Measuring Liver Mitochondrial Oxygen Consumption and Proton Leak Kinetics to Estimate Mitochondrial Respiration in Holstein Dairy Cattle
Summary
Here, we share methods for measuring mitochondrial oxygen consumption, a defining concept of nutritional energetics, and proton leak, the primary cause of inefficiency in mitochondrial generation of ATP. These results can account for 30% of the energy lost in nutrient utilization to help evaluate mitochondrial function.
Abstract
Oxygen consumption, proton motive force (PMF) and proton leak are measurements of mitochondrial respiration, or how well mitochondria are able to convert NADH and FADH into ATP. Since mitochondria are also the primary site for oxygen use and nutrient oxidation to carbon dioxide and water, how efficiently they use oxygen and produce ATP directly relates to the efficiency of nutrient metabolism, nutrient requirements of the animal, and health of the animal. The purpose of this method is to examine mitochondrial respiration, which can be used to examine the effects of different drugs, diets and environmental effects on mitochondrial metabolism. Results include oxygen consumption measured as proton dependent respiration (State 3) and proton leak dependent respiration (State 4). The ratio of State 3 / State 4 respiration is defined as respiratory control ratio (RCR) and can represent mitochondrial energetic efficiency. Mitochondrial proton leak is a process that allows dissipation of mitochondrial membrane potential (MMP) by uncoupling oxidative phosphorylation from ADP decreasing the efficiency of ATP synthesis. Oxygen and TRMP+ sensitive electrodes with mitochondrial substrates and electron transport chain inhibitors are used to measure State 3 and State 4 respiration, mitochondrial membrane PMF (or the potential to produce ATP) and proton leak. Limitations to this method are that liver tissue must be as fresh as possible and all biopsies and assays must be performed in less than 10 h. This limits the number of samples that can be collected and processed by a single person in a day to approximately 5. However, only 1 g of liver tissue is needed, so in large animals, such as dairy cattle, the amount of sample needed is small relative to liver size and there is little recovery time needed.
Introduction
Mitochondria are very sensitive to stress and their cellular environment can contribute to a wide variety of metabolic diseases. Oxygen consumption and proton leak in mitochondria are indicators of mitochondria health. The methods described in this paper estimate mitochondrial energy efficiency using RCR based on oxygen consumption with and without proton leak. These results can account for 30% of the energy lost in nutrient utilization1. Changes in oxygen consumption and proton leak can identify mitochondrial dysfunction which contributes to metabolic disease and results in decreased energy efficiency. These methods can also be used to examine the effect of different treatments on mitochondrial respiration. The overall goal of measuring mitochondrial oxygen consumption and proton leak kinetics is to assess mitochondrial function and energetic efficiency.
Hepatic mitochondrial dysfunction has been associated with several diseases in dairy cattle. The ability of cellular metabolism to switch between carbohydrate and lipid fuels when faced with an energy deficit in early lactation is influenced by the number and function of mitochondria in the cell2. Defects in the ability of mitochondria to adapt to an increased demand for energy and increased β-oxidation can lead to accumulation of intracellular lipid associated with insulin resistance and may lead to the formation of fatty liver in early lactation dairy cows. Mitochondria, as the site of ketone body production and use, can play a key role in ketosis in dairy cows3. A lack of mitochondria or mitochondrial dysfunction will impact fuel availability to the periphery and be reflected in changes in oxygen consumption or RCR.
Mitochondrial oxygen consumption changes in response to inflammation. Seven-day-old broilers were randomly assigned to a group infected with Eimeria maxima and a control group4. Broilers that did not undergo coccidiosis challenge had lower oxygen consumption due to proton leak and higher RCR indicating that liver mitochondria respond to an immune challenge by increasing proton leak. While proton leak and reactive oxygen species production was once considered a sign of mitochondrial membrane dysfunction and detrimental to energetic efficiency, now it is known that it is important for import of proteins and calcium into mitochondria5, and for the generation of heat1.
Electron leak from the respiratory chain makes mitochondria susceptible to reactive oxygen species production and oxidative damage to mitochondrial membrane proteins, lipids and mitochondrial DNA. As mitochondria age, damage can accumulate especially to mtDNA causing further dysfunction in mitochondrial metabolism6 and greater susceptibility of the cow to disease. In practice, many livestock animals are fed high levels of supplements such as Cu, Zn and Mn to boost antioxidant function. However, feeding high levels of Cu, Zn and Mn decreased milk production and increased oxygen consumption due to proton leak (State 4 respiration)7.
Previous research on the role of mitochondrial function in energy efficiency in cattle has focused on changes in mitochondrial oxygen consumption and proton leak. Very few studies have been published in dairy cattle and most papers compare production efficiency in the form of residual feed intake (RFI) to mitochondrial function in beef cattle. Variability in mitochondrial respiration rates were examined by measuring state 3, state 4 and RCR in livers from both lactating Holstein cows and lactating beef cows (Angus, Brangus and Hereford)8. The researchers did not find any correlation in mitochondrial respiration with growth or milking traits for beef cattle but did report a correlation between mitochondrial respiration and milking traits for Holsteins. In two studies, RFI was compared in beef cattle to mitochondrial respiration rates (state 3, state 4 and RCR) in muscle mitochondria9,10. Mitochondrial respiration rates changed in response to DMI and low rates were associated with less efficient beef steers. In another study, RFI of steers from high or low RFI bulls were compared with mitochondrial respiration rates and proton leak kinetics between the two groups of progeny11. Differences were due to gain confirming the conclusion that gain does not impact mitochondrial respiration in beef cattle.
