Separation of Spinach Thylakoid Protein Complexes by Native Green Gel Electrophoresis and Band Characterization using Time-Correlated Single Photon Counting
Summary
Here we present a protocol to separate solubilized thylakoid complexes by Native Green Gel electrophoresis. Green gel bands are subsequently characterized by Time Correlated Single Photon Counting (TCSPC) and basic steps for data analysis are provided.
Abstract
The light reactions of photosynthesis are carried out by a series of pigmented protein complexes in the thylakoid membranes. The stoichiometry and organization of these complexes is highly dynamic on both long and short time scales due to processes that adapt photosynthesis to changing environmental conditions (i.e., non-photochemical quenching, state transitions, and the long-term response). Historically, these processes have been described spectroscopically in terms of changes in chlorophyll fluorescence, and spectroscopy remains a vital method for monitoring photosynthetic parameters. There are a limited number of ways in which the underlying protein complex dynamics can be visualized. Here we describe a fast and simple method for the high-resolution separation and visualization of thylakoid complexes, native green gel electrophoresis. This method is coupled with time-correlated single photon counting for detailed characterization of the chlorophyll fluorescence properties of bands separated on the green gel.
Introduction
Photosynthetic organisms must constantly adjust their physiology to changing environmental conditions to maximize their productivity and successfully compete with neighbors1. This is especially true of the machinery responsible for the light reactions of photosynthesis, as ambient light conditions can fluctuate by three orders of magnitude between shadows and full sunlight. Additionally, environmental factors such as drought, cold, or heat stress can reduce the availability of carbon dioxide for carbon fixation, which is the natural electron sink for the products of the light reactions. Plants must, therefore, harvest and utilize solar radiation as efficiently as possible while retaining the ability to dissipate excess light energy as necessary. While photooxidative damage still occurs routinely under all light conditions2,3, failure to manage absorbed excitation energy successfully can lead to the catastrophic cell damage and death. Several adaptive mechanisms exist that allow the photosynthetic apparatus to be tuned both to changes in prevailing environmental conditions and to transient fluctuations (i.e., over both long and short timescales)4. These include the long-term response (LTR) and non-photochemical quenching (NPQ). NPQ is itself considered to encompass at least three other component phenomena, including state transitions (qT), rapidly inducible energy quenching (qE), and photoinhibition (qI)5.
These processes were originally observed and defined largely in terms of spectroscopic phenomena [e.g., NPQ refers to a drop in observed chlorophyll fluorescence (quenching of chlorophyll fluorescence) that is not due to an increase in the rate of photochemistry]6. The term "state transitions" similarly refers to the observed change in the relative amount of fluorescence from PSI and PSII7. While the spectroscopic techniques that have made enumeration of these phenomena possible [in particular, pulse amplitude modulated (PAM) fluorescence spectroscopy] and continue to be a vital means for observing and dissecting photosynthetic processes in vivo, a great deal of biochemistry is required to elucidate the mechanisms underlying these spectroscopic observations. State transitions, for instance, involves a phosphorylation/dephosphorylation cycle of the LHCII proteins by the STN7 kinase and TAP38/PPH1 phosphatase, respectively8,9,10. This cycle adjusts the physical distribution of the LHCII antenna between the two photosystems by moving a portion of LHCII trimers from PSII to PSI, thereby changing the absorption cross section of the photosystems11,12. The qE component of NPQ rapidly converts excess excitation energy into heat through the actions of the violaxanthin/zeaxanthin epoxidation/de-epoxidation cycle and the PsbS protein. The exact role of PsbS in this process is still not fully understood13. The qI component of NPQ, photoinhibition, is generally ascribed to damage to the D1 protein of PSII. Restoration of full photosynthetic competence requires an elaborate repair process to fix damaged PSII photocenters. The PSII repair cycle involves the migration of PSII complexes out of the granal stacks, dismantling of the complexes, replacement of damaged D1 proteins, reassembly of the PSII complexes, and movement of PSII complexes back into the granal stacks14. The exact nature of photoinhibition and PSII photodamage remains a subject of intense scrutiny15.
The difficulty in studying phenomena like state transitions or PSII repair arises in part from the fact that there is not one simple way to visualize the mechanics of complex biochemical systems. The classic biochemical approach to understanding a process is to first separate its components so that they can be characterized in isolation. Native gel electrophoresis arose from successful efforts in the 1980s to separate and characterize the photosystem complexes from the thylakoid membranes with more preparative methods (namely sucrose gradient centrifugation and chromatography)16. The detergent systems developed to gently solubilize the native complexes from the thylakoid membranes were soon adapted to electrophoretic separation methods, most notably by Allen and Staehelin17 and and Peter and Thornber18, giving rise to native green gel electrophoresis. While representing only one out of a variety of techniques in the experimental arsenal, native PAGE has a number of attractive characteristics that have made it a widely employed method in photosynthesis research. Native PAGE is relatively fast and simple, requiring little specialized equipment, while providing high resolution separation of a large number of thylakoid complexes simultaneously. This makes native PAGE a convenient tool for studying thylakoid dynamics and, when combined with standard PAGE in the second dimension as well as a variety of detergent and buffer systems, a versatile system for finding and characterizing new thylakoid complexes.
