In Vitro Transcribed RNA-based Luciferase Reporter Assay to Study Translation Regulation in Poxvirus-infected Cells
Summary
We present a protocol to study mRNA translation regulation in poxvirus-infected cells using in vitro Transcribed RNA-based luciferase reporter assay. The assay can be used for studying translation regulation by cis-elements of an mRNA, including 5’-untranslated region (UTR) and 3’-UTR. Different translation initiation modes can also be examined using this method.
Abstract
Every poxvirus mRNA transcribed after viral DNA replication has an evolutionarily conserved, non-templated 5'-poly(A) leader in the 5'-UTR. To dissect the role of 5'-poly(A) leader in mRNA translation during poxvirus infection we developed an in vitro transcribed RNA-based luciferase reporter assay. This reporter assay comprises of four core steps: (1) PCR to amplify the DNA template for in vitro transcription; (2) in vitro transcription to generate mRNA using T7 RNA polymerase; (3) Transfection to introduce in vitro transcribed mRNA into cells; (4) Detection of luciferase activity as the indicator of translation. The RNA-based luciferase reporter assay described here circumvents issues of plasmid replication in poxvirus-infected cells and cryptic transcription from the plasmid. This protocol can be used to determine translation regulation by cis-elements in an mRNA including 5'-UTR and 3'-UTR in systems other than poxvirus-infected cells. Moreover, different modes of translation initiation like cap-dependent, cap-independent, re-initiation, and internal initiation can be investigated using this method.
Introduction
According to the central dogma, genetic information flows from DNA to RNA and then finally to protein1,2. This flow of genetic information is highly regulated at many levels including mRNA translation3,4. Development of reporter assays to measure regulation of gene expression will facilitate understanding of regulatory mechanisms involved in this process. Here we describe a protocol to study mRNA translation using an in vitro transcribed RNA-based luciferase reporter assay in poxvirus-infected cells.
Poxviruses comprise many highly dangerous human and animal pathogens5. Like all other viruses, poxviruses exclusively rely on host cell machinery for protein synthesis6,7,8. To efficiently synthesize viral proteins, viruses evolved many strategies to hijack cellular translational machinery to redirect it for translation of viral mRNAs7,8. One commonly employed mechanism by viruses is to use cis-acting elements in their transcripts. Notable examples include the Internal Ribosome Entry Site (IRES) and cap-independent translation enhancer (CITE)9,10,11. These cis-elements render the viral transcripts a translational advantage by attracting translational machinery via diverse mechanisms12,13,14. Over 100 poxvirus mRNAs have an evolutionarily conserved cis-acting element in the 5’-untranslated region (5’-UTR): a 5’-poly(A) leader at the very 5’ ends of these mRNAs15,16. The lengths of these 5’-poly(A) leaders are heterogeneous and are generated by slippage of the poxvirus-encoded RNA polymerase during transcription17,18. We, and others, recently discovered that the 5’-poly(A) leader confers a translation advantage to an mRNA in cells infected with vaccinia virus (VACV), the prototypic member of poxviruses19,20.
The in vitro transcribed RNA-based luciferase reporter assay was initially developed to understand the role of 5’-poly(A) leader in mRNA translation during poxvirus infection19,21. Although plasmid DNA-based luciferase reporter assays have been widely used, there are several drawbacks that will complicate the result interpretation in poxvirus-infected cells. First, plasmids are able to replicate in VACV-infected cells22. Second, cryptic transcription often occurs from plasmid DNA18,23,24. Third, VACV promoter-driven transcription generates poly(A)-leader of heterogeneous lengths consequently making it difficult to control the poly(A)-leader length in some experiments18. An in vitro transcribed RNA-based luciferase reporter assay circumvents these issues and the data interpretation is straightforward.