In this paper, an experiment examining liver RCR in response to feeding 3 antioxidant minerals to lactating dairy cattle illustrates the use of methods to measure oxygen consumption during State 4 and State 3 respiration and PMF.
Protocol
All methods, protocol and studies described here were approved by the Institutional Animal Care and Use Committee (IACUC) of University of California, Davis.
1. Obtaining a Liver Biopsy from a Holstein Dairy Cow
NOTE: A liver biopsy should be performed by a licensed veterinarian. Liver biopsies can be performed on the dairy site where the cows are located. Lactating dairy cows can continue to be milked normally and milk does not need to be withdrawn from the food supply before or after the procedure. It is recommended that at least 4 people are needed to perform the liver biopsy on a dairy cow: a veterinarian to perform the biopsy, an animal handler to stand at the cow's hip to protect the biopsy area and veterinarian, a lab technician on the outside of the pen to transfer tools, materials and biopsy sample to and from the veterinarian and maintain the clean area, which can be in the back of a vehicle (Figure 1), and a technician to retrieve the liver sample and begin mitochondrial isolation.
- One month prior to liver biopsies, give cows a clostridia vaccination. Create surgical packs by autoclaving surgical towels, biopsy instrument, scalpel holders and surgical equipment.
- One day before the liver biopsy, inject the cow with Ceftiofur Hydrochloride 0.044 mL/kg bodyweight subcutaneously in the neck. Monitor cow temperature, intake and fecal scores to use as a baseline for normal function.
- Create mitochondria isolation media (MIM) containining 220 mM mannitol, 70 mM sucrose, 20 mM HEPES, 1 mM EDTA, and 0.1% (w/v) fatty acid free BSA, pH 7.4 at 4 °C. Approximately 30 mL per sample will be needed.
- Restrain cow physically utilizing a headlock with a halter as needed (Figure 2). Using the halter, tie her head to the left side of the stanchion. If necessary, a chemical restraint (Xylazine hydrochloride 100 mg/mL IV at 0.010-0.015 mg / kg bodyweight) can be used.
- The area of the biopsy is found at the right 10 – 11th intercostal space (Figure 3). Draw a straight line from the right tuber coxae to the point of the right shoulder. The biopsy site is where this line intersects with the 10-11th intercostal space. Sterilize the area of the cow to be biopsied by shaving a 10 cm square area (Figure 4). Wash area with 10% providone scrub (Figure 5) using a circular motion. Spray area with 70% ethanol solution (Figure 6). Repeat providone and ethanol washes.
NOTE: The liver is in a slightly different position in Holstein dairy cattle compared to beef cattle. - Inject 2% lidocaine HCl (10-15 mL) locally to the area to provide anesthesia of the skin and underlying muscle and connective tissue (Figure 7). Repeat providone and 70% ethanol washes.
NOTE: The nerve endings are in the skin and muscle, but not internal organs, so only a local anesthestic is needed. At most, the cow should only feel some pressure and no pain during the biopsy procedure. - Make a 1-2 cm stab-incision through the skin of the 10-11th intercostal space (Figure 8). Pass a Schackelford-Courtney bovine liver biopsy instrument through the skin and direct the biopsy instrument in a slight cranial direction while continuing through the diaphragm and into the liver (Figure 9, Figure 10). Obtain a 1 g sample of the liver and remove the instrument (Figure 11). Close the skin with suture placement (Figure 12).
- Place liver sample in a conical tube with enough MIM to cover sample, on ice for immediate mitochondria isolation
- Check incision for any redness, swelling, heat, or pain within 24 hours of biopsy and inject the cow with Ceftiofur Hydrochloride 0.044 mL/kg bodyweight subcutaneously in the neck once a day for the next 3 days (Figure 13). Monitor the cow's temperature, intake and fecal scores daily for 1 week after liver biopsy. If a fever develops, continue antibiotics at the discretion of the veterinarian.
NOTE: If a cow is exhibiting signs of pain, such as kicking at the incision site, recumbancy, redness, heat, or reaction to touch within 1 h after the liver biopsy, a 1 mg/kg bodyweight IV injection of flunixin meglumine can be used to alleviate pain and inflammation. A second injection can be administered if necessary. - Remove sutures 7 days after biopsy.
2. Isolating Mitochondria from Dairy Cow Liver
- As soon as possible after the liver sample is removed from the cow, wash the liver sample in MIM (step 1.3) to remove red blood cells and finely mince the sample with scissors. The liver should be minced in a chilled beaker containing enough isolation media to keep the tissue moist.
- Place the minced liver into a 30 mL glass vial with a teflon pestle of 0.16 mm clearance incubated in ice and containing MIM (1:4 w/v).
- Homogenize liver sample in teflon pestle at 500 rpm for a min with 4 strokes/min.
NOTE: The liver homogenate is kept in an ice-packed beaker in MIM during the entire process, and all of the following centrifugation steps are completed at 4 °C - Centrifuge homogenate at 500 x g for 10 min, discard pellet, transfer supernatant to a chilled centrifuge tube and then centrifuge the resulting supernatant at 10,000 x g for 10 min to obtain the mitochondrial pellet.
- Resuspend and wash the pellet in 10 mL of MIM with fatty acid free BSA and centrifuge at 8100 x g for 10 min. Discard supernatant.
- Resuspend and wash the pellet in 10 mL of MIM without fatty acid free BSA and centrifuge at 8100 x g for 10 min. Discard supernatant.
- Suspend the pellet in 200 µL of isolation media and place on ice until used for oxygen consumption and proton leak kinetic assays.