That being said, native green gels have had a reputation for being an unreliable technique, especially in inexperienced hands, as it is easy to produce poor results consisting of fuzzy, smeary gels with few bands. This problem was solved, in part, with the introduction of blue-native PAGE19. The use of coommassie dye in the BN buffer system makes protein separation more robust. Therefore, BN-PAGE is often an easier and more reliable technique for a relative novice to set up and can provide high resolution separations of thylakoid complexes. For these reasons, BN-PAGE has become the method of choice for most work of this field. While BN-PAGE is generally slower to run than green gel electrophoresis, its main drawback is that the coommassie dye staining interferes with the identification of faint chlorophyll-containing bands, while also making downstream spectroscopic characterization problematic.
The biochemical information provided by native gels and 2D SDS-PAGE can be greatly strengthened when combined with data from spectroscopic techniques. Regardless of the system employed, a central problem with using native gels to identify complexes is that the identification can always be challenged (i.e., the proteins found in a band could always represent comigrating complexes or components, rather than a single physiologically authentic complex). Spectroscopic characterization provides biophysical information about the pigments in green gel bands and can be used to determine what types of complexes they are likely to contain. Chlorophyll fluorescence is especially useful in this regard due to the often dramatically different spectra and fluorescence lifetimes that are characteristic of different photosynthetic pigment-protein complexes. While simple steady-state 77K fluorescence spectra have historically been useful in confirming the identities of native gel complexes, modern time-correlated single photon counting (TCSPC) can provide much more information. TCSPC allows not only the characterization of complexes based on fluorescence lifetimes, but also makes possible the detailed description of energy transfer between spectral components within a complex. This kind of characterization is becoming increasingly necessary as the use of various native gel systems spreads and new putative complexes are discovered, allowing the identification of protein complexes to be better authenticated and providing new biophysical information about how these complexes work.
In this paper we provide a method that allows those having little or no experience with native gel electrophoresis to achieve high quality resolution of native thylakoid complexes for the purpose of investigating the mechanics of the light reactions of photosynthesis. This basic technique can then be augmented at the experimenter's discretion to improve results or extend applicability to other species. We then describe the process for subjecting native green gel bands to TCSPC, as well as some steps for basic analysis and presentation of the data provided by the technique. The coupling of native gel electrophoresis with TCSPC analysis extends the utility of these gel systems by providing authentication and biophysical characterization of protein complexes within the bands. The green gel system described here is based on that developed by Allen and Staehelin17 with some modifications and is the same as that used in Schwarz et al.20. This system is one of many but has specific features that are useful for this methodology. It is rapid enough so that thylakoid isolation, gel electrophoresis, and TCSPC analysis convenient can be performed in one day, obviating potential problems of sample storage and degradation. We also find that this method is robust in the hands of inexperienced users, while still providing results that range from good to superior, depending on the degree of optimization.
It is important to bear in mind that the complexes visualized on a native gel depend on both the detergent and buffer systems used, as well as on the biology of the organism under investigation. Different detergent and buffer systems preferentially separate different kinds of complexes, and a given photosynthetic organism will have different complexes from other organisms, not all of which will be present under any given circumstance. The system described here is particularly suited to the study of PSI megacomplexes, as described in Schwarz et al.20, but it falls on the more destabilizing end of the spectrum for those studying PSII megacomplexes. For a comprehensive study of the various detergent and buffer systems used in native gel electrophoresis of thylakoid proteins, it is recommended to review Järvi et al.21 and Rantala et al.22.
Protocol
1. Stock Solutions Preparation for Pouring Native Green Gels
- Prepare a 4x concentrated buffer solution for resolving gels consisting of 40% glycerol, 200 mM glycine, and 100 mM Tris buffered to pH 8.3.
- Prepare a 4x concentrated buffer solution for stacking gels consisting of 40% glycerol, 200 mM glycine, and 100 mM Tris buffered to pH 6.3.
- Store these buffers at 4 °C to prevent the growth of mold.
Note: The buffers are stable for months at 4 °C, so preparation of 100-200 mL of each buffer for continued use is recommended. - Prepare 1 L of 10x running buffer containing 250 mM Tris HCl pH 8.3, 1.92 M glycine, and 1% SDS. Store the 10x running buffer on the benchtop.
2. Stock Solution Preparation for Isolation and Solubilization of Thylakoids
- Prepare 100 mL of TMK homogenization buffer containing 50 mM Tris buffer (pH 7), 10 mM MgCl2, and 10 mM KCl, and store at 4 °C.
Note: This is the main buffer for homogenizing and manipulating thylakoid samples. Alternatively, concentrated stock solutions can be prepared and diluted as necessary (Tris buffer, MgCl2, and KCl can all be easily stored on the benchtop as 1 M concentrates). - Prepare stock solutions of the thylakoid solubilization detergents, B-decyl maltoside (DM) and n-octyl B-d-glucoside (OG), by dissolving each detergent in TMK buffer at 20% w/v. Freeze at -20 °C in 1 mL aliquots.
- Prepare thylakoid solubilization buffer (SB), which is also the sample loading buffer.
- First, make TMK-glycerol buffer by combining 7 mLs of TMK buffer and 3 mLs of glycerol.
- To 800 μL of TMK-glycerol buffer, add 100 μL of DM stock solution and 100 μL of OG stock solution. Store this SB working solution, containing 2% DM and 2% OG, frozen at -20° C in 1 mL aliquots.
Note: Each aliquot can be thawed and refrozen as needed.
3. Pouring Green Mini Gels for Later Use
- Prepare separate stacking and resolving gel solutions in 15 mL disposable test tubes.
