Assessing the Cellular Immune Response of the Fruit Fly, Drosophila melanogaster, Using an In Vivo Phagocytosis Assay
Summary
This protocol describes an in vivo phagocytosis assay in adult Drosophila melanogaster to quantify phagocyte recognition and clearance of microbial infections.
Abstract
In all animals, innate immunity provides an immediate and robust defense against a broad spectrum of pathogens. Humoral and cellular immune responses are the main branches of innate immunity, and many of the factors regulating these responses are evolutionarily conserved between invertebrates and mammals. Phagocytosis, the central component of cellular innate immunity, is carried out by specialized blood cells of the immune system. The fruit fly, Drosophila melanogaster, has emerged as a powerful genetic model to investigate the molecular mechanisms and physiological impacts of phagocytosis in whole animals. Here we demonstrate an injection-based in vivo phagocytosis assay to quantify the particle uptake and destruction by Drosophila blood cells, hemocytes. The procedure allows researchers to precisely control the particle concentration and dose, making it possible to obtain highly reproducible results in a short amount of time. The experiment is quantitative, easy to perform, and can be applied to screen for host factors that influence pathogen recognition, uptake, and clearance.
Introduction
Innate immune defenses form the first line of defense against pathogenic microbes. These responses can be functionally divided into humoral and cellular innate immunity, both of which are mediated by germline-encoded pattern recognition receptors (PRRs) that sense pathogen-associated molecular patterns (PAMPs)1. Many of the signaling pathways and effector mechanisms of innate immunity are conserved in mammals and invertebrates, such as the nematode, Caenorhabditis elegans, and the fruit fly, Drosophila melanogaster2. The fruit fly has emerged as a powerful system to study host defense against infectious microorganisms3. Drosophila is genetically tractable, easily and inexpensively reared in laboratories, and has a short generation time. Furthermore, the fruit fly exhibits highly efficient defenses against an array of microbes, enabling the examination of host immunity against viral, bacterial, fungal, or parasitic pathogens.
Drosophila immunologists have historically utilized forward genetic screens, genome-wide RNA-mediated interference (RNAi) screening of insect cell lines, and pre-existing mutant fly strains to examine innate immunity – leading to the identification and characterization of several evolutionarily conserved humoral immune pathways4,5,6,7,8. The humoral innate immune response is, arguably, the best characterized immune defense in fruit flies. Following infection, the humoral response leads to the production and systemic release of antimicrobial peptide (AMP) molecules into the hemolymph, the blood equivalent in insects. AMPs are produced by highly conserved Toll and Imd signaling pathways. The Toll pathway is homologous to mammalian TLR/IL-1R receptor signaling, and the Imd pathway is homologous to mammalian Tumor necrosis factor-alpha signaling. In Drosophila, Toll signaling is induced by gram-positive bacteria, fungi, and Drosophila X virus6,9,10 and Imd signaling is induced by gram-negative bacteria11,12.
Cellular immunity, comprised of encapsulation, melanization, and phagocytosis of invasive pathogens, carried out by specialized blood cells called hemocytes13. There are three classes of hemocytes in the fruit fly: crystal cells, lamellocytes, and plasmatocytes13. Crystal cells, which make up 5% of the circulating hemocytes in larvae, release proPhenoloxidase (proPO) enzymes leading to the melanization of pathogens and host tissues at wound sites. Lamellocytes, which are not normally found in healthy embryos or larvae, are adherent cells that encapsulate foreign objects. These cells are induced upon pupariation or when parasitizing wasp eggs are deposited in larvae. Phagocytic plasmatocytes, which make up 95% of circulating hemocytes in larvae and all remaining hemocytes in adults, play a role in tissue remodeling during development and, notably, serve as the main effector cell of Drosophila cellular immunity.
Phagocytosis an immediate and crucial line of innate immune defense; microbes that breach the host epithelial barrier are quickly engulfed and eliminated by phagocytic blood cells (For a comprehensive review of cell biology of phagocytosis see reference 14). This process is initiated when germline-encoded pattern recognition receptors (PRRs) on hemocytes recognize pathogen associated molecular patterns (PAMPs) of microbes. Once bound to their targets, PRRs initiate signaling cascades that lead to the formation of pseudopods through actin cytoskeleton remodeling. The pseudopods surround the microbe, which is subsequently engulfed and internalized into a nascent organelle, the phagosome. Microbes are destroyed as the phagosome undergoes the process of phagosome maturation when the phagosome is trafficked towards the interior of the hemocyte and acidifies through a series of interactions with lysosomes. In vitro and cell biology studies in mammalian primary cells have been instrumental in identifying and characterizing factors that regulate phagocytosis, such as the mammalian Fc-gamma receptor and C3b receptors15,16. Nevertheless, the ability to execute large-scale screens or in vivo studies are limited in mammalian systems.
