Establishment of a Clonal Culture of Unicellular Conjugating Algae
Summary
In this article, we demonstrate the establishment of clonal cultures of unicellular conjugating algal species collected from a natural field site.
Abstract
It is essential to establish clonal cultures of microalgae for use in studies of various topics, such as physiology, genetics, taxonomy, and microbiology. Thus, it is extremely important to develop techniques to establish clonal cultures. In this article, we demonstrate the establishment of clonal cultures of a conjugating alga. Water samples are collected from the field. Subsequently, cells are isolated using a glass capillary pipette, placed in media, and grown under conditions suitable for generating a clonal culture.
Introduction
Conjugating algae (of the order Zygnematales), also known as conjugatophytes, occupy a key phylogenetic position as the closest living relatives of the immediate ancestors of land plants1,2. Established clonal cultures of algae have led to a wide variety of taxonomic, physiological, and genetic studies over the past years. The isolation techniques of microorganisms from a natural field have a long tradition3,4. In recent years, automated isolation techniques using flow cytometry have also been established5. The most common method used to obtain a single cell for the establishment of a clonal culture is single-cell isolation using a glass capillary pipette6. This traditional method requires skill and a precise experimental protocol.
In this article, we demonstrate the sampling of algae from field samples and the establishment of clonal cultures of unicellular conjugating algae, such as the Closterium species, using a glass capillary pipette. A water sample containing vegetative cells is collected from a field site (e.g., a lake, pond, or paddy field). The cells are isolated from the water sample by using a glass capillary pipette and then washed. The cells are cultured in a test tube containing nitrogen-supplemented medium (CA medium) at 24 °C under a 16-h light:8-h dark regime. This method is also suitable for isolating other microalgae to generate clonal cultures.
Protocol
1. Collection of a Water Sample Containing Algae
NOTE: Unicellular conjugatophytes grow in stagnant freshwater areas, such as lakes, marshes, and paddy fields. An algae-containing water sample is collected from one of these environments to produce a culture strain. The following items are needed before starting the process: a plankton net, a disposable pipette, and a bottle or a 50 mL tube for collecting water samples.
- Use a plankton net in a lake environment to collect algae from the water. Use an appropriate net, depending on the purpose.
NOTE: The upper part of the net is permeable to water. The algae caught by the net are concentrated into the bottom container of the plankton net. A coarse mesh allows small algae to pass through. A fine mesh gets clogged easily. - Transfer the water (about 50 mL) to the 50 mL tube or a 100 mL plastic bottle. Bring the concentrated water sample to the laboratory.
- Unicellular conjugatophytes also grow in marshes or paddy fields. In these shallow water areas, water sample containing algae can be directly sampled with a dropper. Then, transfer the water (30 – 50 mL) to the 50 mL tube or a 50 mL plastic bottle.
NOTE: The water is carefully sampled above the soil surface so that the sample does not contain any soil.
2. Confirmation of Algae
NOTE: The following items are required: a loupe or portable microscope and a 60 mm dish for observation.
- To confirm the presence of algae in the sample while it is in the disposable pipette, use the loupe to observe the sample directly.
NOTE: Larger cells of about 400 – 1,000 µm in length (e.g., Closterium ehrenbergii) can easily be observed using this method. - To confirm algae under the portable microscope, pour the water sample in a plastic laboratory dish.
- Put the sample (about 3 mL) on the inner part of the upper half (lid) of the 60 mm Petri dish, then place the lower half, with its bottom part facing the inner part of the lid, on top.
NOTE: This method keeps the sample from moving and makes the observation easier.
- Put the sample (about 3 mL) on the inner part of the upper half (lid) of the 60 mm Petri dish, then place the lower half, with its bottom part facing the inner part of the lid, on top.
3. Preparation of a Glass Capillary Pipette from a Pasteur Pipette for the Isolation
NOTE: Before starting the process, the following items are needed: sterilized Pasteur pipettes, a rack for the Pasteur pipettes, gloves, a rubber bulb, a spirit lamp, methanol for the spirit lamp, a lighter, forceps, a trash can for glass, and a clean room or laminar flow hood (if necessary).
