circle, and box counting examples for each cell analyzed with and without the protocol application are shown in Figure 6C–F. These images help to illustrate the origins of differences in morphology data which are summarized in Figure 6G.
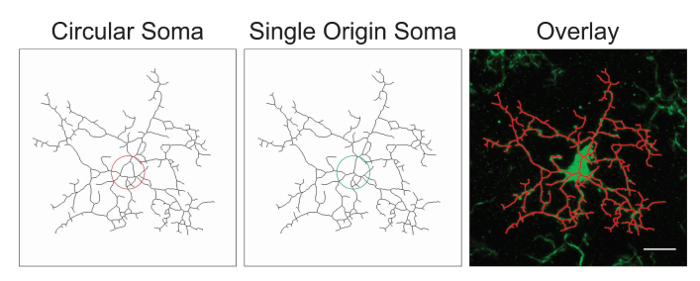
Figure 1. Illustrations of skeletonized microglia with a circular soma (suboptimal) versus a single origin soma (optimal) and the corresponding overlay between the skeletonized cell and the original photomicrograph. Scale bar = 10 µm. Please click here to view a larger version of this figure.
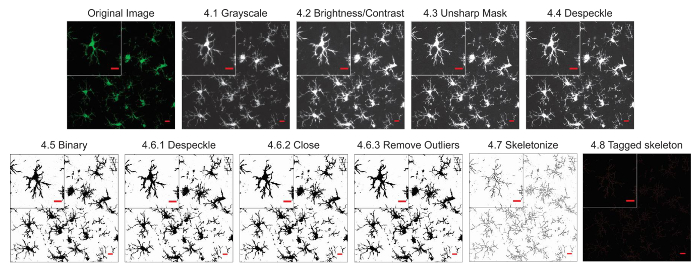
Figure 2. Protocol application to fluorescent photomicrographs. Illustrations of the Skeleton Analysis protocol applied to a fluorescent photomicrograph with a single cell cropped to show details. Scale bar = 10 µm. Please click here to view a larger version of this figure.
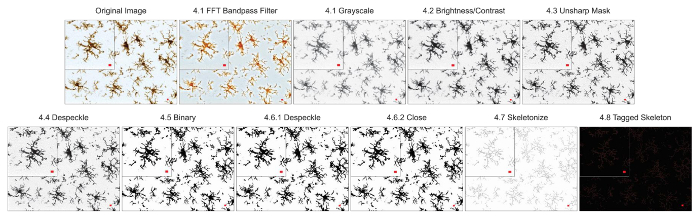
Figure 3. Protocol application to bright-field DAB photomicrographs. Illustrations of the Skeleton Analysis protocol applied to a bright-field DAB photomicrograph with a single cell cropped to show details. Scale bar = 10 µm. Please click here to view a larger version of this figure.
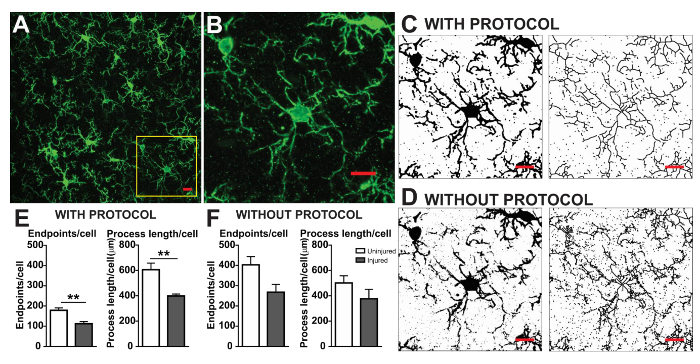
Figure 4. Data analysis with and without the protocol application. (A) An example photomicrograph of fluorescent IHC and cropped cell corresponding to the yellow box in (B). Example binary and skeletonized images with (C) and without (D) the protocol applied as described. Summary data of microglia endpoints/cell and process length/cell in uninjured (white) and injured (grey) cortical tissue with (E) and without (F) the protocol applied. Statistical analysis using student's t-test and n = 3, ** denotes p < 0.01. Scale bar = 10 µm. Please click here to view a larger version of this figure.
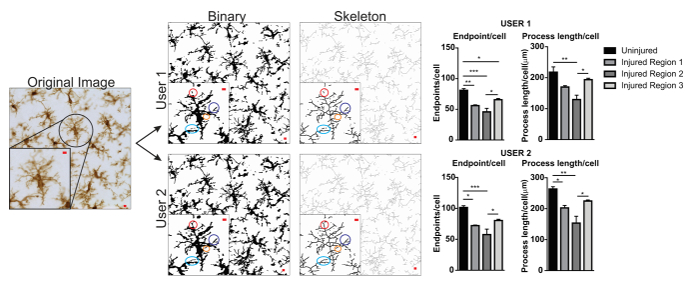
Figure 5. User differences with protocol application. An example of an original image and protocol conversion to binary and skeleton images by User 1 and User 2. Differences between the two images are highlighted with matching colored circles. Summary graphs of microglia endpoints/cell and process length/cell data in uninjured and injured brain regions by User 1 and User 2. Statistical analysis with ANOVA and sample size is n = 3; *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar = 10 µm. Please click here to view a larger version of this figure.