Here we present an in vivo assay for phagocytosis in adult fruit flies, which is based on a procedure first introduced by the laboratory of David Schneider in 200017. The Schneider lab showed that sessile hemocytes clustered along the abdominal dorsal vessel readily phagocytose polystyrene beads and bacteria. To visualize phagocytosis, flies are injected with fluorescently labeled particles (such as E. coli labeled with fluorescein isothiocyanate (E. coli-FITC)), incubated for 30 minutes to allow hemocytes time to engulf the particles, and then injected with trypan blue, which quenches the fluorescence of particles not phagocytosed during the incubation period. Fly dorsal vessels are then imaged using an inverted fluorescent microscope. This seminal paper, using a relatively simple experiment, demonstrated that hemocytes phagocytose bacteria and latex beads, that bacterial phagocytosis can be inhibited by pre-injecting flies with latex beads, and that flies without both cellular and humoral immune responses are susceptible even to E. coli. The assay presented in this report builds on the work of the Schneider lab to quantify in vivo phagocytosis by measuring the fluorescence intensity of particles engulfed by dorsal vessel associated hemocytes.
Similar to the approach taken in mammalian systems, Drosophila geneticists initially used genome-wide in vitro RNAi screens to identify genes required for the cellular immune response18,19,20,21,22,23. However, the development of the adult in vivo phagocytosis assay enabled follow-up experiments to be readily carried out in whole animals, thus allowing researchers to verify the biological the role of factors identified in in vitro studies. Such was the case with the transmembrane receptor Eater, which was first identified as a bacterial receptor in an RNAi screen using S2 cells24 and then later shown to mediate Escherichia coli (E. coli), Enterococcus faecalis, and Staphylococcus aureus (S. aureus) phagocytosis in adults25.
Our lab employed the in vivo phagocytosis assay in forward genetic screens and genome-wide association studies (using the Drosophila Genetic Reference Panel (DGRP)) to identify novel genes that regulate phagocytosis in adult hemocytes. These studies led to the characterization the receptors PGRP-SC1A and PGRP-SA26, the intracellular vesicle trafficking protein Rab1427, the glutamate transporter Polyphemus28, and RNA-binding protein Fox-129.
We anticipate that future screens incorporating the in vivo phagocytosis could lead to the identification of additional genes that are important for the cellular immune response in Drosophila. Screens using fully-sequenced inbred lines, such as the DGRP or the Drosophila Synthetic Population Resource (DSPR), can identify natural variants affecting phagocytosis or hemocyte development. Furthermore, the technique could be adopted in other species of Drosophila or used to screen new community resources, such as the collection of 250 Drosophila species maintained by the National Drosophila Species Stock Center (NDSSC) at Cornell. These experiments can be carried out using fluorescently-labeled bacterial or fungal-wall bioparticles that are available commercially or may be performed using any number of bacterial or fungal species – provided that the microbe expresses fluorescent markers.
Protocol
1. Prepare Fluorescein particles for injection
- Reconstitute 10 mg of commercially available, heat-killed bacteria particles labeled with fluorescein (see Table of Materials) to a stock concentration of 10 mg/mL by adding 990 µL sterile 1x PBS and 10 µL 50 mM sodium azide. Vortex to mix.
- Divide into single-use 8 µL aliquots in 0.2 mL tubes and store in a dark box at 4 °C to minimize light associated sensitivity.
NOTE: Sodium azide preservative is optional and can be omitted if 10 mg/mL stocks are made with 1 mL sterile 1x PBS, aliquoted, and stored at -20 °C.
- Divide into single-use 8 µL aliquots in 0.2 mL tubes and store in a dark box at 4 °C to minimize light associated sensitivity.
- Make a 10 mL solution of 5% food coloring in 1x PBS by mixing 500 µL syringe filtered green food coloring and 9.5 mL sterile 1x PBS.
- Wash particles before injection to remove sodium azide. Mix 42 µL sterile 1x PBS and 8 µL of 10 mg/mL in a 1.7 mL tube. Centrifuge at max speed for 2.5 min at room temperature.
- Remove the supernatant, add 50 µL 1x PBS, and centrifuge at max speed for 2.5 min at room temperature.
- Repeat steps 1.3 and 1.3.1 2x, for a total of 3 washes.
- After the final wash, discard the supernatant and re-suspend particles to 1.6 mg/mL in 50 µL of 5% food coloring in 1x PBS.