There are four key steps in this method: (1) polymerase chain reaction (PCR) to generate the DNA template for in vitro transcription; (2) in vitro transcription to generate mRNA; (3) transfection to deliver mRNA into cells; and (4) detection of luciferase activity as indicator of translation (Figure 1). The resulting PCR amplicon contains the following elements in 5’ to 3’ direction: T7-Promoter, poly(A) leader or desired 5’-UTR sequence, firefly luciferase open reading frame (ORF) followed by a poly(A) tail. PCR amplicon is used as the template to synthesize mRNA by in vitro transcription using T7 polymerase. During in vitro transcription, m7G cap or other cap analog is incorporated in newly synthesized mRNA. The capped transcripts are transfected into uninfected or VACV-infected cells. The cell lysate is collected at the desired time after transfection to measure luciferase activities that indicate protein production from transfected mRNA. This reporter assay can be used to study translation regulation by cis-element present in 5’-UTR, 3’-UTR or other regions of an mRNA. Furthermore, the in vitro transcribed RNA-based assay can be used to study different mechanisms of translation initiation including cap-dependent initiation, cap-independent initiation, re-initiation and internal initiation like IRES.
Protocol
1. Preparation of DNA Template by PCR for In Vitro Transcription
- To prepare the DNA template by PCR, design primers. When designing primers consider crucial characteristics like primer length, annealing temperature (Tm), GC content, 3’ end with G or C etc.
NOTE: Discussed in detail in these literature25,26,27. - Design primers to generate PCR amplicon containing the following elements in 5’ to 3’ direction: T7-Promoter, poly(A) leader, firefly luciferase ORF and a poly(A) tail referred hereafter to as T7_12A-Fluc. Design primers (Forward and Reverse) to encompass all the additional elements not present in the template DNA (Figure 2A).
NOTE: The sequence of all elements can be found in Table 1. - Include several extra nucleotides in forward primer (5’-3’)28, followed by T7-promoter, poly(A) leader or desired 5’-UTR sequence and approximately 20 nucleotides, adjust based on Tm, corresponding to the 5’ end of the reporter gene’s ORF. Ensure the corresponding region in the primer is identical to the sense strand (+ strand) of the gene.
NOTE: For long 5’-UTR, synthesize two DNA fragments: one with T7 promoter followed by long 5’-UTR and second with reporter gene’s ORF. Join these two fragments using overlap extension PCR29. - Design reverse primer (5’-3’) to include poly(A) tail and approximately 20 nucleotides, adjust based on Tm, corresponding to the 3’ end of the reporter gene’s ORF. Ensure the corresponding region in the primer is identical to the anti-sense strand (- strand) of the gene and an in-frame stop codon is present before the poly(A) tail.
NOTE: The desired length of A’s in a poly(A) leader or poly(A) tail can be customized in the primers. For example, to add 50 A’s in the poly(A) tail, the reverse primer should entail 50 T’s. Similarly, to add 20 A’s in the poly(A) leader, the forward primer should entail 20 A’s. - For internal control, design another set of primers containing the following elements in 5’ to 3’ direction: T7 Promoter, a random 5’-UTR coding sequence containing Kozak sequence, Renilla luciferase ORF and poly(A) tail referred hereafter to as T7_Kozak-Rluc.
- In a PCR tube, add the reagents in the following order: DNase free water, 2x high-fidelity DNA polymerase, primers, and sequence confirmed luciferase template DNA (Table 2).
NOTE: Amounts of individual components in the mixture should be adjusted according to the reaction volume. - Use a standard 3-step (Denaturation, Annealing, Extension) PCR cycle to generate a DNA template as shown in Table 3.
NOTE: Annealing temperature X °C depends on the primer set being used and extension time T min depend on the PCR amplicon size and DNA polymerase used. - Detect the PCR product by running 5-10% of PCR reaction in 1% agarose Tris-acetate-EDTA (TAE) gel electrophoresis (containing 0.1µg/ml ethidium bromide) along with commercially available molecular weight standard. Visualize the gel under a UV illuminator to determine the size of the PCR product.