Note: The volumes provided are sufficient for a single mini gel using 1.5 mm plate spacers.- To make the stacking gel solution, combine 1.25 mL of 4x stacking gel buffer, 0.5 mL of 40% acrylamide stock solution (39:1 C), and 3.25 mL of water to give 5 mL of 4% acrylamide in 1x stacking buffer. To make the resolving gel solution combine 1.875 mL of 4x resolving buffer, 0.94 mL of 40% acrylamide stock solution (39:1 C), and 4.7 mL of water to give 7.5 mL of 5% acrylamide in 1x resolving buffer.
- Pour the resolving gel.
- Add 50 μL of 10% ammonium persulfate (APS) to the resolving gel solution, then add 10 μL of TEMED, cap the tube, and gently invert several times to mix. Immediately pour the gel solution between the gel plates, leaving approximately 1 cm between the top of the resolving gel and bottom of the comb teeth for the stacking gel.
- Gently pipette 100% ethanol onto the top of the resolving gel to level the gel.
Note: If the gel does not set within 15 min, fresh APS solution should be made and/or new TEMED should be used. - After the gel has polymerized (the interface between the gel and the ethanol will be readily visible and will not move when the gel is tipped), pour off the ethanol and blot with an absorbent paper.
- Pour the stacking gel.
- Add 25 μL of 10% APS to the stacking gel solution, add 5 μL of TEMED, then cap and invert to mix in the same manner as the resolving gel. Pour the gel solution on top of the resolving gel until the space between the plates is completely filled and insert a 10-well comb.
Note: When ready to be used, remove the comb from the gel and rinse the wells with water, making sure that the wells are straight and unobstructed by gel. The gel can be stored with the comb in place at 4 °C for at least several days.
- Add 25 μL of 10% APS to the stacking gel solution, add 5 μL of TEMED, then cap and invert to mix in the same manner as the resolving gel. Pour the gel solution on top of the resolving gel until the space between the plates is completely filled and insert a 10-well comb.
4. Isolation of Crude Thylakoid Membranes from Spinach Leaves
Note: All steps should be carried out on ice using pre-chilled equipment and buffers. Dim lighting is also recommended. Depending on the experimenter's discretion and the biological processes under study, protease and/or phosphatase inhibitors should be added fresh to TMK buffers before homogenization.
- Completely homogenize spinach leaves in TMK buffer with a glass Dounce homogenizer.
Note: Approximately 1 to 2 mL of buffer is normally sufficient for a small baby spinach leaf. A single baby spinach leaf can generally provide enough material to load several wells on a 1.5 mm mini gel. - Filter the crude leaf homogenate to remove insoluble debris.
- To make a simple filtering device, cut a delicate task wipe in half and fold it into quarters. Pack the delicate task wipe into the bottom of a 5 mL disposable syringe and pre-wet the wipe with TMK buffer.
- Use the syringe plunger to press excess buffer out of the delicate task wipe and be sure that the wipe filter is pressed firmly to the bottom of the syringe after the plunger is removed.
- Pipette the leaf homogenate onto the center of the wipe filter and use the plunger to pass the homogenate through the filter. Collect the filtered homogenate in a 1.5 mL centrifuge tube.
- Centrifuge the homogenate at 5,000 x g for 10 min at 4° C to pellet insoluble material, including thylakoid membranes. Discard the supernatant and resuspend the pellet in 1 mL of TMK buffer.
- Normalize the amount of chlorophyll in each sample by adjusting the volume of each resuspended thylakoid sample so that each sample contains the same total amount of chlorophyll, as described below.
Note: This will allow each sample to be solubilized in the same volume of detergent and minimizes variability in solubilization due to differences in pellet volume.- To extract chlorophyll from each sample of resuspended thylakoid membranes, take a 50 μL aliquot in a 1.5 mL microcentrifuge tube and add 950 μL of methanol to it. Cap the tube and mix by inverting several times.
- Centrifuge the methanol/chlorophyll extract at 10,000 x g for 10 min to pellet precipitated proteins.
- Determine the chlorophyll concentration of the pigment containing supernatant according to Porra et al.23. Take absorbance readings at 652 and 665 nm using a spectrophotometer and a 1 cm cuvette. Determine total chlorophyll concentration using the equation below:
Chls a + b (μg/mL) = 22.12 (Abs 652 nm) + 2.71 (Abs 665 nm) - Using chlorophyll concentration measurements as a guide, remove and discard some volume from each sample, as necessary, so that each tube contains the same total amount of chlorophyll.
- Re-pellet thylakoid membranes by centrifugation at 5,000 x g for 10 min. Remove and discard the supernatant. Be careful to remove all supernatant without aspirating any of the pellet.
5. Solubilization of Thylakoid Membranes for Loading onto Native Gels
- Thaw an aliquot of TMK 30% glycerol detergent solution (SB) and invert several times to mix. Keep it on ice.
- Dissolve the thylakoid pellet by adding the appropriate volume of SB to give a chlorophyll concentration of 1 mg/mL.
Note: This concentration is a starting point for finding the optimal solubilization conditions, which must be determined empirically. The chlorophyll concentration must be kept the same between samples to allow valid comparisons to be made. - Pipette up and down repeatedly while being careful to avoid frothing of the sample. Keep on ice to allow thylakoid samples to solubilize.