- Determine protein concentration of the pellet suspension (1/100 dilution) using Bicinchoninic acid (BCA) kit per manufacturer's protocol with BSA as the standard. All protein is considered to be mitochondrial protein.
3. Measuring Mitochondrial Oxygen Consumption (State 3 and State 4)
- Create oxygen consumption media (OCM) from 120 mM KCl, 5 mM KH2PO4, 5 mM MgCl2, 5 mM Hepes and 1 mM EGTA, pH 7.4 at 30 °C with 0.3% defatted BSA. Approximately 3 mL per sample will be needed. Also prepare a solution of 8 μg/mL oligomycin in ethanol.
- Incubate OCM at 30 °C. Set up respiration chamber, pump and oxygen electrode according to manufacturer directions (oxygraph system). The oxygraph software should already be installed on the computer.
- Place 1 mL of OCM into the respiration chamber and stir vigorously. This will help insure that the solution becomes saturated with air.
- Add 0.35 mg protein of mitochondrial protein to the respiration chamber and maintain temperature at 30 °C.
- Record oxygen consumption for approximately 5 min. The oxygraph system records oxygen concentration so as respiration increases, oxygen concentration decreases. When oxygen consumption becomes constant (a decreasing straight line), record oxygen consumption (slope of line = concentration of oxygen/time). This is baseline oxygen consumption.
- Add 1.25 µL of 4 mM rotenone solution to inhibit Complex I and then add 5 µL of 1 M succinate solution to reach a final concentration in the respiratory chamber of 5 mM succinate. This is state 4 respiration.
- Add 1 µL of 100 mM ADP solution to reach a final concentration in the respiration chamber of 100 μM. Oxygen concentration will decrease (increased respiration) and then after approximately 5 min becomes a straight line. Record oxygen consumption (slope of line = concentration of oxygen/time). This is State 3 respiration.
- Optional: At the end of the run, add FCCP (0.2 μM total volume) to induce maximal respiration. Record respiration for about 5 min (approximately). When oxygen consumption becomes constant, record oxygen consumption. This is maximal oxygen consumption.
- Calculate Respiratory Control Ratio (RCR) using the equation State 3 oxygen consumption / State 4 oxygen consumption.
- Aspirate all of the solutions out of the respiration chamber. Rinse the chamber several times with double deionized water.
4. Measuring Mitochondrial Membrane Potential (MMP) and Proton Motive Force (PMF)
- Prepare solution of 80 ng/mL nigericin in ethanol.
NOTE: These chemicals are dissolved in ethanol, and every effort should be made to limit the amount of ethanol that is added to less than 1 μL, since ethanol can uncouple the electron transport system and cause mitchondrial dysfunction. - After thoroughly rinsing the chamber with double deionized water, place 1 mL of the OCM into the respiration chamber and stir vigorously with a magnetic stir bar. This will help insure that the solution becomes saturated with air. Add methyl-triphenyl-phosphonium (TPMP+) sensitive electrode to chamber setup. TPMP+ electrode should be connected to a pH meter and values are read from the pH meter.
- Add 0.35 mg of mitochondrial protein to the respiration chamber.
- Add 1.25 µL of 4 mM rotenone solution to inhibit Complex I. Record respiration for 2-5 min (approximately). When oxygen consumption becomes constant, record oxygen consumption.
- Add 0.56 μL of 8 μg/mL oligomycin solution for a final concentration of 2.8 µg oligomycin /0.35 mg of mitochondrial protein to inhibit ADP utilization. Record respiration for 2-5 min (approximately). When oxygen consumption becomes constant, record oxygen consumption.
- Add 0.112 μL 80 ng/mL nigericin solution to abolish the pH gradient across the mitochondrial inner membrane. Record respiration for 2-5 min (approximately). When oxygen consumption becomes constant, record oxygen consumption.
NOTE: Rotenone and oligomycin are used to block electron transport chain at Complex I and ATP synthase, respectively. Nigericin is added to convert transmembrane H+ gradient to a K+ gradient that can be measured with an electrode. - Prepare a standard curve for TPMP+ by adding 5 µL of 10 mM TPMP+ solution to the mitochondrial incubation. Repeat this step four more times until a total concentration of 2.5 μM TPMP+ has been added.
- Initiate respiration by adding 5 μL of 1M succinate to the chamber.
- Record respiration until you have achieved a stable trace, and then titrate the system by adding malonate. Additions of malonate should be 0.5 µL, 1 µL, 1.5 µL, 3.0 µL, 6.0 µL, 9.0 µL, then 12.5 µL of 0.1 mM Malonate solution to achieve successive additions of malonate concentrations in the incubation chamber of 0.1, 0.2, 0.3, 0.6, 1.2, 1.8, and 2.5 mM.
- Collect data from the two electrodes (oxygen and TPMP+). Data acquisition software from the oxygraph system can be used to collect simultaneous measurements of mitochondrial oxygen consumption and mitochondrial membrane potential and observe changes in oxygen consumption in real time. Figure 14 shows how the oxygraph system records oxygen consumption as the experiment progresses.
- Calculate MMP in mV based on the Nernst equation:
MMP = 61.5 log ([TPMP+] added – external [TPMP+]) x TPMP+ binding correction/ (0.001 x mg of protein/mL x [TPMP+])
A TPMP+ binding correction of 0.4 µL/mg of mitochondrial protein-1 is used.
Example calculation based on concentrations in protocol:
MMP = 61.5 x log (5 µM – 2 µM) x 0.4/ (0.001 x 0.35 mg mitochondrial protein/mL x 2 µM)
MMP = 198.9 mV - Estimate PMF by plotting a graph of MMP vs. oxygen consumption (Figure 15). PMF is reported as oxygen consumption at a membrane potential of 165 mV.
