Analysis of Fucosylated Human Milk Trisaccharides in Biotechnological Context Using Genetically Encoded Biosensors
Summary
We describe here the high-throughput detection and quantification of fucosylated human milk oligosaccharides (HMOs) using a whole-cell biosensor. We also demonstrate here, the adaptation of this platform towards analysis of HMO production strains, focusing on improving the signal to noise ratio.
Abstract
Human milk oligosaccharides (HMOs) are complex carbohydrate components of human breast milk that exhibit plentiful benefits on infant health. However, optimization of their biotechnological synthesis is limited by the relatively low throughput of detection and quantification of monosaccharide and linkages. Conventional techniques of glycan analysis include chromatographic/mass-spectrometric methods with throughput on the order of hundreds of samples per day without automation. We demonstrate here, a genetically encoded bacterial biosensor for the high-throughput, linkage-specific detection and quantification of the fucosylated HMO structures, 2’-fucosyllactose and 3-fucosyllactose, which we achieved via heterologous expression of fucosidases. As the presence of lactose in milk or in biotechnological processes could lead to false positives, we also demonstrate the reduction of signal from lactose using different strategies. Due to the high throughput of this technique, many reaction conditions or bioreactor parameters could be assayed in parallel in a matter of hours, allowing for the optimization of HMO manufacturing.
Introduction
Human milk oligosaccharides (HMOs) are lactose-derived oligosaccharides, usually comprising three to eight sugar monomers. They have a lactose (Gal-β1,4-Glc) reducing end and are further elongated by glycosidic links (β-1,3- or β-1,6-) to glucose (Glc), galactose (Gal), or N-acetylglucosamine (GlcNAc). In addition, fucose (Fuc, α-1,2- or α-1,3-) or sialic acid (Sia or NeuAc, α-2,3- or α-2,6-) residues are often added1.
Current analysis of oligosaccharides and other carbohydrates is limited in throughput and scope by the need for chromatographic/mass spectrometric (MS) technology2,3,4,5,6,7, which can take roughly an hour per sample, not to mention the necessity for expensive equipment, specialized columns and derivatizing agents, and expertise on the operation of this equipment8. Oligosaccharide linkages are particularly difficult to determine, requiring advanced MS9,10 or nuclear magnetic resonance (NMR) techniques11. Rapid optimization of synthesis of these oligosaccharides is thus limited by the throughput of this slow analytical step.
In this study, we demonstrate linkage-specific detection of fucosylated trisaccharide lactose-based HMOs, focusing on 2’-fucosyllactose (2’-FL) that is the most abundant HMO in human milk, using a genetically encoded Escherichia coli whole cell biosensor with a limit of detection at 4 mg/L. An important feature of this biosensor is its ability to distinguish between isomeric trisaccharides. The design principle is based on the expression of specific fucosidases in E. coli that liberate lactose from HMOs, the presence of which is detected by the lac operon, which in turn generates a fluorescent signal. We achieve this by building a two-plasmid system, one harboring the linkage-specific fucosidase and the other a fluorescent reporter protein. This biosensor platform is suitable for high-throughput screening by flow cytometry or micro-plate reader. We also demonstrate the utilization of the biosensor in quantifying 2’-FL produced by an engineered strain12. Within this study, we also present three strategies on selective removal of lactose that can result in false positive signal from the biosensor, given that the engineered producer strain is grown on lactose.
Taken together, the genetically encoded biosensors allow us to detect and quantify HMOs in a linkage-specific manner, which is difficult even with chromatographic, MS, or NMR techniques. Due to its high throughput and ease of use, this method should have widespread applications in the metabolic engineering and synthesis of HMOs.
Protocol
1. Cell culture and induction conditions
NOTE: In the following experiments, three strains are used: E. coli BL21 (DE3) with an empty vector, E. coli BL21 (DE3) with plasmids pAfcA14 and pET28:green fluorescent protein (GFP), and E. coli BL21 (DE3) with plasmids pAfcB14 and pET28:GFP. All strains are grown in Luria-Bertani broth (LB) or minimal media with appropriate antibiotics. Prepare stock solutions of 1,000x kanamycin (50 mg/mL) and carbenicillin (100 mg/mL) in deionized (DI) water.