Note: Solubilize for at least 10 minutes. Solubilization time should be long enough that the difference in solubilization time between samples is minimized.
Example: If 3 minutes are required to solubilize all samples, then approximately 30 minutes should be allowed for solubilization. - Centrifuge solubilized thylakoids at 10,000 x g at 4 °C to pellet insoluble material.
Note: Solubilized thylakoid samples are stable on ice for hours, but storage at -70 °C should be avoided, as freeze-thaw cycles can result in a loss of megacomplex bands.
6. Separation of Solubilized Thylakoid Proteins by Native Gel Electrophoresis
- Load the solubilized thylakoid supernatant prepared in step 5.4 directly onto the native gel prepared earlier. For a 1.5 mm gel, load 15 μL of solubilized thylakoid per well.
- Run the native green gel in essentially the same manner as SDS-PAGE gels using 1x running buffer. Run the gel at 100 V and place the entire gel tank on wet ice for the duration of the run to mitigate resistive heating of the gel.
Note: The gel should require approximately 2 hours for the free pigment at the migration front to reach the bottom of the gel (Figure 1).
7. Excision of Thylakoid Complex Bands from Native Green Gels
Note: Excising the specific band of interest from the gel is necessary to allow the band to be placed in the beam path and to prevent stray fluorescence from nearby complexes from being collected.
- Remove the gel from the electrophoresis cell and rinse running buffer off of the gel plates with distilled water. Remove the top plate from the gel and rinse the gel with distilled water.
- Keep the gel on the bottom glass plate and place the gel and plate on ice. When not in use, keep the gel in the dark and cover with a plastic wrap to prevent it from drying out.
- With the gel remaining on the glass plate, excise each band of interest when ready for TCSPC analysis. Excise bands cleanly with a sharp scalpel or razor blade and take care that the excised band contains no contaminating band material.
8. Collection of Room Temperature Steady-State Fluorescence Spectra
- For each complex that will be analyzed by TCSPC, a room temperature fluorescence spectrum is taken between 600 and 800 nm using a fluorescence spectrometer.
Note: The excitation wavelength used to collect this spectrum must match the wavelength used for TCSPC.
9. TCSPC of Green Gel Bands
Note: Refer to Figure 2 for a depiction of the TCSPC setup.
- Sandwich the gel slice between two glass microscopy slides. Use masking tape, placed on each end of one of the microscopy slides and folded over several times, to create spacers so that the slides can be held together firmly without compressing the gel slice.
Note: This will create a path for the laser beam to pass between the microscope slides and through the gel slice. - Add a small amount of water to the gel slice at the edge of the glass slides to create a smooth interface that will reduce signal scattering.
- Clamp the gel/slide sandwich in the beam path so that the beam strikes the gel slice through the open edge of the plates, allowing fluorescence emission to be through the side of the glass slides in which the gel is sandwiched, perpendicular to the beam path.
Note: The excitation wavelength used will depend on the experiment. A wavelength of 435 nm will excite both chlorophyll a and chlorophyll b, while 465 nm will preferentially excite chlorophyll b. In this case, 435 nm was used as the excitation wavelength. - Collect 10,000 total data points at regular intervals across the fluorescence emission spectrum for each complex. For example, collect data every 10 nm, starting at 680 nm and ending at 750 nm.
Note: Prepare multiple gel bands for each complex to be studied so that fresh sample is available in the event that photobleaching prevents adequate signal collection.
10. TCSPC (Data Analysis)
- For a given complex, first normalize the peak height of each decay curve for all wavelengths collected.
Note: This step is not necessary for the construction of DAS but allows for decay curves to be overlaid and compared visually with one another as a first inspection of the data. - Tail-match each decay curve to the steady state fluorescence spectrum of the complex as described below.
- Choose a timepoint after which the decay signal has flattened out, usually after a few nanoseconds.
- For each wavelength, normalize the decay curve so that the signal intensity at the selected timepoint is equal to the value of the steady state fluorescence spectrum at that wavelength (i.e., the values of all wavelengths together at the selected timepoint will re-create the steady state fluorescence spectrum).
- Build decay-associate spectra (DAS) from the tail-matched decay curves, as described below.
- Using data points from the tail-matched decay curves construct a series of plots graphing fluorescence intensity vs. wavelength at regular time intervals (e.g., every 10 ps). For example, the DAS at 50 ps is constructed by plotting the value of each decay curve at 50 ps vs. wavelength.
- Overlay all of the decay-associated spectra to create a waterfall style plot that shows the decay of the fluorescence spectrum over time.
Representative Results
Representative results for green gel electrophoresis are presented in Figure 1. Lane 1 provides an example of ideal results for green gel electrophoresis of spinach thylakoids, in which a maximum number of clear, sharp green bands are visible. These results are somewhat atypical, in part because not all of the bands seen in lane 1 are normally present in a given sample. Additional sample cleanup, in the form of chloroplast isolation before thylakoid solubilization, and gradient gel electrophoresis (4-7% acrylamide) are also normally necessary to achieve optimal results. Lanes 2 and 3 present more typical results achieved using the protocol detailed here in conjunction with a 5% non-gradient gel. Lanes 4, 5, and 6 provide an example of poor results due to increasing degrees of under-solubilization of the thylakoid sample. Lane 7 provides an example of typical results achieved with Arabidopsis thylakoids instead of spinach. Note that for Arabidopsis the megacomplex bands at the top of the gel tend to be poorly resolved compared to those in spinach.