NOTE: Titrating the electron transport chain with malonate (0.1 to 2.5 mM) shows the kinetic response of proton leak to MMP. Then, plotting MMP against oxygen consumption determines proton leak kinetics. PMF is determined by calculating oxygen consumption at a common membrane potential (165 mV). - At the end of the last run of the sample, add FCCP (0.2 μM total volume) to induce maximal respiration and release TPMP+ for baseline correction.
- Aspirate all of the solutions out of the respiration chamber. Rinse the chamber several times with double deionized water. At the end of the day, the chamber should also be rinsed a few times with ethanol.
Representative Results
Positive results showing RCR and proton leak kinetics are shown in Table 1 and Figure 15, respectively. In this study7, RCR and protein leak kinetics were measured in Holstein dairy cows at 70 days in milk after cows had been fed 1 of 5 different levels of Cu, Zn and Mn for 28 days. State 4, maximum proton leak-dependent respiration, had a tendency to be affected by mineral intake of Cu, Mn and Zn (p < 0.1). State 3 respiration (maximum ATP stimulated respiration) and RCR = State 3 / State 4 (respiratory control ratio) was not affected by mineral intake. State 4 respiration was highest in LowMn and lowest in Control, indicating that Mn plays an important role in minimizing proton leak dependent respiration. Manganese, through the enzyme Mn Superoxide Dismutase is known to reduce reactive oxygen species in the mitochondrial matrix and reduce proton leak12. Higher State 4 respiration was associated with lower milk and milk protein yield. Since proton leak is an important component of energy efficiency, reducing State 4 respiration through Mn supplementation could improve efficiency.
| Treatments1 | ||||||
| High | Med | Low | LowMn | Control | SEM | |
| Milk, kg | 47.4ab | 50.9a | 46.0ab | 43.6b | 49.7a | 2.9 |
| Milk protein, kg | 1.38ab | 1.44a | 1.40ab | 1.23b | 1.43a | 0.09 |
| State 3 | 75.8 | 64.4 | 78.2 | 73 | 64.1 | 13 |
| State 4 | 26.2ab | 22.6ab | 25.9ab | 27.1a | 22.0b | 3 |
| RCR | 2.89 | 2.76 | 2.98 | 2.65 | 2.83 | 0.27 |
| a b Means within a row not followed by the same superscript letter are significantly different (P < 0.1). | ||||||
| 1 High treatment contains highest levels of Cu, Zn and Mn all well above requirements13, Med treatment contains intermediate levels of Cu, Zn and Mn above requirements, Low treatment contains lower levels of Cu, Zn and Mn but still above requirements, Low Mn treatment contains the lowest levels of Mn (and lower levels of Cu and Zn) but still above requirements and Control treatment contains the lowest levels of Cu and Zn, which are close to requirements. | ||||||
Table 1: Effect of Cu, Mn and Zn supplementation on liver mitochondrial oxygen consumption and milk production from dairy cows at 70 days in milk. This table has been adapted from Acetoze et al. 20177.
Mitochondrial proton leak is a process that dissipates MMP through the movement of protons across the mitochondrial inner membrane without production of ATP14. Proton leak kinetics are assessed by calculating rates of oxygen consumption at a common membrane potential of 165 mV. A lower membrane potential means that protons are 'leaking' across the mitochondrial membrane, which results in lower ATP synthesis (Figure 15). In the Holstein cow study, hepatic proton leak dependent respiration was greatest in LowMn and lowest in Control, which agrees with results in Table 1, that State 4 respiration was greatest in LowMn and lowest in Control.
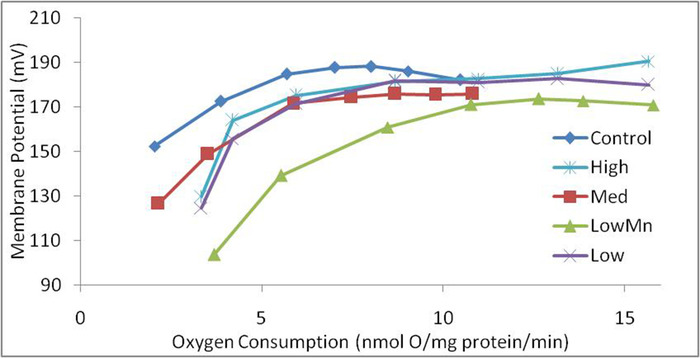
Figure 15. Proton leak kinetics in Holstein cows fed different amounts of Cu, Mn and Zn. This graph is based on data from Acetoze et al. 20177. Please click here to view a larger version of this figure.
Negative results are illustrated in Table 2 and Figure 16. Feed efficiency (RFI) was higher in Angus steers born from low RFI bulls than high RFI bulls, but this was not reflected in mitochondrial RCR (Table 2) or proton leak kinetics (Figure 16). There were no differences in mitochondrial respiration and proton leak kinetics between groups of steers but there was a difference in RFI. There were also no differences (p = 0.88) in hepatic mitochondrial proton leak in high and low RFI steers (Figure 16). There were large standard errors associated with mitochondrial respiration measurements, and proton leak kinetic curves were flat. Liver samples from this study were obtained after steers were slaughtered, a process that delayed liver sample collection and processing by an hour. Variation in mitochondrial respiration measures may reflect mitochondrial respiration degradation due to tissue death. Proton leak kinetic lines were flat because oxygen consumption measurements did not begin until 8 min when plateau had already been reached due to an equipment malfunction.
| Low RFI | High RFI | SEM | P Value | |
| (n=7) | (n=8) | |||
| RFI | -0.58 | -0.01 | 0.1 | 0.05 |
| State 3 | 31.3 | 30.8 | 9.42 | 0.9 |
| State 4 | 9.76 | 10.4 | 3.23 | 0.8 |
| RCR | 3.05 | 3.03 | 0.24 | 0.93 |
Table 2: Performance and mitochondrial respiration of high and low Residual Feed Intake (RFI) Angus bull progeny. This table has been adapted from Acetoze et al. 201511.