- Media preparation
- Prepare LB media by adding 25 g of LB powder stock to 1 L of DI water. Autoclave the solution at 121 °C for 20 min to sterilize. Wait until the sterilized LB media temperature is below 60 °C before addition of 1 µL of antibiotic stock for every 1 mL of LB media.
- To prepare LB plates, add 15 g agar to the LB media before sterilization. Wait until the sterilized LB agar temperature is below 60 °C before addition of antibiotic.
- 1.1.3 For the biosensing cells, prepare media or plates with dual antibiotics, kanamycin and carbenicillin.
- Carbohydrate inducers
- Prepare stock solutions of the carbohydrate inducers at molar equivalents to 20 g/L of lactose: 29 g/L of 2’-FL and 3-FL.
NOTE: Each 200 µL of cells will require 20 µL of 2’-FL or 3-FL. - Induce 200 µL of cells with 2.0 g/L final concentration of lactose or 2.9 g/L final concentration of 2’-FL/3-FL. Incubate at 37 °C with continuous shaking and let cultures grow overnight.
- Prepare stock solutions of the carbohydrate inducers at molar equivalents to 20 g/L of lactose: 29 g/L of 2’-FL and 3-FL.
- Cell culture
- The day before the experiment seed 5 mL of LB medium supplemented with kanamycin and carbenicillin from a fresh bacterial colony, using sterile technique.
- Incubate all cultures at 37 °C with agitation and grow the cultures overnight.
- Transfer 50 µL of overnight culture into 5 mL of fresh LB/M9 media with kanamycin and carbenicillin. Incubate at 37 °C with continuous shaking and grow until culture has reached optical density at 600 nm (OD600) of 0.5–0.7. At this mid-log phase, the cells can be induced.
2. Measurement of fluorescence and limits of detection
- Preparation of cells for flow cytometry
- Transfer the overnight cultures to 1.5 mL tubes and centrifuge at 12,500 x g for 1 min.
- Discard supernatant and re-suspend the pellet in 500 µL of 1x phosphate-buffered saline (PBS; 6 mM Na2HPO4, 1.8 mM NaH2PO4, 145 mM NaCl in DI water, pH 7.2) to prepare a single cell suspension and transfer the single cell suspension to 5 mL flow cytometry tubes.
- Keep the cells in the dark at 4 °C until running the samples on a flow cytometer. For best results, analyze the cells as soon as possible.
- Select 488 nm laser to excite GFP. Collect fluorescein isothiocyanate (FITC) fluorescence levels (20,000–40,000 events, gated for forward and side scatter to avoid aggregated cells and debris) with a 525/50 nm bandpass filter.
- Calibrating the biosensor
- To make a calibration curve, make 8–10 dilutions of the standard 2’-FL in the range 0–2,500 mg/L.
- Culture the 2’-FL biosensing cells (containing pAfcA and pET28:GFP) and induce them with the standard dilutions, as described previously in section 1.3. Carry out three biological replicates for each dilution.
3. Detection and quantification of 2’-FL in a producer strain
- Cultivation of producer strain
- For making minimal media, prepare separate solutions of 1 M K2HPO4, FeSO4·7H2O, sodium citrate, and 1 M thiamine-HCl. Sterilize the solutions using a 0.22 µm filter. Mix M9 media broth powder with 1 L of DI water. Autoclave the M9 solution at 121 °C for 15 min. Cool the solution down to 50 °C. Add MgSO4, CaCl2, K2HPO4, FeSO4·7H2O, sodium citrate, and thiamine to final concentrations of 2 mM, 0.1 mM, 12 g/L, 11 mg/L, 75 mg/L, and 7.5 µg/L, respectively.
- Start a culture of 2’-FL producing strain, e.g., JM109 gwBC-F2, in 5 mL of LB media and let it grow overnight at 37 °C, as described in Baumgartner et al.12.