A graphic depiction of the TCSPC setup used for collecting data from native green gel bands is shown in Figure 2. Figure 3 shows a typical workflow for beginning analysis after TCSPC data has been collected as described in step 10. When TCSPC data is collected, each curve at a given wavelength represents an arbitrary number of data points, or "counts". When these curves are overlaid with one another, as shown in Figure 3A, the curves cannot be compared with one another directly because they are not represented at the same scale and may not all be registered to the same starting time point. The first step in data analysis is therefore to compare each curve to its corresponding instrument response function (IRF). The peak of the IRF for a given curve is set to time t = 0, and the leading edge of the corresponding fluorescence decay curve is set to overlap the leading edge of the IRF, as shown in Figure 3B. This will set all curves to the same time register for later analysis.
While it is not necessary for the construction of DAS, normalizing all curves to the same peak height at this point allows a useful comparison between curves to be made as a first analysis of the data. In Figure 3C, the same decay curves for LHCII presented in Figure 3A are shown after time registration, as in Figure 3B, and peak height normalization. As seen in Figure 3D, peak-normalized decay curves from different complexes can then be overlaid with one another at a given wavelength, allowing the differences in behavior between the complexes to be visualized. For example, LHCII has a characteristically long-lived fluorescence that decays slowly, whereas PSI fluorescence is strongly quenched, decaying very rapidly. The Band 5 fluorescence decay curve provides an interesting example of very suggestive data, in part because it is clearly biphasic. The initial fluorescence decay curve for the Band 5 complex follows the same rate as PSI for approximately 500 ps yet decays even more rapidly thereafter. Intriguing results of this kind can be analyzed in further detail by constructing decay-associated spectra (DAS).
In Figure 4, representative DAS waterfall plots are shown for LHCII and the Band 5 complex. Construction of DAS first requires that the decay curves for a given complex be tail-matched to the room temperature fluorescence spectrum for the complex, as described in step 11.2. The results of tail-matching decay curves for LHCII are shown in Figure 4A. DAS are then constructed from these curves as described in step 11.3. DAS for LHCII were constructed from the decay curves shown in Figure 4A and the results are presented in Figure 4B. DAS between 0 and 100 ps were omitted for clarity, and only presented every 100 ps thereafter due to the characteristically slow decay for isolated LHCII. The DAS for LHCII is notable for the lack of dynamic features, and the shape of the LHCII fluorescence spectrum remains the same as the signal decays over time. The decay of the fluorescence spectrum is also delayed, requiring 100 ps to reach maximum fluorescence. This suggests, as would be expected for isolated light harvesting protein complexes, that energy is not transferred between energetically distinct pigments within the complex as the fluorescence decays. The exception to this is the shift in the spectrum occurring during the first 100 ps, presumably due to the initial redistribution of excitation energy throughout the complex.
DAS for the Band 5 complex are shown in Figure 4C, constructed in a similar fashion to that shown for LHCII. Band 5 provides an instructive contrast to LHCII in several ways. Compared to LHCII, fluorescence from Band 5 decays much more rapidly, reaching maximum intensity in only 30 ps and decaying to less than 20% of initial intensity after 500 ps. The fluorescence spectrum of the Band 5 complex also exhibits a number of interesting dynamics as the emission decays. Comparing the spectra at 0 and 60 ps clearly shows an increase in fluorescence at 720 nm at the expense of fluorescence at 680 and 710 nm. Thereafter, the peak at 680 nm shifts towards 690 nm and broadens, while the peak at 720 nm shifts back toward 710 nm (e.g., compare 60 ps to 500 ps). Data of this type helps rule out the presence of unconnected LHCII antenna proteins while providing evidence for energy transfer from LHCII to the PSI antenna and eventually the PSI core.
These dynamics also suggest that there are likely to be unresolved peaks around 680 nm, 690 nm, 710 nm, and 720 nm. This data therefore provides an example where spectral features could be better resolved by collecting TCSPC data at closer intervals (e.g. every 5 nm rather than every 10 nm). Even in the absence of higher spectral resolution, however, the DAS for the Band 5 complex are an example of evidence for energy transfer between multiple fluorescing species within a complex. There also appears to be a rapidly decaying peak above 740 nm, suggesting that data should also be collected over a broader spectrum to include further spectral features.
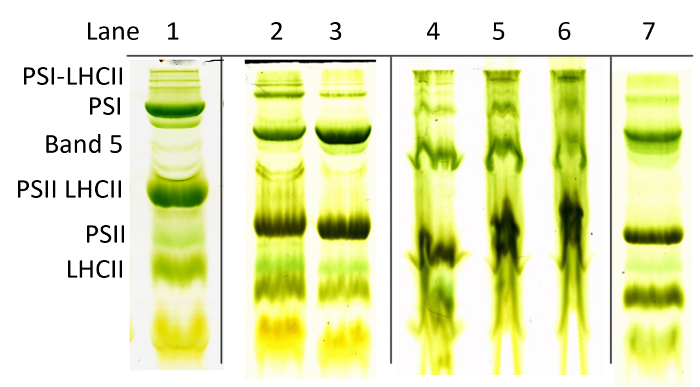
Figure 1: Representative green gel results. Green gel bands subjected to TCSPC analysis are labeled PSI-LHCII (photosystem I LHCII megacomplexes), PSI (photosystem I LHCI), Band 5 (PSI-LHCII complex), PSII-LHCII (photosystem II and associated light-harvesting complex II), PSII (photosystem II core complex), and LHCII (light-harvesting complex II). Lane 1 shows an ideal banding pattern resulting from chloroplast isolation before solubilization and green gel electrophoresis on a 4-7% acrylamide gradient gel. Lanes 2 and 3 show average results from simple thylakoid isolation and electrophoresis on a non-gradient 5% acrylamide gel. Lanes 4-6 show increasing severity of under-solubilization. Lane 7 shows representative results for Arabidopsis using the simple protocol. Please click here to view a larger version of this figure.