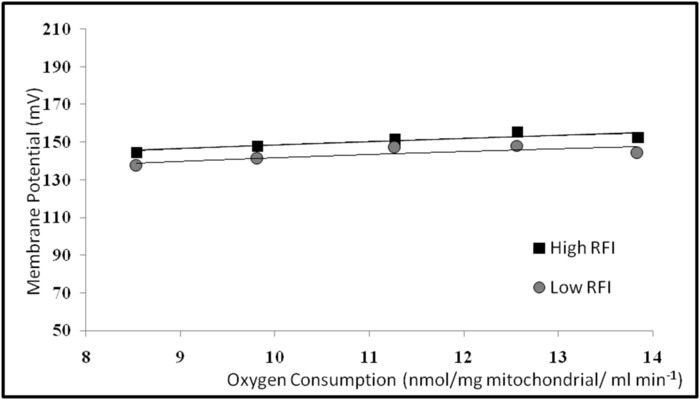
Figure 16. Proton leak kinetics for progeny of high and low RFI Angus bulls. This graph has been adapted from Acetoze et al. 201511. Please click here to view a larger version of this figure.

Figure 1: Clean area for surgical and biopsy materials located in the back of a vehicle and outside of the cow pen. Please click here to view a larger version of this figure.

Figure 2: Restraint of the cow using a halter tied to a cross pole of the head lock. Please click here to view a larger version of this figure.

Figure 3: The area of the cow to clean for the biopsy and location of biopsy at the right 10 – 11th intercostal space found by drawing a straight line from the right tuber coxae to the point of the right shoulder. The biopsy site is where this line intersects with the 10-11th intercostal space. Please click here to view a larger version of this figure.

Figure 4: Shaving a 10 cm area of the cow to prepare to sterilize for the biopsy. Please click here to view a larger version of this figure.

Figure 5: Wash biopsy area of the cow with 10% providone scrub using a circular motion. Please click here to view a larger version of this figure.

Figure 6. Spray biopsy area area with 70% ethanol solution. Please click here to view a larger version of this figure.

Figure 7: Inject 2% lidocaine HCl (10-15 mL) locally to the area to provide anesthesia of the skin. Please click here to view a larger version of this figure.

Figure 8: A 1-2 cm stab-incision through the skin of the 10-11th intercostal space to insert biopsy tool. Please click here to view a larger version of this figure.
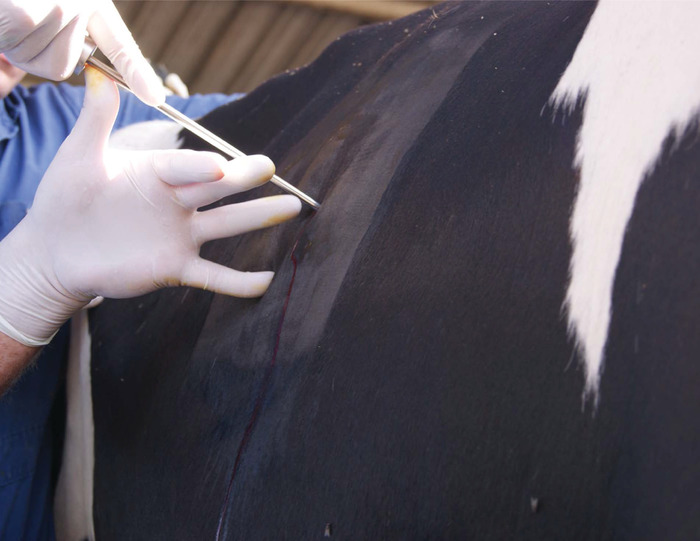
Figure 9: Insertion of bovine liver biopsy instrument through the skin. Please click here to view a larger version of this figure.
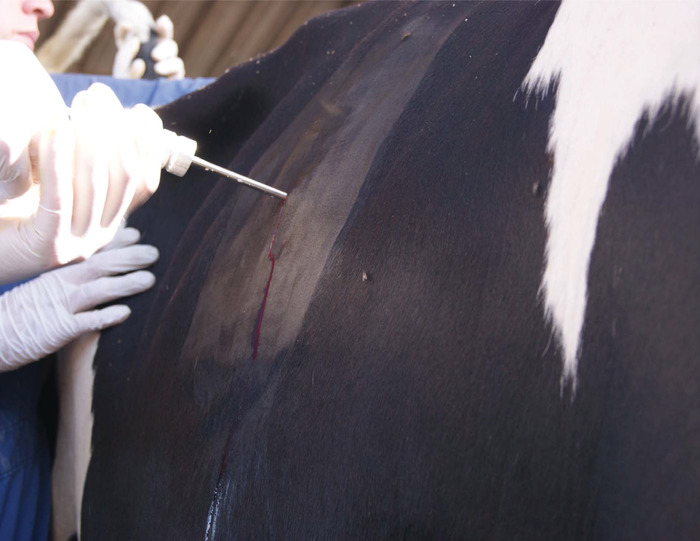
Figure 10: The biopsy instrument should be directed in a slight cranial direction while continuing through the diaphragm and into the liver. Please click here to view a larger version of this figure.