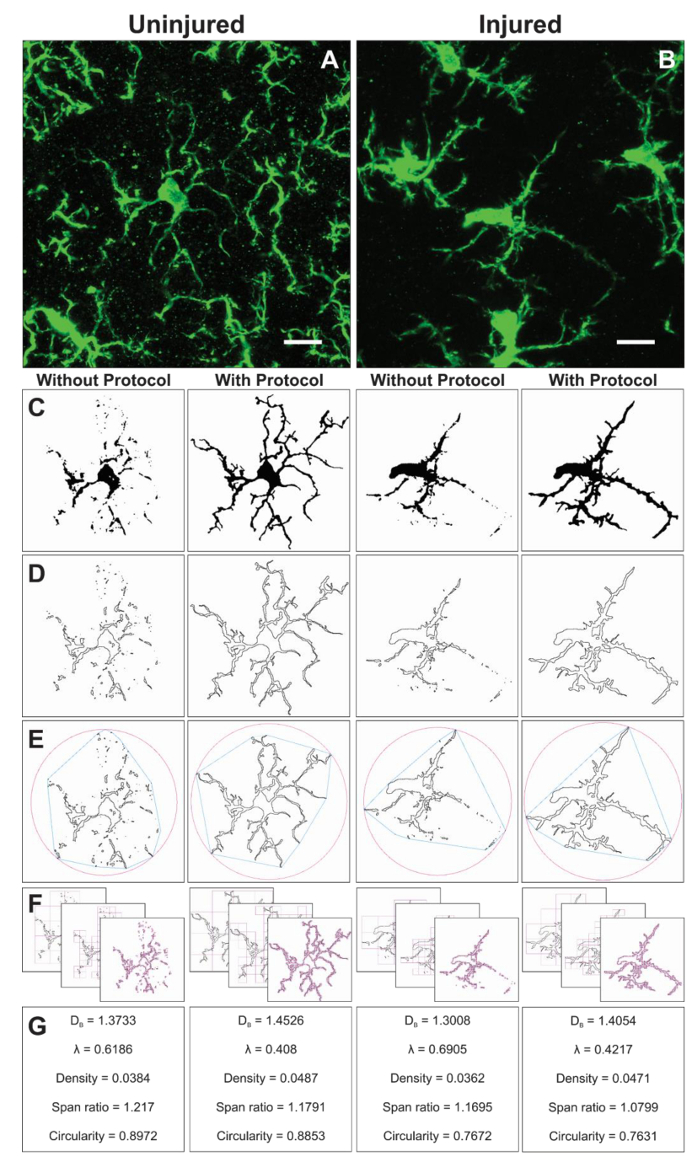
Figure 6. Fractal analysis with and without the protocol application. Example of cropped photomicrographs of microglia in the uninjured (A) and injured (B) cortex with corresponding binary (C), and outline (D) images that result with and without the protocol applied. The associated convex hull (blue) and enclosing circle (pink) for corresponding outline shapes (E) are used to calculate shape density, span ratio, and circularity (G). The box counting method is illustrated in (F), and used for fractal dimension (DB) and lacunarity (λ) calculations (G). Scale bar = 10 µm. Please click here to view a larger version of this figure.
Discussion
Microglia cells are finely tuned to the physiology and pathology within their micro-domains and display a diverse range of morphologies2 in both subtle7,14 and gross injury8. The use of ImageJ protocols makes microglia morphology quantification accessible to all laboratories as the platform and plugins are an open-source image processing software. While the described protocol is focused on image processing and analysis using this software, the consistency of data collection, validity, and reliability begins with excellent IHC and microscopy. This protocol is used to improve binary, skeleton, and outline representations of entire photomicrographs and single cells but cannot take the place of poor IHC staining and microscopy that results in low contrast, blurred, or distorted images. As an additional consideration, care must be taken not to flatten the brain tissue during storage, prior to sectioning, which irrevocably alters microglia morphology. Lastly, within each experiment, microglia must be imaged using the same scale as well as the same microscope. Instrumentation, objectives, and software vary amongst microscopes which will result in different sized photomicrographs despite similar objectives and change the detail as well as the number of cells within each frame. For example, image acquisition using a 40X objective on a Leica SPII results in twice the number of cells and less detail than acquisition using a Zeiss 880. This is particularly important for cell ramification data collected from the entire frame rather than a single cell, as this becomes an issue of data sampling.