- Ignite an alcohol burner.
- To produce a fine glass capillary pipette for the isolation, make sure the flame is not flickering. Sharpen the flame and melt the Pasteur pipette at its tip.
- Heat a sterile glass Pasteur pipette on the flame, supported on the side of the rubber bulb by hand and on the tip side by using forceps.
- Take the Pasteur pipette off the flame and stretch it.
- When the glass is melting, quickly remove the Pasteur pipette from the flame and then pull.
NOTE: In this experiment, the Pasteur pipette was extended by 15 – 20 cm. Make sure that the pipette is off the flame during the pulling.
- When the glass is melting, quickly remove the Pasteur pipette from the flame and then pull.
- Break the tip of the glass capillary pipette to the proper length. Because the bowing part is the finest, it is ideal for the tip.
NOTE: In this study, a glass capillary pipette with a tip of 5 – 10 cm was prepared.- Be sure to discard the capillary after melting it, and confirm that the flame is put out.
- The bubbles released from the glass capillary pipette indicate the size of the tip opening. Select the proper size for the cell to be isolated. About 1 mm of bubbles is proper for the isolation of Closterium species, with cell widths of 10 – 20 µm.
4. Single-cell Isolation by a Glass Capillary Pipette
NOTE: The following items are needed before starting: an inverted light microscope, watch glasses, a plastic dish, and sterile threaded test tubes containing CA medium (Table 1) or conditioned CA medium7.
- Prepare a few watch glasses (22 mm in diameter and 1 mm in depth), each containing 1 mL of sterile CA medium.
- Isolate a target cell from the water sample collected in the field.
- Pour the water sample (about 30 – 40 mL) containing the target cells into the plastic dish.
- While observing the sample in the plastic dish under an inverted light microscope, pick up single cells using the glass capillary pipette.
- Bring the glass capillary pipette near the cell to facilitate the aspiration of the cell by capillary action.
- Transfer the cells into a watch glass containing 1 mL of sterile CA medium (Figure 1a).
- Pick up the cells again using a new glass capillary pipette from the sterile CA medium and transfer it to a second watch glass containing sterile CA medium (Figure 1b).
NOTE: This process is repeated until a single cell is isolated in the spot plate (Figure 1c). - Pick up the single cell using a new glass capillary pipette, and finally transfer it to a test tube containing 14 mL of CA medium or conditioned CA medium (Figure 1d). Incubate the sample at 23 °C under a 16-h light:8-h dark cycle for 2 weeks.
NOTE: The unknown factor(s) for cell proliferation, which are secreted from growing cells into the surrounding environment, are included in the conditioned medium to facilitate cell division4. If a clonal culture is established, prepare the conditioned CA medium from the culture medium4. The culture will turn green if successful.
Representative Results
A water sample (Inba1) of 50 mL was collected from one sampling point in Lake Inba-numa (pH 8.1, 24.7 °C; 35°44′30.2″N 140°12′08.2″E) at Sakura-shi, Chiba, Japan, on June 25, 2016. The water sample was stored at 4 – 8 °C. The next day after the collection, a specimen of Closterium sp. was isolated from the water sample containing vegetative cells. Fifteen vegetative cells of identical morphology (Inba1-1, -2, -3, -4, -5, -6, -7, -8, -9, -10, -11, -12, -13, -14, and -15) were isolated from the sample and then washed using the pipette washing method. Fifteen isolated single cells were cultured in independent test tubes containing 15 mL of conditioned CA medium. After 2 weeks of incubation, cell proliferation was observed in 9 test tubes [Inba1-1, -3, -4, -5 (Figure 2a), -6, -9, -10, -12 (Figure 2b), and -13; Table 2]. Nine clonal cultures were established from the Inba1 sample isolates (Figure 3).