Figure 11: A 1 g sample of the liver being moved from biopsy instrument to Falconer tube for transport to mitochondrial isolation station. Please click here to view a larger version of this figure.

Figure 12: Suturing the skin to close biopsy incision. Please click here to view a larger version of this figure.

Figure 13: Injection of the cow with Ceftiofur Hydrochloride 0.044 mL/kg bodyweight subcutaneously in the neck. Please click here to view a larger version of this figure.
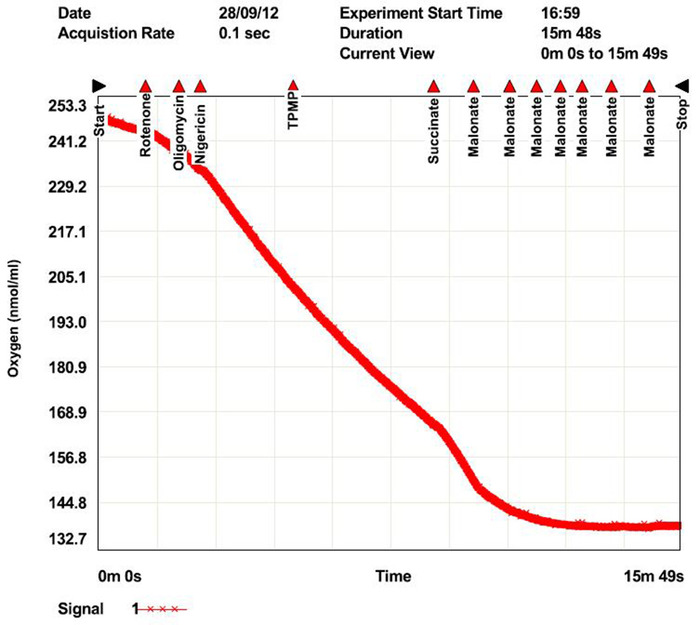
Figure 14: Oxygraph software results showing oxygen consumption responses to addition of each substance to measure mitochondrial membrane potential (MMP) and proton motive force (PMF). Please click here to view a larger version of this figure.
Discussion
The most critical point in the protocol is obtaining a representative liver tissue sample and beginning the isolation of mitochondria as soon as possible after biopsy. Variation in respiration measurements is low (Table 1) due to a short transport time from cow to laboratory. To reduce transport time, a small laboratory was set up in the office of the dairy, and liver samples were driven to the office laboratory as each was collected so that mitochondria were isolated within 10 min of biopsy. Setup and testing of the respiration chamber and electrodes (oxygen, TPMP+) with the pH meter used to record differences in proton gradients the day before collecting and processing samples can avoid malfunctions such as missing early measurements in proton leak kinetics (Figure 16).
Because of the need for fresh liver samples and rapid isolation of mitochondria, the number of samples that can be collected and processed in a day is limited. Each sample takes approximately 5-6 h to complete; therefore, only about 5 samples per day can be collected and analyzed per respiration chamber. This is not a high throughput method; the sample size for treatments is limited and small errors can increase variability associated with results and the ability to detect significance.
The isolation technique may exclude some mitochondria that are associated with some cell components and remain embedded in the pellet or smaller mitochondria may be lost in the pellet washes during the centrifugation steps. This may lead to results that do not reflect the complete population of mitochondria. Mitochondria can change in size and density depending on physiological states such as starvation and exercise (training)15. Estimating mitochondrial number with enzyme activity using citrate synthase16 or succinate dehydrogenase17 to corroborate findings may be needed.
No modifications were made to the original techniques established in rodents for mitochondrial isolation18, respiration19 and proton leak kinetics20. Modifications to this technique can be made depending on tissue source of mitochondria and experimental treatments. BSA (defatted) is used to bind free fatty acids in tissue. If the tissue has a lot of fatty acids (greater than 10%) associated with it, more defatted BSA can be added because free fatty acids will interfere with the mitochondrial measurements.
Measuring mitochondrial oxygen consumption and proton leak kinetics using this technique is the standard procedure. Liver has been the tissue of choice primarily because it has a lot of mitochondria, they are fairly easy to extract, and the liver is the primary site of nutrient processing. Modifications of this technique have been used to measure oxygen consumption in other tissues such as muscle and mammary. However, mitochondrial isolation techniques must be modified to fit the tissue. For instance, in muscle, mitochondria are embedded in muscle fibers and so the isolation procedure must include a protein digestion and the digestion must be controlled to ensure that mitochondrial function is not disrupted.
There are other methods for measuring mitochondrial respiration that require an analyzer specifically designed to measure respiration. Cells must be harvested from tissue and fixed to incubation plates. The analyzer measures whole cell oxygen consumption rate (OCR; basal), ATP linked OCR (associated with mitochondria), nonmitochondrial OCR, and maximal OCR. However, since mitochondria are inhibited during some of the incubation, isolated mitochondria measurements are not possible. This method has been used to examine OCR changes with disease and drug interventions21 in humans.
Current and Future Applications
The contribution of proton leak to energy requirements of the animal can be large and indicative of the physiological state of the animal including growth, lactation and disease. In the past, this technique has been primarily used to examine the association of mitochondrial oxygen consumption and contribution of proton leak to feed or energetic efficiency. However, as our understanding of the role of mitochondria in metabolism expands, the importance of this technique will also increase particularly in combination with other mitochondrial measures such as Electron Transport Chain enzyme activities, calcium dynamics in apoptosis and enzyme activities of the TCA cycle.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by Alltech and USDA Hatch funds through the Center for Food Animal Health at UC Davis School of Veterinary Medicine.