- Subculture at 1% as described earlier in step 1.3.3 in 250 mL of the minimal media prepared in step 3.1.1. Add glycerol to a final concentration of 10 g/L and incubate the culture at 37 °C.
- When the culture reaches an OD600 of 0.5–0.7, add isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and lactose to a final concentration of 2 g/L.
- Incubate at 37 °C with continuous shaking and let cultures grow for 24 h.
- Extraction of 2’-FL
- Transfer the overnight cultures to sterile 50 mL tubes and centrifuge at 900 x g for 15 min.
- 3.2.2 Discard supernatant and re-suspend the pellet in DI water. Repeat the wash step three times to remove residual lactose and finally re-suspend in 5 mL of DI water.
- To lyse the cell suspension, use a sonicator at 30% power, in 30 s pulses. Keep the cells on ice at all times.
- Centrifuge the lysed cells for 25 min at 2,000 x g. Remove the supernatant, pass it through a sterile (e.g., 0.22 µm) filter and store it at 4 °C until analysis.
- Quantification with biosensor
- Inoculate a culture of 2’-FL biosensing cells and at mid-log phase, induce with cell lysate equivalent to estimated concentrations of 2’-FL used to make the calibration curve as described in section 2.2. Perform the calibration in the same milieu as the sample to be analyzed (e.g., cell lysate) by adding dilutions of 2’-FL to control (i.e., non-producer) filtered cell lysate before inducing biosensor cells.
- Run the samples on a flow cytometer. Generate a dose-response curve by plotting the fluorescence output against the oligosaccharide concentration. Compare the response of the standards to the cell lysate.
4. Selective removal of lactose
NOTE: The biosensing cells are sensitive to lactose and it is desirable for the biosensor to selectively detect HMOs over lactose. One strategy would include washing of cells as described in section 3.2. Three other strategies are described below.
- Treatment with commercial purified β-galactosidase
- Dissolve lyophilized β-galactosidase to 1,000 (FCC lactase) units/mL, in 1 mM MgCl2 or use commercial β-galactosidase in solution.
- To determine the optimum concentration of enzyme needed, grow an inoculum of 2’-FL biosensing cells and induce with 2 g/L lactose and 2.9 g/L 2’-FL at mid log phase.
- To 100 µL cultures, add variable amounts of β-galactosidase, up to 12 units, and let the cultures grow overnight at 37 °C with continuous shaking.
- Measure the fluorescence as described in section 2.1 and calculate the optimum units of enzyme needed by determining the minimum enzyme concentration to achieve desired attenuation of signal.
- To 2’-FL producing cells grown for 24 h, add optimum units of β-galactosidase and incubate overnight at 37 °C. Proceed with the protocol for determining the 2’-FL titer as described in section 3.3.
- Pre-treatment with LacZ+ strain
- Start a 25 mL culture of 2’-FL producing strain and once induced at mid-log phase, incubate the culture at 37 °C for 24 h.
- Inoculate an E. coli BL21 (DE3) culture in LB, grow it to mid-log phase at 37 °C and add 50 mL of the culture (2:1) to the 24 h culture.
- Incubate at 37 °C for 3 h. Proceed with the protocol for determining the 2’-FL titer as described in section 3.3.
- Lactose precipitation with ethanol
NOTE: This section is adapted from Matsuki et al.12.- Start a 25 mL culture of 2’-FL producing strain and once induced at mid-log phase, incubate at 37 °C for 24 h.
- Add two volumes of 100% ethanol to the culture, shake well, and incubate at 37 °C for 4 h.
- Centrifuge the suspension at 900 x g for 15 min. Collect the supernatant and incubate overnight at 4 °C, which precipitates the lactose.
- Remove the precipitated lactose by passing the solution through a filter paper (11 µm). Collect the filtrate that is the cell lysate containing the 2’-FL, which can be quantified by following section 3.3.