- After determining the correct size of the PCR product, ~1.7 kb for T7_12A-Fluc and ~1.0 kb for T7_Kozak-Rluc, purify it using a commercially available PCR purification kit. Elute the DNA using 100 µL nuclease free water (Figure 2B).
- Once purified, check the concentration of the DNA using a spectrophotometer and determine the A260/A280 ratio (~1.8-2.0 is acceptable).
- Store purified DNA at -20 °C or use for in vitro transcription immediately.
2. Generate mRNA by In Vitro Transcription
- Synthesize RNA from the PCR product in vitro, using an in vitro transcription kit (Figure 3A).
NOTE: T7_12A-Fluc and T7_Kozak-Rluc DNA templates are used to synthesize 12A-Fluc and Kozak-Rluc mRNAs, respectively.- To do this, take a microcentrifuge tube and add the reagents in the following order: DNase-RNase free water, NTP Buffer Mix, Cap Analog, Template PCR Product, T7-RNA polymerase Mix (Table 4).
NOTE: Other capping systems can also be used to cap RNA sequentially after in vitro transcription following the manufacturer’s instruction. - Mix thoroughly and incubate at 37 °C for 2 h.
- Proceed to the purification of the synthesized RNA using an RNA purification kit.
- To do this, take a microcentrifuge tube and add the reagents in the following order: DNase-RNase free water, NTP Buffer Mix, Cap Analog, Template PCR Product, T7-RNA polymerase Mix (Table 4).
- Run purified RNA in 1.5% agarose Tris-borate-EDTA (TBE) gel (containing 0.5 µg/mL ethidium bromide) to check the RNA. Visualize the gel under a UV illuminator (Figure 3B).
- Check the concentration of the RNA using a spectrophotometer and determine the A260/A280 ratio (~1.8-2.0 is acceptable).
- Aliquot the purified RNA and store at -80 °C.
3. Transfect mRNA to Cells
- Seed HeLa cells in a 24-well plate (to be approx. ~80-90% confluent next day) and incubate overnight in an incubator at 37 °C with 5% CO2.
- Infect HeLa cells with vaccinia virus (VACV) at a Multiplicity of Infection (MOI) of 5 or keep uninfected HeLa cells for comparison.
NOTE: MOI is the number of infectious viral particles per cell. - MOI of X= {[(Number of cells * X) / Virus Titer] * 1000} µl of virus per 1 mL medium.
- After desired h post infection (hpi) (in this experiment at 10-12 hpi), transfect mRNA (500 ng of total mRNA per well of 24-well plates) using a cationic lipid transfection reagent as shown in Figure 3C.
- For one well of a 24-well plate, mix 480 ng of 12A sequence bearing firefly luciferase (12A-Fluc) mRNA and 20 ng of Kozak sequence bearing Renilla luciferase (Kozak-Rluc) mRNA in one microcentrifuge tube. In another microcentrifuge tube add 1.1 µL of cationic lipid transfection reagent.
- Add 55 µL of reduced serum medium in both tubes. Mix and incubate at room temperature for 5 min.
- After 5 min of incubation, add 55 µL cationic lipid transfection reagent containing reduced serum medium in mRNA containing tube.
- Mix gently but thoroughly, and incubate at room temperature for 15 min.
- During the incubation, remove the cell culture medium and add 400 µL of reduced serum medium per well of 24-well plates.
- After incubation, add 100 µL of the mixture dropwise and evenly to one well of 24-well plates.
4. Measure Luciferase Activities
- Five-hours post-co-transfection of 12A-Fluc and Kozak-Rluc mRNA, measure luciferase activity using a luciferase assay system capable of performing two reporter assays (e.g., Dual Luciferase Reporter assay kit).
- Remove the reduced serum medium and lyse the cells by adding 150 µL 1x lysis buffer, a component of the luciferase assay kit.
- After 10 min incubation at room temperature, collect the lysate by scrapping the cells and transfer to a microcentrifuge tube.
- Centrifuge the lysate at 12,000 x g for 10 min at 4 °C to pellet cell debris.