Materials
| Liver Biopsy | |||
| Equipment | |||
| Schackelford-Courtney bovine liver biopsy instrument | Sontec Instruments Englewood CO | 1103-904 | |
| Suture | Fisher Scientific | 19-037-516 | |
| Suture needles | NA | NA | Included with Suture |
| Scalpels | Sigma – Aldrich | S2896 / S2646 | # for handle and blades |
| Surgery towels | Fisher Scientific | 50-129-6667 | |
| Falcon tubes 50 mL | Fisher Scientific | 14-432-22 | |
| Tweezers | Sigma – Aldrich | Z168750 | |
| 50 mL syringes | Fisher Scientific | 22-314387 | |
| Injection needles (22, 2 1/2) | VWR | MJ8881-200342 | |
| Cow halter | Tractor Supply Co. | 101966599 | |
| Cotton swabbing | Fisher Scientific | 14-959-102 | |
| cotton gauze squares (4×4) | Fisher Scientific | 22-246069 | |
| Medical scissors | Sigma – Aldrich | Z265969 | |
| Chemicals | |||
| Coccidiosis Vaccine 0.75 bottle/cow | Provided by Veterinarian | ||
| Clostridia Vaccine | Provided by Veterinarian | ||
| Liver biopsy antibiotics excenel 2 cc/100 lbs for 3 days | Provided by Veterinarian | ||
| Providone Scrub | Aspen Veteterinary Resources | 21260221 | |
| Ethanol 70% | Sigma – Aldrich | 793213 | |
| Xylazine hydrochloride 100 mg/mL IV at 0.010-0.015 mg/kg bodyweight | Provided by Veterinarian | ||
| 2% lidocaine HCl (10-15 mL) | Provided by Veterinarian | ||
| 1 mg/kg IV injection of flunixin meglumine | Provided by Veterinarian | ||
| Isolation of Mitochondria (liver) | |||
| Equipment | |||
| Wheaton vial 30 mL with a Teflon pestle of 0.16 mm clearance | Fisher Scientific | 02-911-527 | |
| Homogenizer Motor | Cole Parmer | EW-04369-10 | |
| Homogenizer Probe | Cole Parmer | EW-04468-22 | |
| Auto Pipette (10 mL) | Cole Parmer | SK-21600-74 | |
| Beaker (500 mL) with ice | Fisher Scientific | FB100600 | |
| Refrigerated microfuge | Fisher Scientific | 75-002-441EW3 | |
| Microfuge tubes (1.5 mL) | Fisher Scientific | AM12400 | |
| Chemicals | |||
| Bicinchoninic acid (BCA) protein assay kit (microplates for plate reader) | abcam | ab102536 | |
| Sucrose | Sigma – Aldrich | S7903-1KG | |
| Tris-HCl | Sigma – Aldrich | T1503-1KG | |
| EDTA | Sigma – Aldrich | EDS-1KG | |
| BSA (fatty acid free) | Sigma – Aldrich | A7030-50G | |
| Mannitol | Sigma – Aldrich | M4125-1KG | |
| Deionized water | Sigma – Aldrich | 38796 | |
| Hepes | Sigma – Aldrich | H3375-500G | |
| Use to create mitochondria isolation media: 220 mM mannitol, 70 mM sucrose, 20 mM HEPES, 20 mM Tris-HCl, 1 mM EDTA, and 0.1% (w/v) fatty acid free BSA, pH 7.4 at 4 °C, will last 2 days in refrigerator | |||
| Mitochondrial Oxygen Comsuption | |||
| Equipment | |||
| Oxygraph Setup + Clark type oxygen electrode | Hansatech (PP Systems) | OXY1 | |
| Thermoregulated Water Pump | ADInstruments | MLE2001 | |
| Clark type Oxygen electrode | NA | NA | |
| Autopipette (1 mL) | Cole Parmer | SK-21600-70 | Included with Oxy1 |
| Small magnetic stir bar | Fisher Scientific | 14-513-95 | |
| Micropipette (10 μL) | Cole Parmer | SK-21600-60 | |
| pH meter | VWR | ||
| Chemicals | |||
| KCl | Sigma – Aldrich | P9333-1KG | |
| Hepes | Sigma – Aldrich | H3375-500G | |
| KH2PO4 | Sigma – Aldrich | P5655-1KG | |
| MgCl2 | Sigma – Aldrich | M1028-100ML | |
| EGTA | Sigma – Aldrich | E3889-100G | |
| Use to make mitochondrial oxygen consumption media: 120 mM KCL, 5 mM KH2PO4, 5 mM MgCl2, 5 mM Hepes and 1 mM EGTA, pH 7.4 at 30 °C with 0.3% defatted BSA | |||
| Rotenone (4 mM solution) | Sigma – Aldrich | R8875-5G | |
| Succinate (1 M solution) | Sigma – Aldrich | S3674-250G | |
| ADP (100 mM solution) | Sigma – Aldrich | A5285-1G | |
| Oligomycin (solution of 8 μg/mL in ethanol) | Sigma – Aldrich | 75351 | |
| FCCP | Sigma – Aldrich | C2920 | |
| Mitochondrial Membrane Potential and Proton Motive Force | |||
| Equipment | |||
| TPMP electrode | World Precision Instruments. | DRIREF-2 | |
| Chemicals-solutions do not need to be fresh but they do need to be kept in a freezer between runs | |||
| Malonate (0.1 mM solution) | Sigma – Aldrich | M1296 | |
| Oligomycin (8 μg/mL in ethanol), keep in freezer | Sigma – Aldrich | 75351 | |
| Nigericin (80 ng/mL in ethanol), keep in freezer | Sigma – Aldrich | N7143 | |
| FCCP | Sigma – Aldrich | C3920 | |
| TPMP | Sigma – Aldrich | T200 | |
| TPMP solution: 10 mM TPMP, 120 mM KCL, 5 mM Hepes and 1 mM EGTA, pH 7.4 at 30 °C with 0.3% defatted BSA | |||
References
- Brand, M. D., Divakaruni, A. S. The regulation and physiology of mitochondrial proton leak. Physiology. 26, 192-205 (2011).