Representative Results
We designed a whole cell biosensor specific to 2’-FL that can be used in conjunction with the biotechnological production of the oligosaccharide. This relies on the specific enzymatic cleavage of modifying terminal sugars generating lactose and thereby activation of the lac operon, leading to expression of a reporter fluorescent protein under a lactose inducible promoter, in proportion to the quantity of 2’-FL. To demonstrate its linkage specificity, 3-fucosyllactose (3-FL), an isomer, was also tested. The genetic circuit contains two parts: a fucosidase gene, afcA or afcB, driven by a constitutive promoter resulting in constitutive expression cloned on to a vector with a signal sequence of pelB encoded upstream of the fucosidase gene to facilitate periplasmic export, as well as a fluorescent reporter system containing the T7 promoter driving expression of GFP. The first part is expressed on plasmid pAfcA and the second reporter system on plasmid pET28:GFP. The final construct was transformed into E. coli BL21 (DE3), a widely available strain that has the T7 RNA polymerase integrated into the genome under control of the lactose-responsive pLacUV5 promoter. Figure 1A shows the schematic design so that the reader can follow the working principle of the biosensor. Figure 1B illustrates the fluorescent signal under different conditions of induction as analyzed on a flow cytometer. Consistent with our hypothesis, over a 100-fold increase in fluorescence was observed with the 2’-FL biosensor compared to the vector-only control or with the biosensor for 3-FL. This shows the high linkage-specificity of the biosensor. To ensure that the signal is indeed due to defucosylation, we ran a control by adding commercial α-1,2-fucosidase to the culture during induction, which is confirmed as shown by the bars on the right. The sensitivity of the assay can be determined by generating a dose-response curve with varying carbohydrate levels (Figure 2A). From the plot, the dynamic linear range of 2’-FL detection is between 40 and 400 mg/L and the limit of detection is 4 mg/L. This assay can be carried out over the course of a working day, as shown by the snapshot measurements of the dynamics of reporter expression (Figure 2B).
Finally, we present how the biosensing cells can be used to detect and quantify 2’-FL from an engineered 2’-FL producer strain. Since this strain uses lactose as a substrate, we developed strategies to reduce signal from exogenous lactose so that the signal readout would directly correspond to the 2’-FL concentration (Figure 3A). One strategy is to enzymatically degrade the lactose using a β-galactosidase that does not act on modified lactose. This can be achieved using commercial β-galactosidase that preferentially acts on lactose, reducing the lactose signal to 20% of the original (Figure 3B) or via incubation with a LacZ+ strain, wild-type BL21 (DE3), which in three hours drops the lactose signal to background levels (Figure 3C). The last strategy utilizes ethanol, which not only selectively precipitates lactose but also lyses the producer cells, cell lysis being a necessary step downstream (Figure 3D). This is a simple extraction process, but care should be taken due to lower recovery of 2’-FL compared with other methods, possibly from losses due to some crystallization of 2’-FL. In spite of the addition of ethanol, we did not observe significant inhibition of growth of the biosensing cells, likely because the final ethanol concentrations are low (<2%, v/v) in the cultures. Finally, the most effective but time-consuming method we demonstrate is to pellet and wash the cells prior to cell lysis to release the intracellular 2’-FL (Figure 3E). We advise the reader to use discretion for selecting the appropriate method for lactose removal based on efficiency and availability of time and reagents.
Figure 3E also demonstrates our ability to reliably quantify 2’-FL in biotechnological context. We cultured either an engineered 2’-FL-producing strain11 or a fucosyltransferase negative mutant (negative control) with lactose to monitor production of 2’-FL in cell lysate. After culturing the cells, we measured the concentration using both the biosensor and high-performance liquid chromatography (HPLC) for validation (Supplementary Figure 1). The dose response curves for producer lysate and control lysate spiked with 2’-FL are very similar, which demonstrates quantitative measurement of 2’-FL in the context of cell lysate.
