Subcellular Fractionation from Fresh and Frozen Gastrointestinal Specimens
Summary
Here, we present a protocol to perform a simple cellular fractionation for the subcellular separation of cytoplasmic and nuclear proteins in human fresh and frozen intestinal biopsies.
Abstract
The purpose of this protocol is to fractionate human intestinal tissue obtained by endoscopy into nuclear and cytoplasmic compartments for the localization analysis of specific proteins or protein complexes in different tissue states (i.e., healthy vs. disease). This method is useful for the fractionation of both fresh and frozen intestinal tissue samples; it is easily accessible for all laboratories and not time consuming.
Introduction
Proteins participate in almost all biological functions within a cell and any variation in their structure, quantity or location can lead to a pathogenic scenario. Tissue sample fractionation methods are a useful approach to reduce the complexity of disease related protein analyses. Some studies use protein localization information or enrich a protein from a specific cellular compartment, so a protocol for reproducible fractionation of intact proteins is useful to answer certain biological questions. Determining the subcellular localization of proteins and monitoring their compartmental redistribution or interactions at basal and disease conditions will help identify disease related functional differences1,2. Thus, this method aims to reproducibly fractionate intestinal tissue biopsies, both fresh and frozen, into cytoplasmic and nuclear subcellular compartments.
Intestinal biopsy samples are routinely obtained during endoscopy procedures3 (Figure 1) and can be effectively used for protein quantification or immunoprecipitation studies. Since tissue samples from intestinal epithelial will contain proteins that may be directly involved in the disease pathogenesis4, intestinal biopsy samples are a very valuable source for the successful identification of intestinal disease-specific proteins. Stored frozen, patient tissue samples together with the clinical information are useful resources for protein analysis, and a simple and reproducible sample preparation is a key issue to provide cell compartmental information using limiting amounts of frozen tissue5.
There are several commercially available kits for the separation of cellular fractions, but these are more expensive and generally more time consuming than the protocol presented here. Protocols based on multiple-step gradient centrifugation and continuous gradients have also been previously used for the fractionation of diverse cellular compartments in different tissues6,7. However, maintaining the consistency of continuous gradients is often a difficult task. Similar protocols based on the sequential lysis of membranes have been previously described but they generally need more than two buffers and the hands on time is longer8 .
When fractionating tissue samples, bear in mind that tissue samples present cell-cell and cell-matrix interactions that are both not present in cultured cells and important when proceeding to protein extraction. The proper breakdown between cells and extracellular matrix without affecting the protein quality will be a critical factor in the intestinal tissue protein isolation9. In this protocol, cell-cell and cell-matrix contacts are first broken, releasing the cells and allowing the buffer to reach the individual cells. To avoid protein-compartment mixing prior to fractionation, the breakdown of the contacts must occur without altering the integrity of the cells or the nuclear membranes.
Due to the enrichment of various proteases in the intestinal mucosa10, it is important to control the protein degradation during extraction. To minimize the potential protein degradation, multiple protease inhibitor must be included in the protein extraction buffers. Additionally, if extracts are going to be used for functional assays, it is essential to avoid the denaturation of proteins or proteolysis as this will cause a loss of protein activity11. For this purpose, the protocol is performed at 4 °C and fresh phenylmethylsulfonyl fluoride (PMSF) is added to the buffers at a final concentration of 0.1 mM just prior to use to further inhibit proteolysis.
The first step in cell fractionation is tissue disruption and cell lysis. As mentioned previously, the objective is to disaggregate the cells and break them open with minimum damage. Intestinal tissue samples must be homogenized, and the cells lysed to achieve maximum breakage of the cell membrane. In this protocol, we use a motorized pestle mixer to break the intestinal tissue which is homogenized within seconds by means of high speed vortexing action. After homogenization, tissue cells are incubated in a hypotonic buffer that will burst the cell membrane but will keep the nuclei intact, followed by addition of a non-denaturing detergent (nonyl phenoxypolyethoxylethanol) and a short vortex to separate nuclei from the cytoplasmic fraction. After the removal of the cytoplasm, the cell nuclei membrane is burst into a hypertonic buffer with shaking.