In general, skeleton analysis which utilizes the entire photomicrograph precedes the single cell fractal analysis for two reasons. Determining cell ramification of all cells in a photomicrograph is rapid when compared to the single cell fractal analysis and may be considered as a screening tool if time is a factor. In addition, the binary images derived during skeleton analysis are used for fractal analysis. Once imaged, there are a number of critical steps that may influence skeleton analysis results and introduce user-influence. The protocol steps that are most variable between users are step 4.2 (increasing image brightness) and step 4.5 (determining the threshold). Where possible, an optimal number to increase brightness (max or min slider between 0-255) is determined and held constant for all images and users. Where image variability is great, the user can instead choose a brightness that will vary between images. Alternatively, if images are bright and contrast is high, then increasing brightness can be omitted and thresholding can be standardized by using a specialty threshold filter (e.g., Huang) rather than the more variable default. Once optimized, the parameters should be adhered to in order to minimize additional user-influence.
An example of user variability is presented in Figure 5. Data values were increased in User 1 versus User 2 and therefore variability would be increased if both User 1 and User 2 contributed to data collection. An example of the differences in User 1 and User 2 binary and skeletonized images are highlighted by colored circles (Figure 5). In this instance, both users were briefly trained undergraduate students with limited expertise in microglia. Regular oversight and mentoring by a microglia expert along with increased protocol training2 can reduce inter-user variability. Although not assessed here, fractal analysis is less subject to inter-user variability because binary cells are manually and individually isolated from a photomicrograph rather than relying solely on thresholding to determine microglia shapes. However, all methods possess some variability between users. Therefore, a single user (ideally, trained by some expertise in microglia cells) should complete the data collection for an entire dataset.
Additional modifications can be easily made to this protocol and will depend on image quality, and the efforts taken to reduce noise and ensure process connectivity. For example, if contrast is adequate, then unsharp mask is not necessary and can be omitted. It is prudent to optimize and finalize the protocol for a specific set of images, both experimental cases and controls, prior to collecting data from an entire set. Lastly, additional plugins may be used in place of others to clarify or sharpen images that were not described in this protocol such as dilate or sharpen.
Advantages of this protocol are its universal availability and adaptability. In addition, assessing cell ramification using AnalyzeSkeleton is quick and applicable to an entire photomicrograph. A benefit of multi-cell analysis approach is the focus on an entire region rather than single cells. Therefore, it is possible to quickly assess the average ramification (in terms of endpoints and process length) of all microglia within the image. Skeleton analysis provides an analysis of multiple cells: a data sampling in terms of cell numbers that cannot be matched by fractal analysis due to the required time investment to isolate single cells from photomicrographs. An instance where this might be best suited would be in screening microglia morphologies in proximities to a focal injury. One limitation is the whole field image rendering to create skeleton models of IHC photomicrographs is imperfect when compared to the more time consuming single cell approach. In addition, a region analysis is not appropriate to circumstances where microglia morphologies are drastically different within the same field. Lastly, this analysis method is dependent on cell count, a parameter that may differ between experimental conditions.
Fractal analysis is conducted on a single cell and therefore complements the average cell ramification data output resulting from the skeleton analysis. Although much more time consuming, this investment yields a broad range of morphometric data. For example, cell density, span ratio, and circularity data describe the size, elongation, and shape of the cell outline, respectively. Fractal dimension and lacunarity summarize the cell complexity and shape heterogeneity, respectively. A more in-depth summary of how each parameter is calculated and how data may be interpreted is provided in the interactive manual16 and such detail should be considered in relation to the specific research question. The described protocol results in sensitive tools to quantify small changes in 2D microglia morphologies that may occur in physiologic and pathologic conditions. Additional morphometric analysis such as solidity, convexity, and form factor16,20 may be possible if generating 3D shapes.
Protocol development and adaptation is continuous and user-driven. It has been extended from fluorescence8 to DAB/bright-field images7 but not yet to paraffin embedded tissue. It addition, it can be used in conjunction with proprietary software such as Imaris for additional analysis. This protocol can be applied to a variety of physiology and is not limited to microglia but may be applied to any cell or tissue with particular patterns or shapes that can be identified using IHC methods. Lastly, with sufficient sample size, a multivariate or cluster analysis can be applied to stratify microglia according to morphology12,21; this is meaningful information as microglia morphology is a vital indicator of microglia functions and responses to their surroundings. The appreciation for microglial morphologic diversity is expanding and important to fully understand neuron-glia-vascular interactions during health and disease. Growth in this field is enhanced by well-developed, easy to use, and reproducible protocols to quantify and summarize microglia morphology using multiple continuous variables.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study received financial support from NINR (F32NR013611). We would like to further acknowledge and thank the developers of AnalyzeSkeleton(2D/3D) and FracLac (Arganda-Carreras et al. and Karperien et al., respectively) without which the analysis described herein would not be possible.