- Stephenson, E. J., Hawley, J. A. Mitochondrial function in metabolic health: A genetic and environmental tug of war. Biochimica et Biophysica Acta. 1840, 1285-1294 (2014).
- Bartlett, K., Eaton, S. Mitochondrial B oxidation. European Journal of Biochemistry. 271, 462-469 (2004).
- Acetoze, G., Kurzbard, R., Klasing, K. C., Ramsey, J. J., Rossow, H. A. Oxygen Consumption, Respiratory Control Ratio (RCR) and Mitochondrial Proton Leak of broilers with and without growth enhancing levels of minerals supplementation challenged with Eimeria maxima (Ei). Journal of Animal Physiology and Animal Nutrition. 101, e210-e215 (2016).
- Wallace, D. C., Fan, W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 10, 12-31 (2010).
- Paradies, G., Petrosillo, G., Paradies, V., Ruggiero, F. M. Oxidative stress, mitochondrial bioenergetics and cardiolipin in aging. Free Radicals in Biology and Medicine. 48, 1286-1295 (2010).
- Acetoze, G., Champagne, J., Ramsey, J. J., Rossow, H. A. Liver mitochondrial oxygen consumption and efficiency of milk production in lactating Holstein cows supplemented with Copper, Manganese and Zinc. Journal of Animal Physiology Animal Nutrition. 102, e787-e797 (2017).
- Brown, D. R., DeNise, S. K., McDaniel, R. G. Mitochondrial respiratory metabolism and performance of cattle. Journal of Animal Science. 66, 1347-1354 (1988).
- Golden, M. S., Keisler, J. W., H, D. The relationship between mitochondrial function and residual feed intake in Angus steers. Journal of Animal Science. 84, 861-865 (2006).
- Lancaster, P. A., Carstens, G. E., Michal, J. J., Brennan, K. M., Johnson, K. A., Davis, M. E. Relationships between residual feed intake and hepatic mitochondrial function in growing beef cattle. Journal of Animal Science. 92, 3134-3141 (2014).
- Acetoze, G., Weber, K. L., Ramsey, J. J., Rossow, H. A. Relationship between liver mitochondrial respiration and proton leak kinetics in low and high RFI steers from two lineages of RFI Angus bulls. ISRN Vet Sci. 2015 (194014), (2015).
- Halliwell, B., Gutteridge, J. M. C. Protection against oxidants in biological systems: The superoxide theory of oxygen toxicity. Free Radicals in Biology and Medicine. , 186-187 (1989).
- National Research Council. . Nutrient Requirements of Dairy Cattle. , (2001).
- Ramsey, J. J., Harper, M. E., Weindruch, R. Restriction of energy intake, energy expenditure, and aging. Free Radical Biology and Medicine. 29, 946-968 (2000).
- Mehta, M. M., Weinberg, S. E., Chandel, N. S. Mitochondrial control of immunity: beyond ATP. Nature. 17, 608-620 (2017).
- Kirby, D. M., Thorburn, D. R., Turnbull, D. M., Taylor, R. W. Biochemical assays of respiratory chain complex activity. Methods in Cell Biology. 80, 93-119 (2007).
- Alex, A. P., Collier, J. L., Hadsell, D. L., Collier, R. J. Milk yield differences between 1x and 4x milking are associated with changes in mammary mitochondrial number and milk protein gene expression, but not mammary cell apoptosis or SOCS gene expression. Journal of Dairy Science. 98, 4439-4448 (2015).
- Lossa, S., Lionetti, L., Mollica, M. P., Crescenzo, R., Botta, M., Barletta, A., Liverini, G. Effect of high-fat feeding on metabolic efficiency and mitochondrial oxidative capacity in adult rats. British Journal of Nutrition. 90, 953-960 (2003).
- Boily, G., Seifert, E. L., Bevilacqua, L., He, X. H., Sabourin, G., Estey, C., Moffat, C., Crawford, S., Saliba, S., Jardine, K., Xuan, J., Evans, M., Harper, M. E., McBurney, M. W. SirT1 regulates energy metabolism and response to caloric restriction in mice. PloS One. 3 (3), e1759 (2008).
- Chen, Y., Hagopian, K., Bibus, D., Villaba, J. M., Lopez-Lluch, G., Navas, P., Kim, K., McDonald, R. B., Ramsey, J. J. The influence of dietary lipid composition on liver mitochondria from mice following 1 month of calorie restriction. Bioscience Reports. 33, 83-95 (2013).
- Chacko, B. K., Kramer, P. A., Ravi, S., Benavides, G. A., Mitchell, T., Dranka, B. P., Ferrick, D., Singal, A. K., Ballinger, S. W., Bailey, S. M., Hardy, R. W., Zhang, J., Zhi, D., Darley-Usmar, V. M. The bioenergetic health index: a new concept in mitochondrial translational research. Clinical Science. 127, 367-373 (2014).