The method presented here is an appropriate method for the subcellular fractionation of fresh and frozen intestinal biopsies that have been obtained during endoscopic procedures and range between 2 and 10 mg in weight. The protocol is easy and reproducible and could be performed using basic laboratory equipment and reagents in under an hour.
Protocol
This study was approved by the Cruces University Hospital Ethics Board and analyses were performed after informed consent was obtained from all subjects or their parents.
1. Biopsy Collection
NOTE: Biopsy specimens from the distal duodenum of patients are obtained during routine diagnosis endoscopy managing biopsies that range between 2 and 10 mg in weight. Biopsied tissue is a complex tissue composed of different cell types including epithelial, immune and endothelial cells. Clinical gastroenterologists perform the endoscopy following established clinical guidelines.
- Place the biopsy on a sterile filter paper just after endoscopy.
- Use forceps to transfer the biopsies in 1.5 mL tubes, soak them with PBS, and place it into a cryotube.
- Keep tubes on ice until they arrive at the laboratory.
- Start processing of fresh biopsies immediately.
- For frozen biopsies, flash freeze the biopsy containing tubes and store in liquid nitrogen until use.
NOTE: It is important to keep the sample frozen until the cold supplemented buffer 1 is added.
2. Buffer Preparation
Note: Before starting, supplement the buffers as follows.
- Add 1 mM DTT (final concentration) and protease and phosphatase inhibitor cocktail (1x final concentration) to the hypotonic buffer 1 and hypertonic buffer 2 (Table 1).
- Prepare 1.5 mL of buffer 1 and 150 µL of buffer 2 per sample. DTT will prevent the oxidation of the samples.
- Prepare nuclei wash buffer by adding 1% detergent (nonyl phenoxypolyethoxylethanol) to a final concentration of 0.1% to the already supplemented buffer 1. Prepare 1 mL of nuclei wash buffer per sample.
3. Biopsy Homogenization
- Fresh Biopsies
- Place the disposable pestle in the motorized mixer. Ensure that the pestle reaches the bottom of the tube.
- Take the biopsy out from the cryotube and place it in a 1.5 mL tube with a pipette tip.
- Add 200 µL of supplemented buffer 1 to the 1.5 mL tube.
- Homogenize the biopsy with the pestle until the tissue is completely disrupted and keep it on ice.
- Once all the biopsies are homogenized proceed with step 4.
Note: Disposable pestles must be changed between biopsies.
- Frozen Biopsies
CAUTION: Handle liquid nitrogen carefully; contact of liquid nitrogen with the skin or eyes may cause serious freezing injury. Protect hands and eyes always when working with liquid nitrogen .- Take some liquid nitrogen in a box to store the cryotubes retrieved from the nitrogen tank.
- Find the biopsy in the nitrogen tank and place the cryotube in the box with the liquid nitrogen. Repeat this step for all the biopsies.
- Transfer the biopsy from the cryotube to a 1.5 mL tube.
NOTE: Biopsies will be attached to the frozen tube. Push the biopsy with a sterile pipette tip so it will detach from the tube. - Add 200 µL of supplemented buffer 1.
- Homogenize the tissue using the pestle and mortar until the tissue is completely disrupted, maintaining the temperature at 4 °C throughout all the procedure.
Note: Do not let biopsies thaw before adding the buffer.
4. Cytoplasm Isolation
- Incubate the homogenized biopsy on ice for 10 min.
- Add 10 µL of 1% detergent (nonyl phenoxypolyethoxylethanol) to each sample to a final concentration of 0.05%.
NOTE: Cells will swell and burst by osmotic lysis as the mild detergent will disrupt the cell membrane. - Incubate on ice for 5 minutes.
- Vortex briefly and centrifuge at 400 x g at 4 ˚C for 2 min.
- After vortexing, remove the supernatant and transfer it to a new clean 1.5 mL tube ; this will be the cytoplasmic fraction. Do not discard the pellet.
5. Nuclear Isolation
- Resuspend the pellet from the previous step with 200 µL of nuclei wash buffer.
- Centrifuge the tube for 2 min at 400 x g and 4 °C.
- Discard the supernatant and repeat the washing procedure two more times.
NOTE: The washes will remove cytoplasmic contamination in the nuclear fraction. - After the third wash, resuspend the pellet in 100 µL of buffer 2.