Materials
| primary antibody anti-IBA1 | Wako | 019-19741 | rabbit host |
| Vectashield soft mount | Vector Labs | H-1000 | |
| Secondary antibody | Jackson ImmunorResearch | 711-545-152 | donkey host |
| 4 mL glass vial | Wheaton | UX-08923-11 | |
| Triton X-100 | Fisher Scientific | BP151 | |
| Sodium Azide (NaN3) | Sigma | S-8032 | |
| glass coverslip | Fisher Scientific | 12-544-G |
References
- Davalos, D., et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 8 (6), 752-758 (2005).
- Karperien, A., Ahammer, H., Jelinek, H. F. Quantitating the subtleties of microglial morphology with fractal analysis. Front Cell Neurosci. 7 (3), eCollection (2013).
- Taylor, S. E., Morganti-Kossmann, C., Lifshitz, J., Ziebell, J. M. Rod microglia: a morphological definition. PLoS One. 9 (5), e97096 (2014).
- Nimmerjahn, A., Kirchhoff, F., Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 308 (5726), 1314-1318 (2005).
- Abiega, O., et al. Neuronal Hyperactivity Disturbs ATP Microgradients, Impairs Microglial Motility, and Reduces Phagocytic Receptor Expression Triggering Apoptosis/Microglial Phagocytosis Uncoupling. PLoS Biol. 14 (6), e1002466 (2016).
- Wyatt-Johnson, S. K., Herr, S. A., Brewster, A. L. Status Epilepticus Triggers Time-Dependent Alterations in Microglia Abundance and Morphological Phenotypes in the Hippocampus. Front Neurol. 8 (700), eCollection (2017).
- Morrison, H., Young, K., Qureshi, M., Rowe, R. K., Lifshitz, J. Quantitative microglia analyses reveal diverse morphologic responses in the rat cortex after diffuse brain injury. Sci Rep. 7 (1), 13211 (2017).
- Morrison, H. W., Filosa, J. A. A quantitative spatiotemporal analysis of microglia morphology during ischemic stroke and reperfusion. J Neuroinflammation. 10 (4), (2013).
- Gyoneva, S., Traynelis, S. F. Norepinephrine modulates the motility of resting and activated microglia via different adrenergic receptors. J Biol Chem. 288 (21), 15291-15302 (2013).
- Xu, H., et al. Environmental Enrichment Potently Prevents Microglia-Mediated Neuroinflammation by Human Amyloid beta-Protein Oligomers. J Neurosci. 36 (35), 9041-9056 (2016).
- Rodriguez, J. J., Noristani, H. N., Verkhratsky, A. Microglial response to Alzheimer’s disease is differentially modulated by voluntary wheel running and enriched environments. Brain Struct Funct. 220 (2), 941-953 (2015).
- Soltys, Z., et al. Quantitative morphological study of microglial cells in the ischemic rat brain using principal component analysis. J Neurosci Methods. 146 (1), 50-60 (2005).
- Orlowski, D., Soltys, Z., Janeczko, K. Morphological development of microglia in the postnatal rat brain. A quantitative study. Int J Dev Neurosci. 21 (8), 445-450 (2003).
- Morrison, H. W., Filosa, J. A. Sex differences in astrocyte and microglia responses immediately following middle cerebral artery occlusion in adult mice. Neuroscience. 339, (2016).
- Arganda-Carreras, I., Fernandez-Gonzalez, R., Munoz-Barrutia, A., Ortiz-De-Solorzano, C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc Res Tech. 73 (11), 1019-1029 (2010).
- Davis, B. M., Salinas-Navarro, M., Cordeiro, M. F., Moons, L., De Groef, L. Characterizing microglia activation: a spatial statistics approach to maximize information extraction. Sci Rep. 7 (1), 1576 (2017).
- . ImageJ User Guide Available from: https://imagej.nih.gov/ij/docs/guide/ (2014)
- Karperien, A. L., Jelinek, H. F., , Fractal, Multifractal, and Lacunarity Analysis of Microglia in Tissue Engineering. Front Bioeng Biotechnol. 3 (51), eCollection (2015).
- Martyanova, E. K., Tishkina, A. O. 3D quantitative analysis of microglial morphology. available as conference preceedings SkoltechOn. , (2015).
- Fernandez-Arjona, M. D. M., Grondona, J. M., Granados-Duran, P., Fernandez-Llebrez, P., Lopez-Avalos, M. D. Microglia Morphological Categorization in a Rat Model of Neuroinflammation by Hierarchical Cluster and Principal Components Analysis. Front Cell Neurosci. 11 (235), eCollection (2017).