- Wrap the tube in aluminum foil to protect from light. Store at 4 °C, discard after 1 week.
2. Prepare the injection station and flies
- Prepare the injection pad. To inject up to 4 genotypes of flies at the same time, use laboratory tape to divide a rectangular CO2 fly pad into 4 sections. On the bench near the microscope, designate areas to place the vials once flies have been lined up on the pad (one for each corner of the pad).
- Prepare vials of age-matched, 4-7 days-old, flies for injection. For each strain to be tested, transfer 5 males and 5 females into a fresh, labeled vial of prepared fly food and keep at 25 °C.
- Prepare the pneumatic injector (see Table of Materials) by setting the instrument to a 100 ms (short bursts of gas pressure to expel the liquid – allowing the delivery of sub-nanoliter volumes) TIMED mode.
- Prepare the microscope slides. Cut 1.5-inch strips of electrical tape, fold into a loop with the adhesive side out, and place onto a labeled microscope slide.
3. Prepare glass capillary needles
- Pull glass needles (thin wall glass capillaries) using a needle puller.
- Hold the needle under the microscope with a micrometer and break the tip using #5 fine point stainless steel tweezers. 100 µm tips are sufficient to pierce the fly’s cuticle while minimizing wounding.
- Measure the volume of liquid that will be injected into each fly. Load the needle with sterile 5% food coloring in 1x PBS and expel the liquid onto a drop of mineral oil on a 0.01 mm stage micrometer.
NOTE: If the liquid droplet is spherical, the volume in picoliters is calculated as (size)3/1910.30 A needle with a 100 µm diameter will eject ~2 nL in 100 ms.
4. Inject flies
- Pipette 10 µL of 1.6 mg/mL particles onto a small square of parafilm.
- Pull the liquid into the needle and mount in the injector nozzle (see Table of Materials).
- Anesthetize flies with CO2 and line them up in their designated area on the flypad, with the ventral side up and the heads oriented towards the front of the pad. Place vials in corresponding areas on the bench.
- Inject flies at the upper corner of the abdomen with 5, 100 ms pumps of liquid (~10 nL total).
- Transfer the injected flies to the appropriate vials, note the time on the vial. Keep at 25 °C.
- Load a new needle with 0.4% Trypan Blue Solution.
- Set the pneumatic injector to GATED, which allows a constant flow of air to push the liquid out of the needle.
- Anesthetize flies after they have rested for 30 min and inject with Trypan Blue until the abdomen is full and distended.
NOTE: When examining phagosome maturation with particles labeled with a pH-sensitive dye, allow flies to rest for 1 h and do not inject with trypan blue before mounting flies. - Mount flies on microscope slides with electrical tape, ventral side down. Push the wings to the side of the fly and secure them to the tape. Also, gently push the head into the tape to ensure that the fly will not move.
- Immediately move to step 5.
5. Imaging flies
- Image flies, one at a time, at 25x or 32x magnification using an inverted fluorescence microscope attached to a digital camera and computer (see Table of Materials). Focus on the dorsal vessel of the fly using computer software for the digital camera.
- Record the exposure time and magnification between experiments.
NOTE: Losing track of genotypes is a potential source of error when photographing multiple strains in a single sitting. To avoid mislabeling flies, make a note of the image number of the first and last fly photograph for each genotype.
6. Quantifying and normalizing the fluorescence
- Open the software and open one image at a time.
- Measure the fluorescence intensity of the dorsal vessel. Draw a polygon around the dorsal vessel. Select Measure and record the fluorescence intensity inside the polygon.
- Determine the background fluorescence intensity. Copy the first polygon and move it to an area adjacent to the dorsal vessel of the fly. Select Measure and record fluorescence intensity of the background area.
- Normalize the dorsal vessel fluorescence by the background fluorescence:
Dorsal vessel ÷ background. - Calculate the average normalized dorsal vessel fluorescence intensity of all flies in a strain.
- Repeat the experiment 2 more times.
- Use a Student’s unpaired t-test to compare the mean relative fluorescence intensities of control flies and test flies from the three experiments. Calculate the effect size using the formula: Cohen’s d = (M1-M2) ÷ SDpooled, where M1 is the mean of genotype 1 and M2 is the mean of genotype 2 &
SDpooled =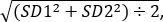
where SD1 is the standard deviation of genotype 1 and SD2 is the standard deviation of genotype 2.