- Shake the sample vigorously at 4 ˚C for 30 min. Alternatively, incubate the sample on ice and vortex every 5 min.
- After shaking, centrifuge the sample for 10 min at top speed (> 12,000 x g) and 4 °C.
- After vortexing, transfer the supernatant into a new clean 1.5. mL tube. This will be the nuclear fraction.
6. Protein Quantification
Note: Quantify the proteins with a bicinchoninic acid (BCA) protein assay kit. The concentration of proteins in the fractions will range between 0.5 and 5 µg/µL, being lower in the nuclear fraction (see Table 2 for protein concentrations resultant from 2 mg biopsies).
- Prepare a standard curve with the following bovine serum albumin (BSA) amounts (µg)12: 0-0.5-1-2-3-4-5-6.
- Prepare the reagents needed to use 100 µL of final BCA mix per sample.
- Pipette 2 µL of each protein fraction in a 96 well plate, add the BCA mix and follow the manufacturer's instructions for the reading.
Representative Results
Figure 1 shows a hematoxylin eosin staining of an intestinal biopsy section with intact villi and crypt structures.
Representative results of nuclear and cytoplasmic fractionation using this protocol are shown in Figures 2 and 3. In the experiment shown in Figure 2, a fresh (Fh) and a frozen (Fz) biopsy were simultaneously fractionated following the protocol presented here and a western blot was performed using 20 μg of each sample. Protein concentrations in each fraction and biopsy are shown in Table 2, and the protein concentrations are not very different between the fresh and the frozen tissue. The yield of protein extracted from the 2 mg biopsies is around 3-4 µg/µL in the cytoplasmic fraction and approximately 1.5 µg/µL in the nuclear fraction. To check the purity of the fractions, HSP90 and tubulin were used as a cytoplasmic controls and HDAC1 and H3 as a nuclear controls13. It can be observed that HSP90 and tubulin are primary located in the cytoplasm and HDAC1 and H3 are located in the nucleus with very minimal mixing between the two fractions. To assess whether the protocol affects cell death, we also blotted for the presence of caspase 3. From Figure 2, both samples present a very minimal staining for caspase 3 confirming that the procedure does not affect the cell death pathway even after flash freezing the biopsies. It was also analyzed if protein modifications are maintained after the procedure, so proteins can be used for downstream functional assays. PhosphoSTAT1 was used for staining as the frozen biopsy used in this fractionation came from an individual with an intestinal inflammatory condition; this is not true of the fresh biopsy. As expected, phosphoSTAT1 is located in the cytoplasm of the frozen biopsy, confirming that the protein modifications are maintained after fractionation.
The reproducibility of the method using frozen biopsies is shown in Figure 3. A western blot was performed after fractionation of three independent frozen biopsies, and the fractions are clean with minimal compartment protein mixing (Figure 3A): HSP90 located in the cytoplasmic fraction and HDCA1 in the nuclear fraction. Additionally, the XPO1 protein that is present in both nuclear and cytoplasmic fractions14 was also blotted. It can be observed that the levels of XPO1 quantified by ImageJ are stable in each fraction (Figure 3B), confirming reproducibility of the results using different biopsies.
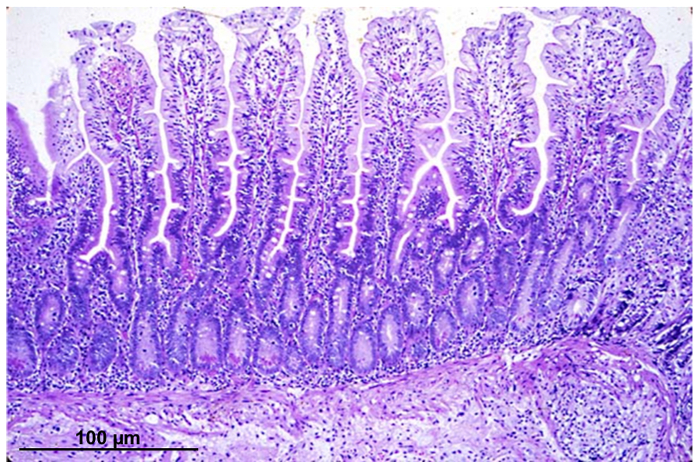
Figure 1: Light micrograph of an intestinal epithelial section, stained with hematoxylin eosin, from a biopsy acquired by endoscopic procedure. Please click here to view a larger version of this figure.