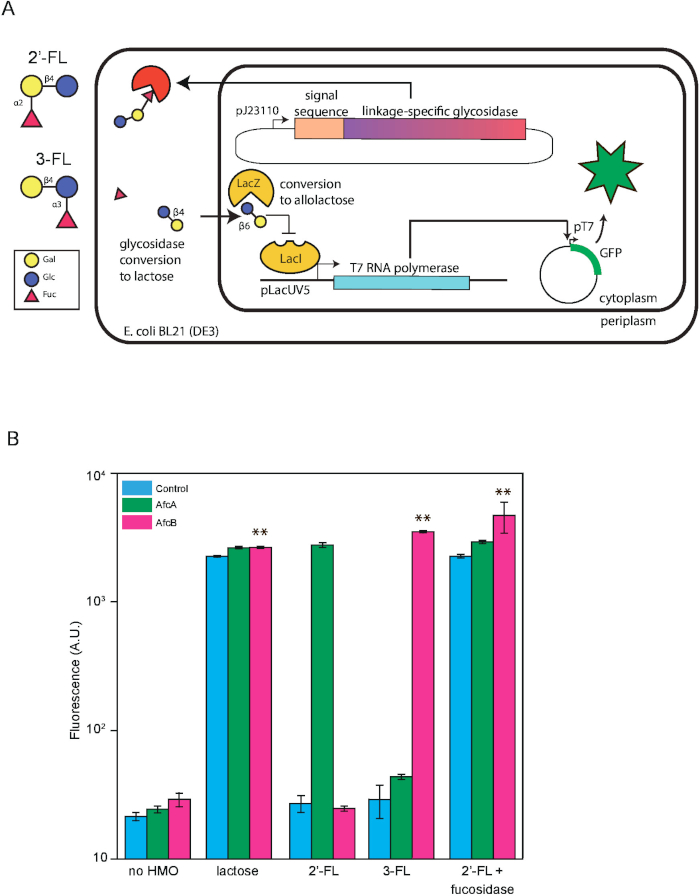
Figure 1: Schematic of 2’-FL biosensor design and representative data. (A) 2’-Fucosyllactose (2’-FL) enters the bacterial periplasm and is converted to lactose by heterologously expressed fucosidase. Lactose is transported to the cytoplasm and converted to allolactose. This activates the native LacUV5 promoter in E. coli BL21 (DE3), producing T7 RNAP, which transcribes GFP under the T7 promoter, leading to fluorescent protein expression. (B) Detection of 2’-FL and 3-FL. Cells containing pET28:GFP and either pAfcA (green), pAfcB (magenta), or an empty vector control (blue) were induced overnight with no sugar, 2’-FL, 2’-FL with exogenously added α-1,2-fucosidase, 3-FL, or lactose. All data are an average of three biological replicates each analyzed in triplicate, where error bars represent standard deviation from the mean. Statistical significance was determined by Tukey’s multiple comparison test and ** is indicative of p < 0.01. This figure first appeared in Enam and Mansell14 and is reused with permission. Please click here to view a larger version of this figure.