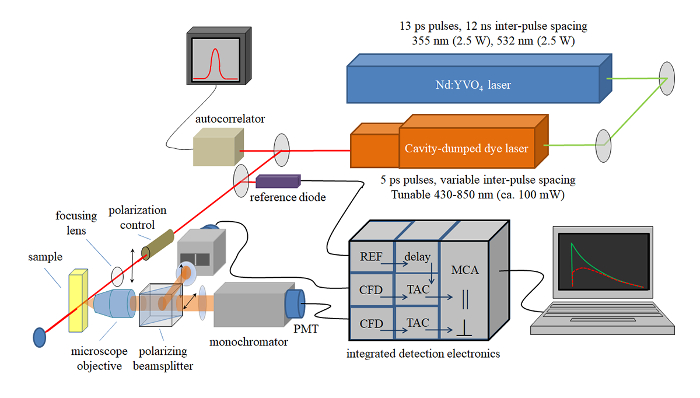
Figure 2: Schematic of time-correlated single photon counting instrument. Light source is a passively mode-locked NdYVO4 laser that pumps a cavity dumped dye laser. 5-ps pulses at variable repetition rates and wavelengths are characterized using an autocorrelator. The pulses are divided, with one portion going to a reference photodiode and the rest exciting the sample. Sample emission is collected using a microscope objective, and polarized components are detected using two detection channels. Sample emission is time resolved using the detection electronics indicated, where CFD = constant fraction discriminator, TAC = time-to-amplitude converter, and MCA = multi-channel analyzer. Time resolution is determined from the instrument response function (ca. 40 ps). Please click here to view a larger version of this figure.
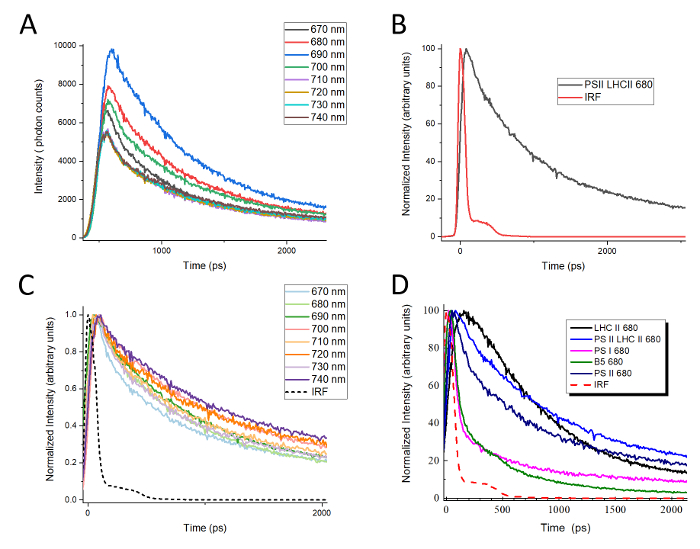
Figure 3: Representative raw TCSPC data and normalized fluorescence decay curves. (A) Overlay of raw TCSPC data for LHCII. TCSPC data was collected every 10 nm between 670 nm and 740 nm (B) A representative curve showing time registration of the fluorescence data to the instrument response function (IRF). (C) The LHCII data from (A) normalized to a maximum peak height of 1 after all curves were registered to their respective IRFs. (D) Overlay of fluorescence decay curves at 680 nm for PSI, PSII, PSII-LHCII, LHCII, and Band 5. All decay curves were normalized to a maximum peak height of 1. Please click here to view a larger version of this figure.
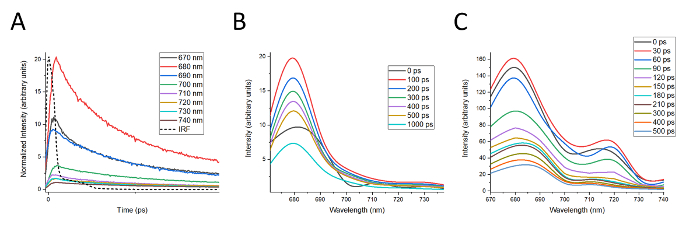
Figure 4: Construction of waterfall-style DAS. (A) Tail matching of the decay curves for LHCII in preparation for construction of DAS plots. (B) DAS plots for LHCII for time t = 0, for every 100 ps from 100 to 500 ps, and at 1000 ps. (C) DAS plots for Band 5 every 30 ps from t = 0 to 210 ps and every 100 ps thereafter to 500 ps. For both (B) and (C), time = 0 is defined by the IRF, which is 40 ps. Please click here to view a larger version of this figure.