- Add 30 µL of supernatant in opaque-walled 96 well white assay plate with a solid bottom.
- Measure the dual luminescence using the luciferase assay kit and a multimode plate reader luminometer.
- Perform the measurement using kinetics function (on a per-well basis) using the settings described in Table 5.
NOTE: The reading can also be taken using manual luminometer. Add an equal volume of lysate and substrate for Fluc in a cuvette. Wait for 2 s and measure for 10 s using luminometer. Following Fluc measurement, quickly take out cuvette from luminometer and add an equal volume of the substrate for Rluc manually. Again, wait for 2 s and measure for 10 s using luminometer. - Export the luminescence reading data into a desirable file format.
- Determine relative translation rate from 12A-Fluc mRNA in uninfected and VACV infected HeLa cells by dividing Fluc value by internal control Rluc value.
NOTE: Supplementary Figure 1 shows the step-by-step analysis of raw data to get relative Fluc activity.
Representative Results
The four steps of in vitro transcribed RNA-based luciferase reporter assay: PCR to generate DNA template for In vitro transcription, in vitro transcription to generate mRNA, mRNA transfection, and luciferase measurement, can be seen in the schematic diagram (Figure 1). Designing of primers for both DNA templates (Fluc and Rluc) and the general scheme of overhang extension PCR is illustrated in the schematic (Figure 2A). After PCR, the correct sized PCR product was detected by TAE agarose gel electrophoresis (Figure 2B). Subsequently, the PCR product is used as the template to synthesize RNA in vitro (Figure 3A), which is purified and run in TBE gel electrophoresis to verify the size (Figure 3B). The purified and verified mRNA is transfected into cells using cationic lipid transfection reagent (Figure 3C). Primers used in this protocol are listed in Table 6.
The in vitro transcribed RNA-based luciferase reporter assay was developed to understand the role of 5'-poly(A) leader in mRNA translation during poxvirus infection. Using this assay, we tested the translation efficiency of a Fluc mRNA that contains a 5'-poly(A) leader (12 nt) in uninfected and VACV-infected cells. The Fluc value was normalized using Rluc value in both uninfected and VACV-infected cells to determine the relative Fluc activity (i.e. Fluc activity/Rluc activity) (Figure 4A). The division of Fluc by Rluc normalized the transfection efficiency and RNA stability in a particular well. Using this analysis approach, we determined that a 5'-poly(A) leader containing mRNA has a translational advantage during VACV infection (Figure 4B). The advantage in infected cells was not due to differential transfection efficiency or mRNA stability as the RNA level was similar in uninfected and VACV infected cells 5 h post mRNA transfection19.
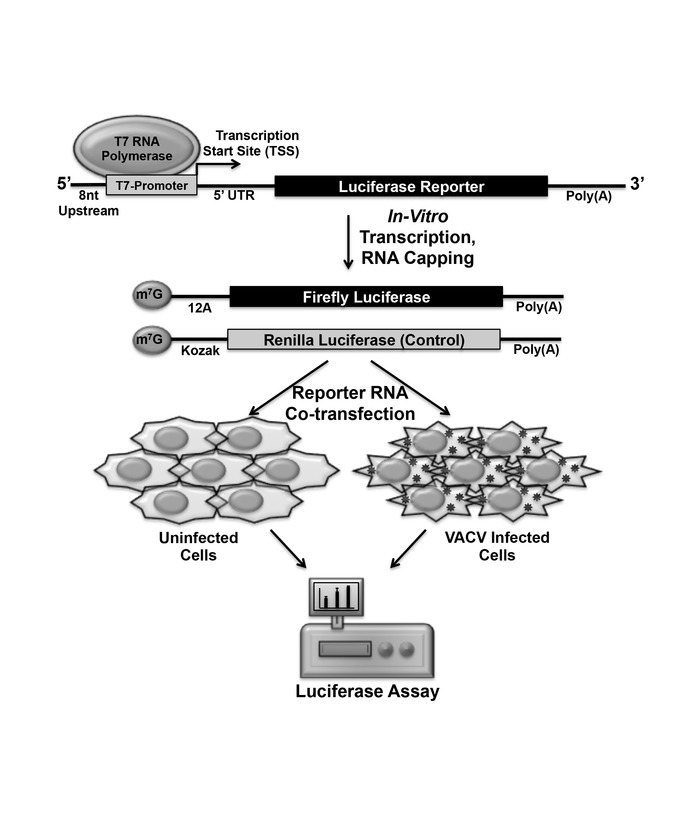
Figure 1: Schematic of the experimental procedure. PCR is used to generate a DNA template with the desired elements. mRNA encoding a luciferase reporter gene is synthesized in vitro using a T7 RNA polymerase-based system. A Firefly luciferase (Fluc) mRNA is co-transfected with a Renilla luciferase (Rluc) mRNA into uninfected or VACV-infected cells. Luciferase activities are measured using a luminometer with dual luciferase capability. Please click here to view a larger version of this figure.