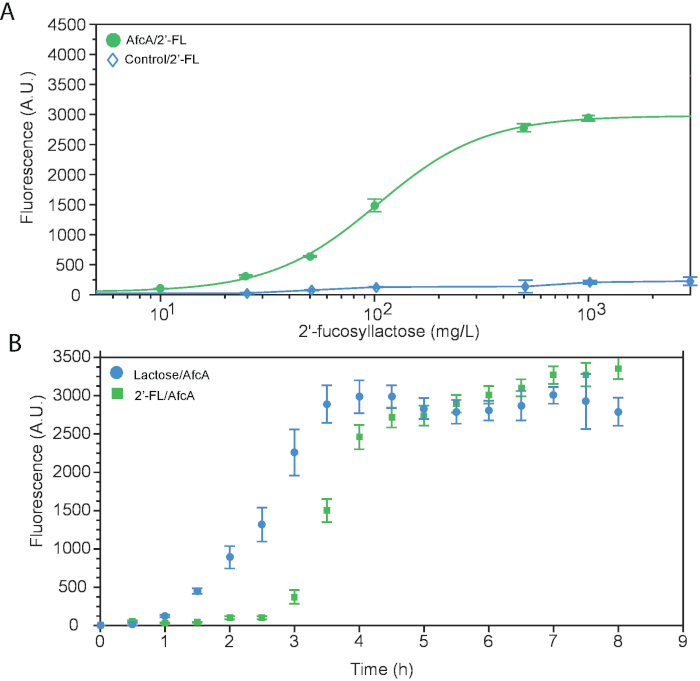
Figure 2: Characterization of biosensor for 2’-FL detection. (A) Limits of detection and transfer functions of biosensors. Response curves depicting mean fluorescence for pAfcA/2’FL (green circle) or empty control vector (blue diamond) at varying amounts of 2’-FL cultured. (B) Time course of reporter expression. Strain pAfcA was incubated with 2’-FL (green square) and fluorescence was monitored over 8 h of induction. Strain pAfcA incubated with lactose is included as a control. All data are an average of three biological replicates each analyzed in triplicate, where error bars represent standard deviation from the mean. This figure first appeared in Enam and Mansell14 and is reused with permission. Please click here to view a larger version of this figure.
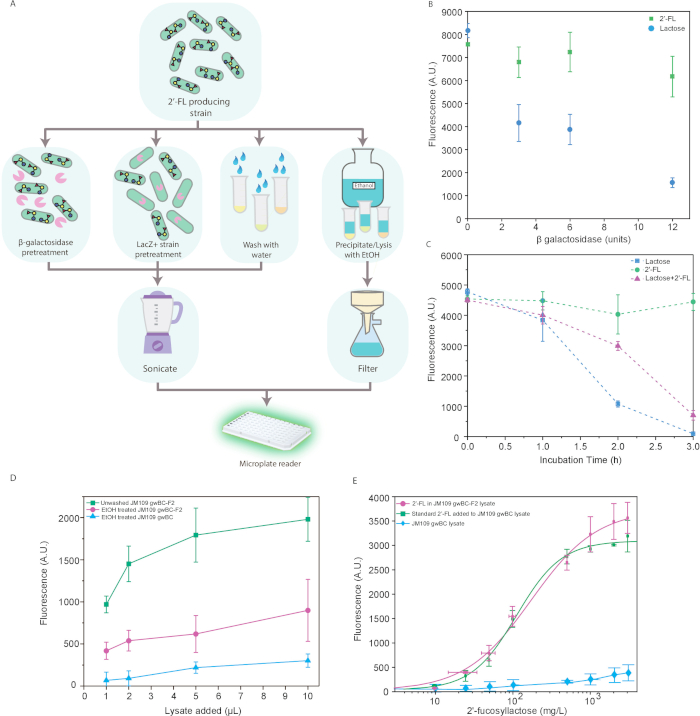
Figure 3: Detection and quantification of 2’-FL from a producer strain. (A) Workflow demonstrating the use of the biosensor in a simple protocol with key focus on removing noise from residual lactose. (B) Removal of lactose by β-galactosidase enzymatic digestion. Cultures containing 0.3% 2’-FL (green square) or 0.2 % lactose (blue circle) were incubated with biosensor cultures containing varying amounts of exogenous β-galactosidase at induction time were assayed for fluorescence after incubation overnight. (C) Removal of lactose by incubation with wild-type LacZ+ strain. LB containing 0.3% 2’-FL (green circle), 0.2% lactose (blue square), or a 1:1 mixture (0.2% lactose equivalent, i.e., 0.1% lactose + 0.15% 2'-FL, magenta triangle) was incubated for various times with BL21 (DE3) cells, and then added to biosensor cultures for fluorescence measurement after incubation overnight. (D) Precipitation of lactose and cell lysis by ethanol pretreatment. Fermentations of JM109 gwBC-F2 (magenta circle) or JM109 gwBC (blue triangle) cells were incubated with 2 volumes of ethanol and filtered before incubation with fluorescent biosensor cells and compared with fluorescence from untreated JM109 gwBC-F2 culture (green square). (E) Quantification of 2’-FL in cell lysate. The biosensor was evaluated for its ability to detect 2’-FL from a metabolically engineered producer strain. Fluorescence measurements generated by addition of either crude lysate of JM109 gwBC-F2 (magenta circle, 2’-FL as quantified by LC-MS), 2’-FL in defined amounts to nonproducer strain JM109 gwBC cell lysate (green square), or crude lysate of JM109 gwBC strain (blue diamond). Results were validated by HPLC quantification of 2’-FL in producer strain. All data are an average of three biological replicates each analyzed in triplicate, where vertical error bars represent standard deviation from the mean fluorescence and horizontal error bars represent the standard deviation in 2’-FL measured by HPLC in the triplicates. This figure first appeared in Enam and Mansell14 and is reused with permission. Please click here to view a larger version of this figure.