Discussion
A successful thylakoid solubilization and native gel run will result in the resolution of multiple distinct visible green bands on the gel without significant distortion or smearing of the bands. Overloading the gel, a high detergent concentration, an incorrect sample pH, undissolved material, running the gel too rapidly or at too high a temperature, and an improperly poured gel are all factors that may contribute to poorly resolved thylakoid complexes. While optimizing the conditions of the gel itself (e.g., acrylamide gradient concentrations), it can help to maximize the resolution of bands of interest. It is our experience that solubilization conditions and the biology of the sample material itself are the most important factors contributing to the quality and quantity of thylakoid complexes resolved on native green gels. It is important to remember that not all complexes will be present under all biological conditions.
Native green gels are poured in essentially the same manner as normal SDS-PAGE mini gels. Gels can be poured in the morning and are ready to use later the same day, or they can be stored at 4 °C for several days with the comb in the gel and no significant changes in performance. Standard, readily available 39:1 C acrylamide solutions can be used to pour both the stacking and resolving gels. So-called "large pore" gels can be made using higher acrylamide:bis-acrylamide ratios (e.g., 100:1 C) in order to increase separation of megacomplexes at the top of the gels, but we find that this also tends to result in less sharply resolved bands. In our experience, the highest band resolution (with the greatest number of bands separated) is achieved at the lower C ratio and with a linear acrylamide concentration gradient of 5-7%. However, if a gradient former is not available, very satisfactory band separation and resolution can be achieved with a resolving gel concentration of around 5% acrylamide. It is important that electrophoresis be carried out at relatively low voltage to reduce heating of the gel, which could denature the complexes, and to allow native complexes to separate from one another with high resolution.
The method for thylakoid isolation given here is rapid but relatively "dirty" (i.e., it does not attempt to separate chloroplasts from other organelles or thylakoid membranes from other cell membranes). This is sufficient for the purposes of visualizing thylakoid complexes and subsequent fluorescence-based measurements, since only chlorophyll-containing proteins are visualized. It should be noted that for other downstream applications (e.g., second dimension SDS-PAGE and protein detection), non-thylakoid proteins will be present. It should also be noted that the best resolution is achieved when chloroplasts are isolated before solubilization, presumably due to the removal of insoluble materials before solubilization. When following the method described here, other homogenization methods such as a blender may be used, but a glass Dounce homogenizer is rapid and effective and allows all homogenized leaf material to be retained quantitatively. The ratio of buffer-to-leaf material does not appear to be important, to our knowledge. Approximately 2 mL of buffer for one half of a baby spinach leaf is a convenient starting point but may be adjusted based on experimental needs.
The appropriate ratio of solubilization buffer to chlorophyll is crucial for optimization of green gel performance and should be determined by the experimenter for a given plant species or experimental setup. Proper solubilization will result in a clear, deep emerald-green supernatant above a white starch pellet. A layer of green material on top of the white pellet indicates that the thylakoids have been under-solubilized. Under-solubilization must be avoided, as it will result in poor migration on the gel, including smearing of the bands, and will not provide an accurate banding pattern. The thylakoid pellets produced by this method should be easy to resuspend and solubilize. In case of compact pellets which cannot be resuspended quickly and homogeneously an alternative method is recommended. Samples can first be resuspended in half the volume of TMK-glycerol buffer, to which is then added an additional half-volume of TMK-glycerol buffer containing a 2X concentration of detergents.
Over-solubilization should also be avoided, as it may result in the loss of density of some megacomplex bands. Additionally, it is not possible to make up for over-solubilization by loading a larger volume of sample onto the gel – we have found that loading more than 15 μL of the sample per well on a 1.5 mm gel reduces the quality of the separation; although, doing so may be desirable in order to visualize complexes with low abundance. Conversely, loading less than 15 μL can improve band resolution and reduce smearing but will reduce the visibility of some low-density bands. In general, both increased protein concentration and increased detergent concentration contribute to band distortion and gel smearing.
For time-correlated single photon counting (TCSPC) analysis of the green gel complexes, the excitation and emission wavelengths must be chosen by the experimenter at their discretion. For our purposes, the chosen excitation wavelength was 435 nm, which will excite both chlorophyll a and chlorophyll b. Different wavelengths can, however, be used to excite specific chlorophylls or other pigments more selectively (e.g., an excitation wavelength around 465 nm will preferentially excite chlorophyll b). Similarly, a fluorescence emission spectrum covering a region from 680 to 740 nm was selected based on the reported emission spectra for LHCII, PSI, and PSII. Therefore, this region covers all the relevant spectral features of interest for our purposes. The wavelength intervals at which data are collected are also up to the discretion of the experimenter. Shorter intervals, such as every 5 nm instead of every 10 nm, will make it possible to construct a more detailed decay-associated spectrum, but this requires more time and may not be necessary. Conversely, longer intervals may fail to resolve relevant spectral details.
Simply overlaying normalized fluorescence decay curves from different complexes can provide revealing information about those complexes. Fluorescence from LHCII, for example, is characteristically long-lived, while fluorescence from PSI decays much more rapidly. Care should be taken at this point to examine data against the instrument response function (IRF) to ensure that the observed decay kinetics are temporally resolved from the response time of the instrument.