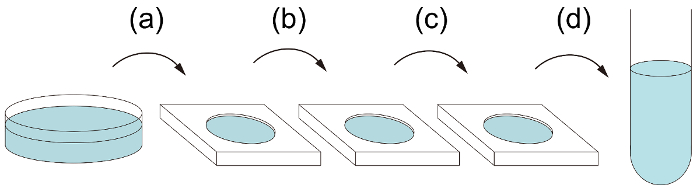
Figure 1: The flow of single-cell isolation. (a) Transfer the single target algal cell from the original water sample to the sterile medium in the first watch glass. (b, c) Transfer the cell again to the next watch glass to wash. This procedure should be repeated until there are no other cells/contaminants than the target in the medium; three watch glasses are shown in the diagram here as an example. (d) Finally, transfer the washed cell to a test tube of the appropriate medium for growth. Please click here to view a larger version of this figure.

Figure 2: Photographs of vegetative cells of Closterium sp. (a) Inba1-5 and (b) Inba1-12 are shown here. The scale bar = 50 µm. Please click here to view a larger version of this figure.

Figure 3: Establishment of a clonal culture from the Inba1 sample. The test tubes cultured for 2 weeks were photographed from the bottom. An asterisk indicates cell proliferation. The scale bar = 2 cm. Please click here to view a larger version of this figure.
| Name of the media | Reference | Comments | |
| CA | Ichimura and Watanabe12 | Ca(NO3)2·4H2O | 2 mg |
| KNO3 | 10 mg | ||
| NH4NO3 | 5 mg | ||
| β–Na2glycerophosphate·5H2O | 3 mg | ||
| MgSO4·7H2O | 2 mg | ||
| Vitamin B12 | 0.01 μg | ||
| Biotin | 0.01 μg | ||
| Thiamine HCl | 1 μg | ||
| PIV metals | 0.1 mL | ||
| Fe (as EDTA; 1:1 molar) | 0.1 mL | ||
| HEPES | 40 mg | ||
| Add water to make 100 mL | |||
| pH adjust with NaOH to 7.2 | |||
| P IV metals | Provasoli and Pintner13 | Na2EDTA·2H2O | 100 mg |
| FeCl3·6H2O | 19.6 mg | ||
| MnCl2· 4H2O | 3.6 mg | ||
| ZnCl2* | 1.04 mg | ||
| CoCl2·6H2O | 0.4 mg | ||
| Na2MoO4·2H2O | 0.25 mg | ||
| Add water to make 100 mL | |||
| Fe (as EDTA; 1:1 molar ) | Provasoli14 | Fe(NH4)2(SO4)2·6H2O, | 70.2 mg |
| Na2EDTA·2H2O, | 66 mg | ||
| Add water to make 100 mL | |||
| Conditioned CA medium | Abe et al.7 | Incubate cells in fresh CA medium for 14–20 days. Collect the cultured medium by filtration using qualitative filter paper and sterilize the filtered medium by autoclaving (121 °C for 15 min). | |
| *In the NIES, 1.04 mg ZnCl2 is replaced by 2.2 mg ZnSO4·7H2O. | |||
Table 1: Formulations of the media used in this study.
| Water sample | Name of cell | Status after culture | Strain name |
| Inba1 | -1 | Cells proliferated | Inba1-1 |
| -2 | Not changed | ||
| -3 | Cells proliferated | inba1-3 | |
| -4 | Cells proliferated | inba1-4 | |
| -5 | Cells proliferated | inba1-5 | |
| -6 | Cells proliferated | inba1-6 | |
| -7 | Not changed | ||
| -8 | Not changed | ||
| -9 | Cells proliferated | inba1-9 | |
| -10 | Cells proliferated | inba1-10 | |
| -11 | Not changed | ||
| -12 | Cells proliferated | inba1-12 | |
| -13 | Cells proliferated | inba1-13 | |
| -14 | Not changed | ||
| -15 | Not changed |
Table 2: The isolation trial summary.
Discussion
Using the present method, 9 clonal culture strains of Closterium sp. were established from 15 cells isolated from the water sample (Inba1), representing a 60% success rate. Species identification will be carried out in the future by morphological observation as well as DNA analyses, such as molecular phylogenetic analysis8.