Representative Results
A schematic of the in vivo phagocytosis assay using fluorescein-labeled particles is shown in Figure 1A. Flies are mounted ventral side down on a piece of electrical tape and the first two segments of the abdomen, where the dorsal vessel is located, is clearly visible (Figure 1B). Key sources of experimental error arise at the injection and imaging steps of the procedure (Figure 1C). Using the same needle to inject multiple flies may cause it to become clogged with fly tissue or particles (top left panel in Figure 1C). Because Drosophila auto-fluoresces under GFP light, flies injected with a clogged needle will fluoresce faintly but lack the clear, punctate fluorescence of particles internalized by hemocytes. Another potential source of experimental error can occur during the Trypan Blue injections. Thirty minutes after the first injection with fluorescently-labeled particles, flies receive a second injection with Trypan Blue, which quenches the fluorescence of extracellular, un-phagocytosed particles. Relative fluorescence intensity for each fly is calculated by dividing the fluorescence of the dorsal vessel by that of an equally sized adjacent area on the abdomen. Flies that do not receive enough Trypan Blue fluoresce brightly throughout their entire abdomen (bottom left panel of Figure 1C). This bright fluorescence can reduce the ratio of the dorsal vessel to background fluorescence, thus decreasing the true fluorescence intensity ratio of the animal. Finally, flies should be photographed while immobilized by CO2, as actively moving flies produce blurry images that cannot be quantified (top and bottom, right panels of Figure 1C).
The Zuker collection is a panel of ethyl methanesulfonate (EMS) treated flies with autosomal mutations.31 Originally established as a community resource in 2004, the lines have been used to carry out forward and reverse genetic screens. Our lab identified a line from the Zuker collection that is unable to phagocytose gram-negative (E. coli K-12 strain) and gram-positive bacteria (S. aureus Wood strain without protein A) (Figure 2). This mutant, called argus, shows almost no dorsal vessel fluorescence in the in vivo phagocytosis assay. When compared to the Zuker isogenic background strain, cn bw, argus shows significantly less uptake of E. coli (p-value = 0.019; Cohen’s d = 1.677) and S. aureus (p-value = 0.0083; Cohen’s d = 1.672). Argus bacterial phagocytosis is also significantly impaired, E. coli (p-value = 0.045; Cohen’s d = 0.850) and S. aureus (p-value = 0.0136; Cohen’s d = 1.422) when compared to another common laboratory control strain, Canton-S.
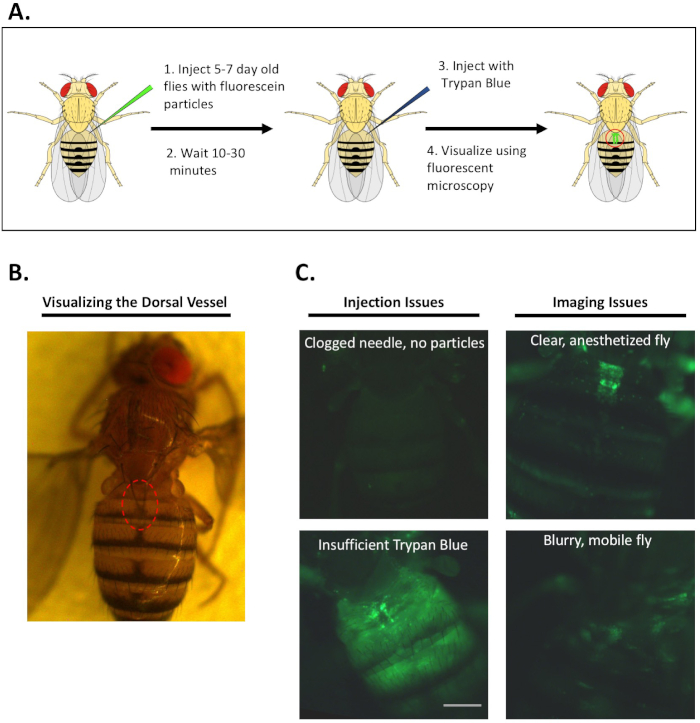
Figure 1: Overview and representative images of in vivo phagocytosis assay. A. Schematic of the in vivo phagocytosis assay using fluorescein-labeled particles. Two injections, 30 minutes apart, are used to visualized particle recognition and uptake by dorsal vessel-associated blood cells. B. White light image showing a fly mounted on tape, with clearly visible anterior abdominal segments. C. Example in vivo phagocytosis images. (top left) A fly injected with a clogged needle has no particle fluorescence. (bottom left) A fly with high extracellular fluorescence because of insufficient trypan blue. (top right) An immobilize fly with no extracellular fluorescence – ideal image. (bottom right) A fly moving as CO2 anesthesia wears off creates a blurry image. Scale bar, 0.2 mm. Please click here to view a larger version of this figure.