Constructing decay-associated spectra (DAS) from the TCSPC data creates a much more detailed picture of energy transfer between chromophores within a complex than what is possible from simply looking at isolated decay curves. When constructing DAS, tail matching of the TCSPC decay curves to the steady state fluorescence spectrum is necessary because the intensity of signal collected by TCSPC is arbitrary. At long time points such as 5,000 ps, however, the fluorescence intensity at a given wavelength (the "tail" of the decay) is the same as the steady state fluorescence intensity for that wavelength. The steady-state fluorescence spectrum can therefore be used to normalize all of the TCSPC decays so that their tails are equivalent to the steady state spectrum. This gives the true relative intensity of fluorescence signal for each decay curve. The entire series of DAS, when overlaid together in waterfall plot style, provides a graphic depiction of the decay of the fluorescence spectrum over time. If the plots are carried out to long enough time points (to the tails of the decay curves) the waterfall plot will decay all the way to the steady-state fluorescence spectrum. The earliest time point, on the other hand, is determined by the response function of the TCSPC instrument.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Funding and support were provided by the Department of Chemistry at Michigan State University.
Materials
| Glycine | Sigma | G8898 | |
| Tris base | Sigma | #648310 | |
| SDS | Sigma | L3771 | |
| Decyl Maltoside | Sigma | D7658 | n-decyl beta d maltopyranoside, not dodecyl maltoside or alpha decyl maltoside |
| Octyl Glucoside | Sigma | O8001 | |
| Acrylamide | BioRad | 161-0148 | 37.5/1 C 40% solution |
| TEMED | BioRad | 161-0800 | |
| Ammonium Persulfate | BioRad | 161-0700 | |
| Magnesium Chloride | Sigma | M2670 | |
| Potassium Chloride | Sigma | P9333 |
References
- Yamori, W. Photosynthetic response to fluctuating environments and photoprotective strategies under abiotic stress. Journal of Plant Research. 129 (3), 379-395 (2016).
- Kim, J. H., Nemson, J. A., Melis, A. Photosystem II reaction center damage and repair in Dunaliella salina (Green Alga) (analysis under physiological and irradiance-stress conditions). Plant Physiology. 103 (1), 181-189 (1993).
- Sundby, C., McCaffery, S., Anderson, J. M. Turnover of the photosystem II D1 protein in higher plants under photoinhibitory and nonphotoinhibitory irradiance. Journal of Biological Chemistry. 268 (34), 25476-25482 (1993).
- Dietze, l. L., Bräutigam, K., Pfannschmidt, T. Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry-functional relationships between short-term and longterm light quality acclimation in plants. FEBS Journal. 275 (6), 1080-1088 (2008).
- Horton, P., Ruban, A. V., Walters, R. G. Regulation of light harvesting in green plants. Annual Review of Plant Physiology and Plant Molecular Biology. 47, 655-684 (1996).
- Baker, N. R. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annual Review of Plant Biology. 59, 89-113 (2008).
- Williams, W. P. Spatial organization and interaction of the two photosystems in photosynthesis. Nature. 225 (5239), 1214-1217 (1970).
- Bellafiore, S., Barneche, F., Peltier, G., Rochaix, J. D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 433 (7028), 892-895 (2005).
- Shapiguzov, A., et al. The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proceedings of the National Academy of Sciences USA. 107 (10), 4782-4787 (2010).
- Pribil, M., Pesaresi, P., Hertle, A., Barbato, R., Leister, D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. Public Library of Science Biology. 8 (1), (2010).
- Larsson, U. K., Jergil, B., Andersson, B. Changes in the lateral distribution of the light-harvesting chlorophyll-a/b-protein complex induced by its phosphorylation. European Journal of Biochemistry. 136 (1), 25-29 (1983).
- Minagawa, J. State transitions–the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochimica et Biophysica Acta. 1807 (8), 897-905 (2011).
- Ruban, A. V. Nonphotochemical Chlorophyll Fluorescence Quenching: Mechanism and Effectiveness in Protecting Plants from Photodamage. Plant Physiology. 170 (4), 1903-1916 (2016).
- Rokka, A., Suorsa, M., Saleem, A., Battchikova, N., Aro, E. M. Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochemical Journal. 388 (Pt 1), 159-168 (2005).
- Li, L., Aro, E. M., Millar, A. H. Mechanisms of Photodamage and Protein Turnover in Photoinhibition. Trends in Plant Science. , (2018).
- Dunahay, T. G., Staehelin, L. A. Isolation of photosystem I complexes from octyl glucoside/sodium dodecyl sulfate solubilized spinach thylakoids: characterization and reconstitution into liposomes. Plant Physiology. 78 (3), 606-613 (1985).
- Allen, K. D., Staehelin, L. A. Resolution of 16 to 20 chlorophyllprotein complexes using a low ionic strength native green gel system. Analytical Biochemistry. 194 (1), 214-222 (1991).
- Peter, G. F., Thornber, J. P. Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. Journal of Biological Chemistry. 266 (25), 16745-16754 (1991).
- Schägger, H., von Jagow, G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Analytical Biochemistry. 199, 223-231 (1991).
- Schwarz, E. M., Tietz, S., Froehlich, J. E. Photosystem I-LHCII megacomplexes respond to high light and aging in plants. Photosynthesis Research. 136 (1), 107-124 (2018).
- Suorsa, M., Paakkarinen, V., Aro, E. M. Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochemical Journal. 439 (2), 207-214 (2011).
- Rantala, M., Tikkanen, M., Aro, E. M. Proteomic characterization of hierarchical megacomplex formation in Arabidopsis thylakoid membrane. Plant Journal. 92 (5), 951-962 (2017).
- Porra, R. J., Thompson, W. E., Kriedemann, P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) – Bioenergetics. 975 (3), 384-394 (1989).