Supplemental Figure 1: HPLC-MS analysis of 2’-FL. (A) Extracted ion chromatograms of 2’-FL (m/z 511). Chromatogram of a commercial standard is shown on the top. Chromatogram of 2’-FL from the JM109gwBC-F2 strain (10-fold dilution) is shown at the bottom. The peak at 511 m/z is the sodium adduct. (B) An enlargement of mass spectrum m/z from 300−800, where the most abundant signals were concentrated is shown for the commercial standard (top) and the 2’-FL from the producer strain (bottom). (C) A standard curve was created by LC-MS analysis with varying levels of commercial 2’-FL. Error bars represent standard deviations from triplicate measurements. Please click here to download this file.
Discussion
We present a high-throughput strategy for the linkage-specific detection of fucosylated human milk oligosaccharides. This was accomplished by building whole cell biosensors by genetically engineering E. coli which upon induction with specific glycans respond with a fluorescent signal. The protocol also details on how the biosensor can be used to detect and quantify HMOs in a metabolically engineered bacterial strain.
Our protocol offers advantages over alternative methods due to its high throughput in addition to its linkage-specificity compared to chromatographic/MS methods. The low limit of detection and broad dynamic range allows for the direct measurement of HMOs.
Critical steps in this protocol occur in the preparation of the cell lysate of producer strains. For ensuring maximum titer of 2’-FL, it is crucial to provide the optimum conditions. Variabilities in batches may arise due to sonication parameters or induction times. Care should be taken during characterization of the biosensor. It is also recommended to run proper controls for all steps in the experiment to ensure consistent results.
One limitation of our protocol is its reliance on lactose for induction, which becomes a bottleneck when lactose naturally present, as in breast milk, or used as the substrate in biotechnological production of HMOs. We have addressed this with four distinct strategies to remove the signal to noise ratio so that the biosensor can selectively detect HMOs over lactose.
The genetically encoded whole-cell biosensor presented here will allow optimization for the manufacture of HMOs, as many reaction conditions or bioreactor parameters can be assayed in parallel in a matter of hours. When performed in 96- or 384-well plates, the method shows the potential to screen thousands of samples per day.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by Iowa State University Startup Funds. F.E. was partially funded by the NSF Trinect Fellowship and Manley Hoppe Professorship. T.J.M. was partially supported by the Karen and Denny Vaughn Faculty Fellowship. The authors thank the Iowa State University Flow Cytometry Facility and the W.M. Keck Metabolomics Research Laboratory for assistance with fluorescence and LC-MS studies.