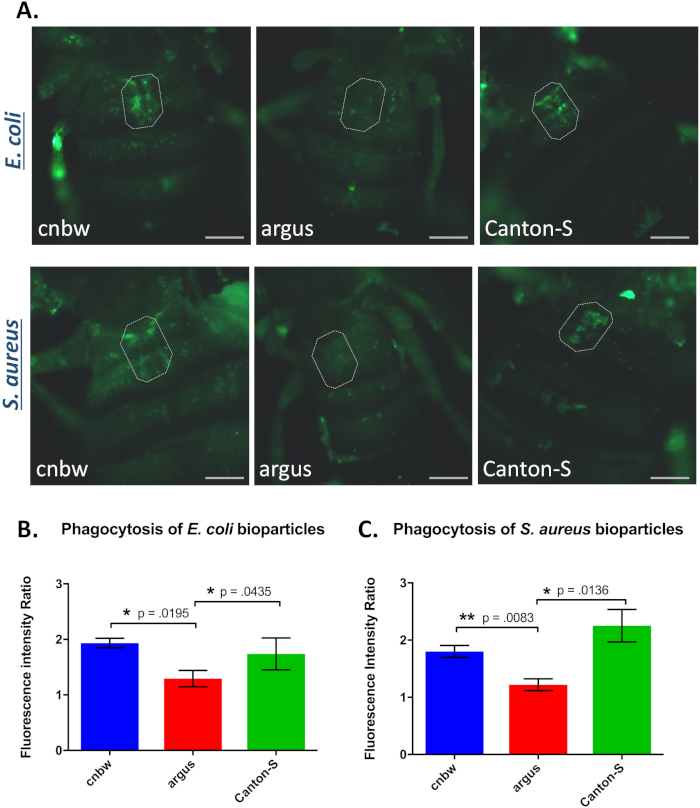
Figure 2: Phagocytosis of bacteria in mutant and control flies. A. Representative images of phagocytosis bacteria labeled with fluorescein dye; E. coli (top panels) and S. aureus (bottom panels). B. The fluorescence intensity ratio of flies injected with fluorescein-E. coli particles. C. The fluorescence intensity ratio of flies injected with fluorescein-S. aureus particles. n = 3 experiments for each type of bacteria, with 6-8 flies per experiment. Error bars, ± SEM. Statistical analysis = Two-tailed t-test. Scale bar, 0.2 mm. Please click here to view a larger version of this figure.
Discussion
Commercially available, fluorescently labeled particles are used to assess phagocytosis in general (0.2 µm carboxylate-modified microspheres) or phagocytosis of microbes (fluorescently-labeled heat- or chemically killed bacteria or yeast). To assess phagosome maturation, researchers can select particles labeled with a pH-sensitive dye that fluoresces when pH decreases from neutral to acidic, as in the phagolysosome. Alternatively, to examine the initial steps of phagocytosis, pathogen recognition and uptake, researchers should choose particles labeled with fluorescent tags that fluoresce inside and outside of cells.
Carefully controlled experimental conditions are key to ensure the in vivo phagocytosis assay is carried out in a reproducible manner. First, Drosophila undergoes cellular immunosenescence32, so flies used in the experiment should be age-matched, optimally within a range of 4-7 days old. Second, the period between injections should be consistent. Phagocytosis occurs rapidly, within minutes of the host cell sensing the microbe14. Over the next hour, phagosome maturation takes place, leading to the acidification of the phagosome and destruction of the internalized microbe. Designating a set interval to wait between injections reduces experimental variability and allows comparisons to be made between strains and experiments conducted on different days. We recommend waiting thirty minutes when using fluorescein particles and one hour for the in vivo phagosome maturation assay using particles labeled with pH-sensitive fluorescent dye.
Using the same needle to inject multiple strains ensures that all flies receive the same dose of particles and experimental variability can arise if glass needles break or become clogged. So, setting up the injection station and flies beforehand is key to ensuring that the in vivo phagocytosis assay is carried out quickly and efficiently. Before starting the experiment, measure the volume of liquid expelled by several needles and retain a set of needles of the same size. Thus, if one needle breaks between injections, it can be replaced immediately. Furthermore, clogged needles can lead to false negative results, making it difficult to distinguish between flies whose hemocytes are unable to phagocytose microbes and flies that did not receive fluorescent particles. Dye solutions made with green food coloring, make it easier to track which flies have been injected and to identify issues with clogged needles. Ideally, the experiment should be performed multiple times, with least 6-8 flies per strain in each experiment, and the results of experiments compared to identify outliers. Finally, enough Trypan Blue should be administered to fully quench the fluorescence of extracellular particles so that the background fluorescence is comparable between flies (Figure 1C).