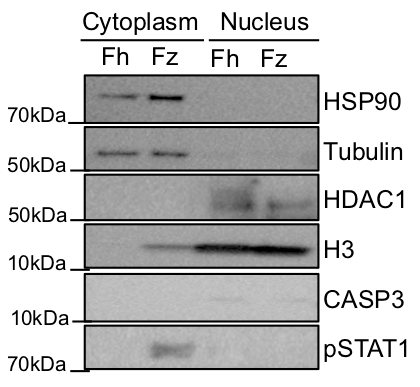
Figure 2: Representative Western blot of a subcellular fractionation performed in a fresh (Fh) and a frozen (Fz) biopsy. HSP90 and tubulin were used as cytoplasmic controls and HDAC1 and H3 as nuclear controls. The presence of CASP3 and phosphoSTAT3 were also assessed.
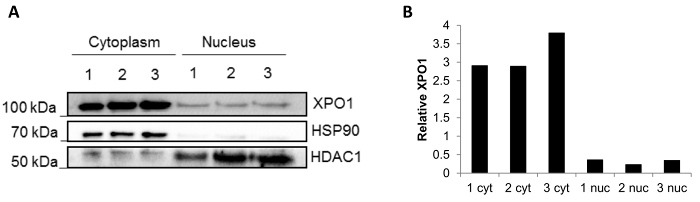
Figure 3: Fractionation results of three independent frozen biopsies. (A) Representative Western blot of a subcellular fractionation performed in three frozen biopsies (1, 2 and 3). HSP90 was used as a cytoplasmic control and HDAC1 as a nuclear control. The localization of XPO1 was also assessed. (B) Relative quantification of XPO1 in each of the fraction done using ImageJ software. Please click here to view a larger version of this figure.
| Buffer | Components | Notes |
| Hypotonic buffer 1 | 10 mM HEPES pH 7.9 | 1.5 ml buffer/sample |
| 10 mM KCl | ||
| 0.1 nmM EDTA | ||
| Hypertonic buffer 2 | 20 mM HEPES pH 7.9 | 150 µl buffer/sample |
| 400 mM NaCl | ||
| 0.1 mM EDTA | ||
| *Note: Buffers can be stored at 4ºC for a month. | ||
| **Note: Buffers have to be supplemented with proteinase and phosphatase inhibitors and DTT before use | ||
Table 1: Buffer composition.
| Fresh | Frozen | |
| Cytoplasm | 2.975 | 4.612 |
| Nucleus | 1.516 | 1.385 |
Table 2: Protein concentration of each of the fractions in a fresh and frozen 2 mg biopsy used in Figure 2; concentration is presented in μg/μL.
Discussion
The protocol described here is used for the nuclear and cytoplasmic fractionation of intestinal biopsies. The purified proteins are not denatured and can be used not only in western blot analysis as shown in Figure 2 and 3 but also in assays requiring native-folded proteins such as immunoprecipitation, electrophoretic mobility shift assay (EMSA) or native polyacrylamide gel electrophoresis (PAGE).
The described method relies on mechanical homogenization of the tissue together with osmotic lysis to burst the cell membrane without affecting the nuclear membrane. The result is a clean fractionation of both the cytoplasmic and nuclear proteins to be used for downstream analyses. It is important to control the degradation of the proteins during the process by the addition of protein and phosphatase inhibitors. Moreover, if extracted proteins are to be used for downstream functional assays, it is also important to avoid denaturation and proteolysis of the samples. The processes should be performed at 4 °C and PMSF should be also added to the buffers.
The procedure described in this protocol is simple and affordable, and the sample processing time is minimal. Problems such as long processing times and the use of costly fractionation kits are not constraints in this protocol.
Fresh and frozen biopsies can both be used in this protocol, with frozen biopsies giving quite clean and reproducible fractionation results as seen in Figure 2 and 3. If there is compartment contamination, the cell lysis time can be reduced to avoid the release of nuclear to the cytoplasm and washing steps can be added to avoid cytoplasmic contamination in the nuclear compartment.