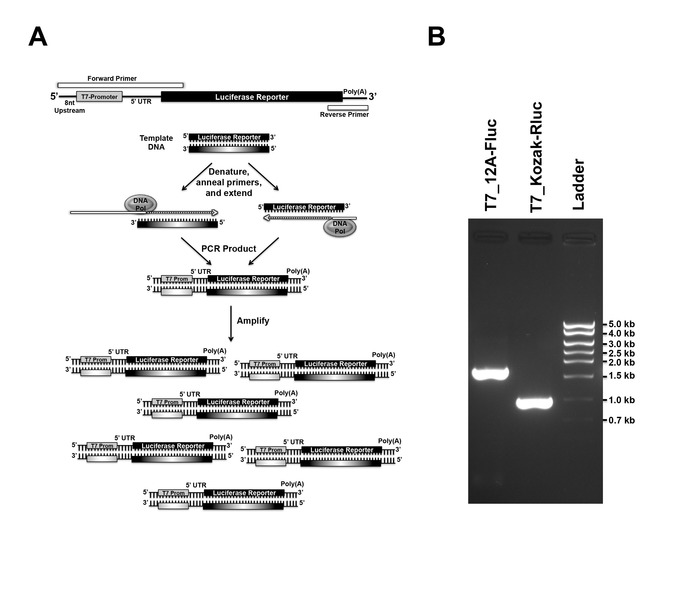
Figure 2: Primer design and PCR-based DNA amplification. (A) The forward primer is synthesized to include an 8nt random sequence, T7 promoter followed by a desired 5'-UTR and part of the 5' end of the luciferase reporter gene, while the reverse primer includes a T-tract to generate poly(A) tail and the 3' end of the luciferase reporter gene. By overhang extension PCR using a plasmid template containing a luciferase gene, a DNA template is generated. (B) DNA band of the desired size from PCR reaction was detected using 1% agarose TAE gel electrophoresis. Please click here to view a larger version of this figure.