In the present method, the freshness of the water sample is important to the success rate. It is better to isolate the cells from the water samples as soon as possible after the field collection. Although cells of the Closterium species (e.g., C. ehrenbergii, C. peracerosum-strigosum-littorale complex) can be maintained in the water sample for about 7 d by storing it at 4 – 8 °C, the isolation of the cells is desirable on the day of the collection or the next day. It is also important to remove any contaminants other than the target cells, so increasing the number of washing repetitions (i.e., > 3x) may prevent a contamination of the cultures by microorganisms in the soil and cells.
A limitation of this technique is that the culture media used in this protocol (CA or conditioned CA medium) are not always appropriate for all conjugating algae. In some cases, it may be necessary to consider using another medium, as well as different light and temperature conditions. Also, as it is difficult to prove whether a culture has grown from one single cell, it is necessary to accurately pick up only 1 cell when transferring cells to a test tube. Therefore, the transferring should be done with a new glass capillary pipette each time.
Establishing a clonal culture with this pipette washing method is a classical/standard method and is the most reliable method to use to isolate the desired cells for a study. The clonal cultures of conjugating algae used in many studies8,9,10,11 were established using the present method. It is difficult to analyze conjugating algae without a clonal culture. Furthermore, in recent years, de novo whole genome sequences became easy to obtain (e.g., by using the MinION nanopore sequencer), and establishing clonal cultures will further facilitate de novo sequencing. In other words, this method is the first step in the future study of these algae to better understand their biodiversity.
In order to establish a stable culture strain, it is necessary to perform certain critical steps accurately within the protocol. The most important step is the preparation of a glass capillary pipette with an optimal aperture to permit the passage of a single cell only. The aperture of the glass capillary pipette must be slightly more than the minor axis length of the cell of interest. The optimum glass capillary pipette can aspirate a single cell by capillary action. However, if the diameter of the aperture is too large, the capillary will also draw in non-living contaminant materials or even (an)other organism(s), in addition to the target cell.
To obtain a glass capillary pipette with an optimum aperture, the following points are important to note. First, the pillar of flame must be narrow when heating the glass capillary pipette. Next, when pulling the Pasteur pipette, it has to be removed from the flame; otherwise, the aperture of the Pasteur pipette will collapse.
The equipment and containers shown in this protocol are basic and can be modified. For example, by using multi-well plates instead of watch glasses with one well, the cell washing can be made more efficient. In addition, the containers can be substituted for other similar ones. By transferring a single cell to the culture medium at the last step, a clonal culture can be established.
Preparing a sterilized container and working in a hood will establish axenic (uncontaminated) strains. To prevent contamination, it is necessary to repeat the washing step with fresh aliquots more than 3x. Sometimes, bacteria are also sterilized by adding antibiotics15.
This method focused on desmid (Zygnematales) isolation, but by changing the size of the glass capillary pipette aperture, it can be applied to the isolation of differently sized plankton.
Acknowledgements
We thank Hisayoshi Nozaki (University of Tokyo), Takashi Nakada (Keio University), Yuka Marukawa Hashimoto (Japan Women's University), and Hiroyuki Sekimoto (Japan Women's University). We wish to thank the reviewers for their comments. This study was supported by a Grant-in-Aid for Scientific Research (No. 26440223 and 15H04413) from the Japan Society for the Promotion of Science, Japan, and by a grant from the New Technology Development Foundation to Yuki Tsuchikane.