When imaging flies, care should also be taken to obtain clear, consistent images of the dorsal vessel. Black electrical tape is used to create a dark background and flies should be mounted ventral side down, with their wings and heads secured to the adhesive side of the tape. If possible, flies should remain immobilized under CO2 while images are taken. Finally, to limit the possibility of experimental bias, we recommend using a blinded experimental setup; where vials and strains are coded by one researcher and the experiment is carried out and quantified by another researcher.
The in vivo phagocytosis assay is a useful tool to identify global defects in phagocytosis. However, it does not distinguish between hemocyte phagocytosis efficiency and differences in hemocyte number or location, due to age-related decline or developmental defects.32 Moreover, the procedure does not provide information on differences in the size or phagocytic capacity of individual hemocytes. Thus, once flies with reduced phagocytosis have been identified using the in vivo phagocytosis assay, researchers should carry out follow-up studies, perhaps using alternative particles, in order to discriminate between immune-related phagocytosis defects and general phagocytic defects. Ultimately, determining the molecular mechanisms that underlie phagocytic defects in mutant strains requires an examination of phagocytosis in single hemocytes. This can be accomplished using, for example, confocal microscopy, fluorescence activated cell sorting or magnetic bead isolation of adult hemocytes27,28,29,32,33,34.
Overall, Drosophila has proven to be an effective model to study innate immunity in live animals. Here we have provided an overview of the in vivo phagocytosis assay, a method to study the global phagocytosis and the cellular immune response in adult flies. The procedure can be applied to screen for new genes that may be important for host defense, validate genes identified in large-scale in vitro RNAi screens, and further characterize mutants with defects in the humoral, gut or antiviral immune responses. It is easy to set-up and can yield a large amount of reliable data about multiple strains of flies in a short amount of time.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Dr. Beth Gonzalez and Dr. Aprajita Garg for support in carrying out the in vivo phagocytosis experiments. An NSF UMD ADVANCE Seed Grant and UMD NIH T32 Training Grants, Cell and Molecular Biology (CMB) and the Host-Pathogen Interactions (HPI), funded this work.
Materials
| 0.2μm Red Fluorescent Carboxylate Modified FluoSpheres | Invitrogen | F8810 | Fluorescently-labeled latex beads to test general phagocytic capacity of phagocytes. (~580/~605 nm) Inject a 1:20 dilution in PBS with 5% dye. |
| 5430-10 PicoNozzle Kit | World Precision Instruments | 5430-10 | Holder for 1.0mm pipette |
| E. coli (K-12 Strain) BioParticles, Alexa Fluor 488 conjugate | Invitrogen | E13231 | Killed E. coli labeled with Alexa Fluor 488. Use to test phagocyte recogntion and uptake of gram-negative bacteria. (~495/~519 nm) |
| E. coli (K-12 Strain) BioParticles, Alexa Fluor 594 conjugate | Invitrogen | E23370 | Killed E. coli labeled with Alexa Fluor 594. Use to test phagocyte recogntion and uptake of gram-negative bacteria. (~590/~617 nm) |
| E. coli (K-12 Strain) BioParticles, Fluorescein conjugate | Invitrogen | E2861 | Killed E. coli labeled with FITC (Fluorescein). Use to test phagocyte recogntion and uptake of gram-negative bacteria. (~494/~518 nm) |
| E. coli (K-12 Strain) BioParticles, Texas Red conjugate | Invitrogen | E2863 | Killed E. coli labeled with Texas Red. Use to test phagocyte recogntion and uptake of gram-negative bacteria. (~595/~615 nm) |
| E. coli (K-12 Strain) BioParticles, Texas Red conjugate | Invitrogen | E2863 | Killed E. coli labeled with Texas Red. Use to test phagocyte recogntion and uptake of gram-negative bacteria. (~595/~615 nm) |
| Needle Pipette Puller | David Kopf Instruments | Model 725 | |
| pHrodo Red E. coli BioParticles Conjugate for Phagocytosis | Invitrogen | P35361 | Killed E. coli labeled with pHrodo Red. Use to test phagocyte reconition, uptake, and phagosome maturation of gram-negative bacteria. (~560/~585 nm). No need to quench with Trypan Blue. |