Materials
| 2’-Fucosyllactose | Carbosynth | 41263-94-9 | |
| 3-Fucosyllactose | Carbosynth | 41312-47-4 | |
| Agar | Fisher Scientific | BP9744500 | |
| Calcium Chloride, Dihydrate | Fisher Scientific | C79-500 | |
| Carbenicillin | Fisher Scientific | BP26481 | |
| Dextrose (D-Glucose), Anhydrous | Fisher Scientific | D16-1 | |
| Flow Cytometer | BD | FACSCanto Plus RUO | |
| HPLC | Agilent Technologies | 1100 Series HPLC system | |
| HPLC Column | Luna | C18 reversed phase column | |
| Kanamycin | Fisher Scientific | 11815024 | |
| LB Broth, Miller | Fisher Scientific | 12-795-027 | |
| Lactose | Fisher Scientific | 64044-51-5 | |
| M9, Minimimal Salts, 5x | Sigma-Aldrich | M6030 | |
| Magnesium Sulfate, Anhydrous | Fisher Scientific | M65-500 | |
| MS | Agilent Technologies | Mass Selective Trap SL detector | |
| Sodium chloride | Sigma-Aldrich | 7647-14-5 | |
| Sodium phosphate dibasic | Sigma-Aldrich | 7558-79-4 | |
| Sodium phosphate monobasic | Sigma-Aldrich | 13472-35-0 |
References
- Ninonuevo, M. R., et al. A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry. 54 (20), 7471-7480 (2006).
- Kailemia, M. J., Ruhaak, L. R., Lebrilla, C. B., Amster, I. J. Oligosaccharide analysis by mass spectrometry: a review of recent developments. Analytical Chemistry. 86 (1), 196-212 (2014).
- Royle, L. Chapter 8 – Glycans and Monosaccharides. Liquid Chromatography. , 185-202 (2013).
- Shubhakar, A., Reiding, K. R., Gardner, R. A., Spencer, D. I. R., Fernandes, D. L., Wuhrer, M. High-Throughput Analysis and Automation for Glycomics Studies. Chromatographia. 78 (5-6), 321-333 (2015).
- Goubet, F., Jackson, P., Deery, M. J., Dupree, P. Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Analytical Biochemistry. 300 (1), 53-68 (2002).
- Evangelista, R. A., Liu, M. -. S., Chen, F. -. T. A. Characterization of 9-Aminopyrene-1,4,6-trisulfonate Derivatized Sugars by Capillary Electrophoresis with Laser-Induced Fluorescence Detection. Analytical Chemistry. 67 (13), 2239-2245 (1995).
- Jiao, J., Zhang, H., Reinhold, V. N. High Performance IT-MS Sequencing of Glycans (Spatial Resolution of Ovalbumin Isomers). International Journal of Mass Spectrometry. 303 (2-3), 109-117 (2011).
- Doherty, M., et al. An automated robotic platform for rapid profiling oligosaccharide analysis of monoclonal antibodies directly from cell culture. Analytical Biochemistry. 442 (1), 10-18 (2013).
- Hsu, H. C., Liew, C. Y., Huang, S. -. P., Tsai, S. -. T., Ni, C. -. K. Simple Method for De Novo Structural Determination of Underivatised Glucose Oligosaccharides. Scientific Reports. 8 (1), 5562 (2018).
- Mank, M., Welsch, P., Heck, A. J. R., Stahl, B. Label-free targeted LC-ESI-MS2 analysis of human milk oligosaccharides (HMOs) and related human milk groups with enhanced structural selectivity. Analytical and Bioanalytical Chemistry. 411 (1), 231-250 (2019).
- Chai, W., Piskarev, V. E., Zhang, Y., Lawson, A. M., Kogelberg, H. Structural determination of novel lacto-N-decaose and its monofucosylated analogue from human milk by electrospray tandem mass spectrometry and 1H NMR spectroscopy. Archives of Biochemistry and Biophysics. 434 (1), 116-127 (2005).
- Baumgärtner, F., Seitz, L., Sprenger, G. A., Albermann, C. Construction of Escherichia coli strains with chromosomally integrated expression cassettes for the synthesis of 2’-fucosyllactose. Microbial Cell Factories. 12 (1), 1-13 (2013).
- Matsuki, T., et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nature Communications. 7, 11939 (2016).
- Enam, F., Mansell, T. J. Linkage-Specific Detection and Metabolism of Human Milk Oligosaccharides in Escherichia coli. Cell Chemical Biology. 25 (10), 1292-1303 (2018).