Materials
| Sampling | |||
| Plankton net | RIGO, Japan | N-NO380T | Mesh size: 32 µm |
| Portable microscope (e.g., Nature Scope FABRE) | Nikon, Japan | JAN: 4960759 206725 | Magnification: 20× |
| Loupe (e.g., Peak 1985 Steinheil System Magnifier) | Tohkai Sangyo Co. Ltd., Japan | 1985-20 | Magnification: 20× |
| Spuit | EIKEN CHEMICAL CO. LTD. Japan | Sterile spuit No.4 | |
| 50 mL Tube (50 mL centrifuge tubes with screw caps) | Labcon, USA | 3181-345-008 | |
| Bottle (100 mL Polycarbonate bottle) | AS ONE Corporation, Japan | 1-7403-01 | |
| 60 mm dish | Asahi glass Co. Ltd., Japan | 1010-060 | For observation of algae. 60 mm/non-treated dish |
| Preparation of a micropipette | |||
| Pasteur pipettes (cotton plugged 9” Pasteur Pipettes) | Fisher Scientific, USA | 13-678-8B | 7 × 225 mm |
| Rubber bulb (SPOID SILICONE, 1 cc) | AS ONE Corporation, Japan | 5-5669-01 | 10 × 40 mm |
| Stainless forceps | AS ONE Corporation, Japan | PT-09 | 110 mm |
| Single-cell isolation | |||
| Watch glass (Blood reaction board) | Sekiya Rika Co., Ltd, Japan | F14-155-030 | to rinse cells during isolation. 22 mm diameter and 1 mm depth |
| Threaded test tubes | Fujimotorika, Co., Ltd, Japan | XX142 | 18 Ø × 170 mm |
| Inverted light microscope | Olympus, Tokyo, Japan | CKX41N | for isolation of algae. |
| Plastic dish | Asahi glass Co. Ltd., Japan | SH90-15E | 90 × 15 mm |
References
- Wickett, N. J., et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences. 111, E4859-E4868 (2014).
- Delwiche, C. F., Cooper, E. D. The evolutionary origin of a terrestrial flora. Current Biology. 25, R899-R910 (2015).
- Daste, P., Neuville, D., Victor-Baptiste, B. A simple procedure for obtaining clonal isolation of diatoms. British Journal of Psychology. 18, 1-3 (1983).
- Pringsheim, E. G. . Pure Cultures of Algae: Their Preparation and Maintenance. , 119 (1946).
- Sieracki, M., Poulton, N., Crosbie, N., Andersen, R. A. Automated isolation techniques for microalgae. Algal Culturing Techniques: Automated isolation techniques for microalgae. , 101-116 (2005).
- Andersen, R. A., Kawachi, M., Andersen, R. A. Traditional microalgae isolation techniques. Algal Culturing Techniques: Traditional micro algae isolation techniques. , 83-100 (2005).
- Abe, J., et al. Stable nuclear transformation of the Closterium peracerosum-strigosum-littorale complex. Plant and Cell physiology. 52, 1676-1685 (2011).
- Tsuchikane, Y., Tsuchiya, M., Kokubun, Y., Abe, J., Sekimoto, H. Conjugation processes of Penium margaritaceum (Zygnemophyceae, Charophyta). Phycological Research. 59, 74-82 (2011).
- Kanda, N., et al. CRISPR/Cas9-based knockouts reveal that CpRLP1 is a negative regulator of the sex pheromone PR-IP in the Closterium peracerosum-strigosum-littorale complex. Scientific Reports. 7, 17873 (2017).
- Ichimura, T. Mating types and reproductive isolation in Closterium-ehrenbergii Meneghini. Botanical Magazine-Tokyo. 94, 325-334 (1981).
- Sekimoto, H., Satoh, S., Fujii, T. Biochemical and physiological properties of a protein inducing protoplast release during conjugation in the Closterium peracerosum-strigosum-littorale complex. Planta. 182, 348-354 (1990).
- Ichimura, T., Watanabe, M. The Closterium calosporum complex from the Ryukyu Islands-Variation and taxonomical problems. Memoirs of the National Science Museum. 7, (1974).
- Provasoli, L., Pintner, I. J. Artificial media for fresh-water algae: problems and suggestions . The Ecology of Algae, a symposium held at the Pymatuning Laboratory of Field Biology on June 18 and 19, 1959. , 84-96 (1960).
- Provasoli, L., Tryon, C. A., Hartmann, R. T. Media & prospects for the cultivation of marine algae. Proceedings of U. S.-Japan Conference in Hakone, Japan. , 63-75 (1968).
- Guillard, R. R. L., Andersen, R. A. Purification methods for microalgae. Algal Culturing Techniques: Purification methods for microalgae. , 117-132 (2005).