| pHrodo Red S. aureus BioParticles Conjugate for Phagocytosis | Invitrogen | A10010 | Killed S. aureus labeled with pHrodo Red. Use to test phagocyte reconition, uptake, and phagosome maturation of gram-positve bacteria. (~560/~585 nm). No need to quench with Trypan Blue. |
| Pneumatic PicoPump PV820 | World Precision Instruments | SYS-PV820 | The World Precision Instruments Pneumatic PicoPump PV820 uses differential pressures to hold liquid in the glass needle between injections. The user manually controls short bursts of gas pressure to expel the liquid – allowing delivery of sub-nanoliter volumes. The amount of liquid delivered depends on two main variables – the size of the glass needle opening and the amount of time injection pressure is applied. set the instrument to 100 ms “TIMED” mode. |
| S. aureus (Wood Strain without protein A) BioParticles, Alexa Fluor 488 conjugate | Invitrogen | S23371 | Killed S. aureus labeled with Alexa Fluor 488. Use to test phagocyte recogntion and uptake of gram-positive bacteria. (~495/~519 nm) |
| S. aureus (Wood Strain without protein A) BioParticles, Alexa Fluor 594 conjugate | Invitrogen | S23372 | Killed S. aureus labeled with Alexa Fluor 594. Use to test phagocyte recogntion and uptake of gram-positive bacteria. (~590/~617 nm) |
| S. aureus (Wood Strain without protein A) BioParticles, Fluorescein conjugate | Invitrogen | E2851 | Killed S. aureus labeled with FITC (Fluorescein). Use to test phagocyte recogntion and uptake of gram-positive bacteria. (~494/~518 nm) |
| Thin Wall Glass Capillaries | World Precision Instruments | TW100F-3 | Needles for injection. OD = 1.0 mm |
| Trypan Blue Solution (0.4%) | Sigma | T8154 | Used to quench extracellular fluorescence of Fluorescein, Alexa Fluor, or Texas Red labeled particles. |
| ZEISS SteREO Microscope (Discovery.V8) | Zeiss | SteREO Discovery.V8 | Inverted fluorescence microscope for imaging flies. Use a digital camera (example: AxioCam HC camera) and the accompanying software (example: AxioVision 4.7 software) to take pictures. |
| Zymosan A (Saccharomyces cerevisiae) BioParticles, Alexa Fluor 488 conjugate | Invitrogen | Z23373 | Killed labeled with Alexa Fluor 488. Use to test phagocyte recogntion and uptake of yeast. (~495/~519 nm) |
| Zymosan A (Saccharomyces cerevisiae) BioParticles, Alexa Fluor 594 conjugate | Invitrogen | Z23374 | Killed labeled with Alexa Fluor 594. Use to test phagocyte recogntion and uptake of yeast. (~590/~617 nm) |
| Zymosan A (Saccharomyces cerevisiae) BioParticles, Fluorescein conjugate | Invitrogen | Z2841 | Killed labeled with FITC (Fluorescein). Use to test phagocyte recogntion and uptake of yeast. (~494/~518 nm) |
| Zymosan A (Saccharomyces cerevisiae) BioParticles, Texas Red | Invitrogen | Z2843 | Killed labeled with Texas Red. Use to test phagocyte recogntion and uptake of yeast. (~595/~615 nm) |
References
- Akira, S., Uematsu, S., Takeuchi, O. Pathogen recognition and innate immunity. Cell. 124 (4), 783-801 (2006).
- Kim, D. Studying host-pathogen interactions and innate immunity in Caenorhabditis elegans. Disease Models & Mechanisms. 1 (4-5), 205-208 (2008).
- Lemaitre, B., Hoffmann, J. The host defense of Drosophila melanogaster. Annual Review Immunology. 25, 697-743 (2007).
- Wu, L. P., Choe, K. M., Lu, Y., Anderson, K. V. Drosophila Immunity: Genes on the Third Chromosome Required for the Response to Bacterial Infection. Genetics. 159 (1), 189-199 (2001).
- De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M., Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO Journal. 21 (11), 2568-2579 (2002).
- Michel, T., Reichhart, J. M., Hoffmann, J. A., Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 414 (6865), 756-759 (2001).
- Choe, K. M., Werner, T., Stoven, S., Hultmark, D., Anderson, K. V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 296 (5566), 359-362 (2002).
- Wu, J., Randle, K. E., Wu, L. P. ird1 is a Vps15 homologue important for antibacterial immune responses in Drosophila. Cellular Microbiology. 9 (4), 1073-1085 (2007).
- Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M., Hoffmann, J. A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 86 (6), 973-983 (1996).
- Zambon, R. A., Nandakumar, M., Vakharia, V. N., Wu, L. P. The Toll pathway is important for an antiviral response in Drosophila. Proceedings of the National Academy of Science of the United States of America. 102 (20), 7257-7262 (2005).