The possibility of using frozen tissues for the independent analysis for nuclear and cytoplasmic protein fractions can increase our knowledge about intestinal inflammatory diseases in which frozen biopsies are stored. Moreover, this technique could be adapted to other kinds of human tissues acquired by biopsies. The volume of buffers used must be modified depending on the size of the biopsy.
The protocol described here is designed to generate reproducible tissue fractions; however, some intrinsic variability within the sample, including the size and the status of the tissue can lead to deviations in the protein amount and distribution managed in each fraction.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Authors acknowledge the assistance of Ander Lopez and Miren Telletxea for video recording and editing. ACR is funded by an Ikerbasque fellowship and a research project from Asociación de Celiacos Madrid (ACM). JRB is funded by Project ISCIII-PI16/00258 and co-funded by the European Union ERDF/ESF “A way to make Europe”. IS is funded by a research project grant 2015111068 of the Basque Department of Health. IRG and AJM are supported by predoctoral fellowship grants from the UPV/EHU and the Basque Department of Education, respectively.
Materials
| HEPES | Sigma Aldrich | H4034-1kg | |
| KCl | Sigma Aldrich | P9333-1kg | |
| EDTA | Sigma Aldrich | E9884-100G | |
| NaCl | Sigma Aldrich | 5588886-1kg | |
| DTT | Sigma Aldrich | 10197777001 | |
| PMSF | Thermo Scientific | 36978 | |
| Proteinase and phosphatase inhibitors | Thermo Scientific | A32959 | |
| NP-40 | Sigma Aldrich | CA-630 | Detergent |
| BCA assay | Thermo Scientific | 23227 | Protein quantification kit |
| Disposable plastic pestles | Sigma Aldrich | Z359947-100EA | |
| Dounce homogeneizer | VWR | 431-0100 | |
| Microcentrifuge | Eppendorf | 5415-R | |
| Shacker | IKA | MS3 basic |
References
- Cox, B., Emili, A. Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nature Protocols. 1 (4), 1872-1878 (2006).
- Itzhak, D. N., Tyanova, S., Cox, J., Borner, G. H. Global, quantitative and dynamic mapping of protein subcellular localization. eLife. 5, (2016).
- Preedy, V. R., Watson, R. R., Ronald, R. . Methods in disease investigating the gastrointestinal tract. , (1998).
- Alex, P., Gucek, M., Li, X. Applications of proteomics in the study of inflammatory bowel diseases: Current status and future directions with available technologies. Inflammatory bowel diseases. 15 (4), 616-629 (2009).
- Ericsson, C., Franzén, B., Nistér, M. Frozen tissue biobanks. Tissue handling, cryopreservation, extraction, and use for proteomic analysis. Acta oncologica (Stockholm, Sweden). 45 (6), 643-661 (2006).
- Hoffmann, K., et al. New application of a subcellular fractionation method to kidney and testis for the determination of conjugated linoleic acid in selected cell organelles of healthy and cancerous human tissues. Analytical and Bioanalytical Chemistry. 381 (6), 1138-1144 (2005).
- Foster, L. J., et al. A Mammalian Organelle Map by Protein Correlation Profiling. Cell. 125 (1), 187-199 (2006).
- Baghirova, S., Hughes, B. G., Hendzel, M. J., Schulz, R. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX. 2, e440-e445 (2015).
- Börner, A., et al. Subcellular protein extraction from human pancreatic cancer tissues. BioTechniques. 46 (4), 297-304 (2009).
- Antalis, T. M., Shea-Donohue, T., Vogel, S. N., Sears, C., Fasano, A. Mechanisms of disease: protease functions in intestinal mucosal pathobiology. Nature clinical practice. Gastroenterology. 4 (7), 393-402 (2007).
- Cutler, P. . Protein purification protocols. , (2004).
- Walker, J. M. The Bicinchoninic Acid (BCA) Assay for Protein Quantitation. The Protein Protocols Handbook. , 11-14 (1996).
- McLane, L. M., et al. Differential localization of T-bet and Eomes in CD8 T cell memory populations. Journal of immunology (Baltimore, Md.: 1950). 190 (7), 3207-3215 (2013).
- Nguyen, K. T., Holloway, M. P., Altura, R. A. The CRM1 nuclear export protein in normal development and disease. International journal of biochemistry and molecular biology. 3 (2), 137-151 (2012).