- Lemaitre, B., et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proceedings of the National Academy of Science of the United States of America. 92 (21), 9465-9469 (1995).
- Leulier, F., Rodriguez, A., Khush, R. S., Abrams, J. M., Lemaitre, B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Reports. 1 (4), 353-358 (2000).
- Meister, M., Lagueux, M. Drosophilablood cells. Cellular Microbiology. 5 (9), 573-580 (2003).
- Flannagan, R. S., Jaumouille, V., Grinstein, S. The cell biology of phagocytosis. Annual Review Pathology. 7, 61-98 (2012).
- Anderson, R. A., Sando, G. N. Cloning and expression of cDNA encoding human lysosomal acid lipase/cholesteryl ester hydrolase. Similarities to gastric and lingual lipases. Journal of Biological Chemistry. 266 (33), 22479-22484 (1991).
- Ross, G. D., Reed, W., Dalzell, J. G., Becker, S. E., Hogg, N. Macrophage cytoskeleton association with CR3 and CR4 regulates receptor mobility and phagocytosis of iC3b-opsonized erythrocytes. Journal of Leukocyte Biology. 51 (2), 109-117 (1992).
- Elrod-Erickson, M., Mishra, S., Schneider, D. S. Interactions between the cellular and humoral immune responses in Drosophila. Current Biology. 10, 781-784 (2000).
- Ramet, M., Pearson, A. M., Manfruelli, P., Li, X., Koziel, H., Gobel, V. Drosophila Scavenger Receptor CI Is a Pattern Recognition Receptor for Bacteria. Immunity. 15 (6), 1027-1038 (2001).
- Ramet, M., Manfruelli, P., Pearson, A. M., Mathey-Prevot, B., Ezekowitz, R. A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 416 (6881), 644-648 (2002).
- Philips, J. A., Rubin, E. J., Perrimon, N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 309, 1251-1253 (2005).
- Agaisse, H., Burrack, L. S., Philips, J. A., Rubin, E. J., Perrimon, N., Higgins, D. E. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 309 (5738), 1248-1251 (2005).
- Stuart, L. M., et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. Journal of Cell Biology. 170 (3), 477-485 (2005).
- Stroschein-Stevenson, S. L., Foley, E., O’Farrell, P. H., Johnson, A. D. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biology. 4 (1), e4 (2006).
- Kocks, C., et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 123 (2), 335-346 (2005).
- Nehme, N. T., et al. Relative roles of the cellular and humoral responses in the Drosophila host defense against three gram-positive bacterial infections. PLoS One. 6 (3), e14743 (2011).
- Garver, L. S., Wu, J., Wu, L. P. The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proceedings of the National Academy of Science of the United States of America. 103 (3), 660-665 (2006).
- Garg, A., Wu, L. P. Drosophila Rab14 mediates phagocytosis in the immune response to Staphylococcus aureus. Cellular Microbiology. 16 (2), 296-310 (2014).
- Gonzalez, E. A., Garg, A., Tang, J., Nazario-Toole, A. E., Wu, L. P. A glutamate-dependent redox system in blood cells is integral for phagocytosis in Drosophila melanogaster. Current Biology. 23 (22), 2319-2324 (2013).
- Nazario-Toole, A. E., Robalino, J., Okrah, K., Corrada-Bravo, H., Mount, S. M., Wu, L. P. The Splicing Factor RNA-Binding Fox Protein 1 Mediates the Cellular Immune Response in Drosophila melanogaster. Journal of Immunology (Baltimore, Md: 1950). 201 (4), 1154-1164 (2018).
- Guille, M. . Molecular Methods in Developmental Biology: Xenopus and Zebrafish. , (1999).
- Koundakjian, E. J., Cowan, D. M., Hardy, R. W., Becker, A. H. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 167 (1), 203-206 (2004).
- Horn, L., Leips, J., Starz-Gaiano, M. Phagocytic ability declines with age in adult Drosophila hemocytes. Aging Cell. 13 (4), 719-728 (2014).
- Brennan, C. A., Delaney, J. R., Schneider, D. S., Anderson, K. V. Psidin is required in Drosophila blood cells for both phagocytic degradation and immune activation of the fat body. Current Biology. 17 (1), 67-72 (2007).
- Akbar, M. A., Tracy, C., Kahr, W. H., Kramer, H. The full-of-bacteria gene is required for phagosome maturation during immune defense in Drosophila. Journal of Cell Biology. 192 (3), 383-390 (2011).