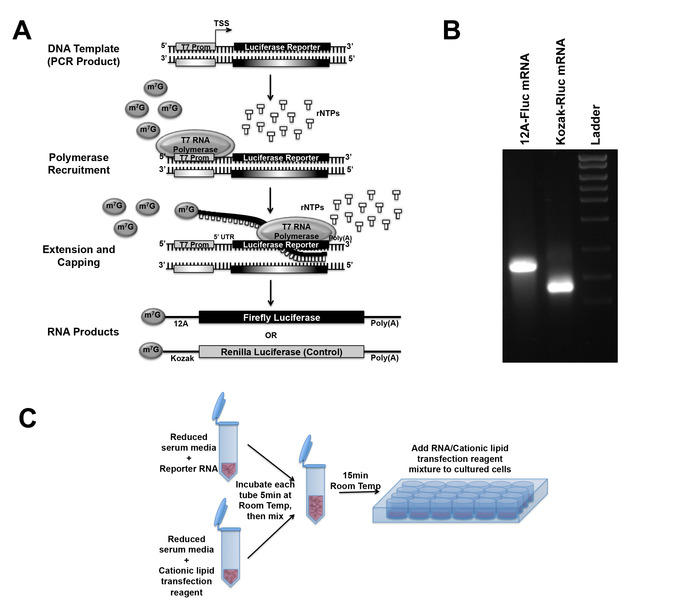
Figure 3: mRNA synthesis and transfection. (A) Schematic of in vitro transcription. DNA amplified by PCR containing the luciferase gene downstream from the 5'-UTR of interest and the T7 promoter is used as the template. The T7 RNA polymerase is recruited to the promoter and adds ribonucleotides, shown in white, from 5' to 3' direction. Once mRNA is 25-30 nt long, m7G cap is added using an anti-reverse cap analog, ARCA. (B) RNA bands from in vitro transcription ware detected using 1.5% agarose TBE gel electrophoresis. (C) Schematic demonstrating the transfection of reporter mRNA into cells. Medium containing either the reporter mRNA or cationic lipid transfection reagent in separate tubes is allowed to equilibrate at room temperature for 5 min. The solutions are then mixed followed by incubation at room temperature for 15 min after which the RNA/transfection reagent mixture is added into cells in culture plates. Please click here to view a larger version of this figure.
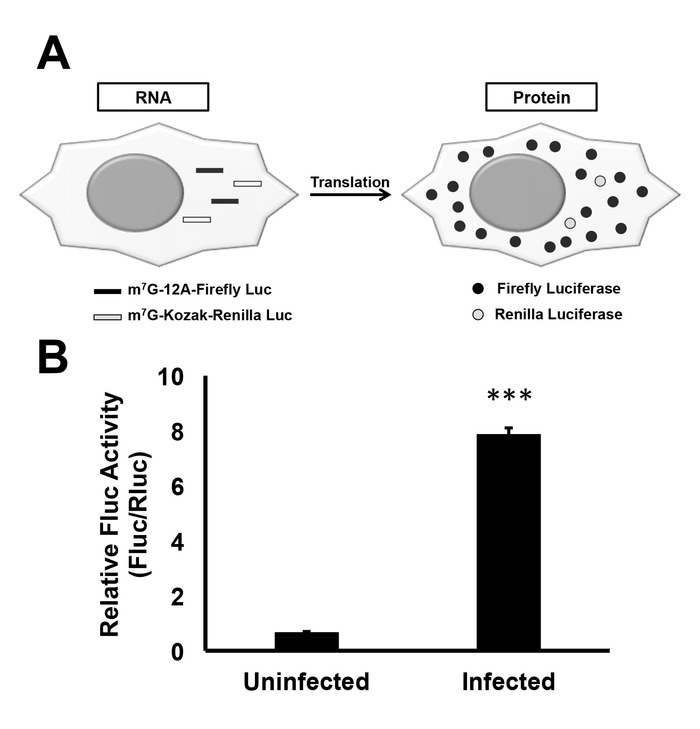
Figure 4: Increased translational efficiency of mRNA containing a 5'-poly(A) leader. (A) Fluc mRNA containing a poly(A) leader in the 5'-UTR and Rluc mRNA with the Kozak consensus sequence in the 5'-UTR are co-transfected into cells. (B) Fluc mRNA with 5'-poly(A) leader was transfected in uninfected and VACV-infected cells along with Rluc mRNA. 5 hpi, luciferase activity was measured using a luminometer. Rluc normalized Fluc activity is represented in uninfected and VACV-infected cells. Error bars indicate the standard deviations (SD) of at least three repeats. Student's t-test was used to determine P-values; ***P-value < 0.001. Please click here to view a larger version of this figure.
| Elements | Sequence |
| T7 Promoter | TAATACGACTCACTATAGGG |
| Poly(A) leader | AAAAAAAAAAAA, ranging from 3 to 51 As |
| Kozak sequence | GCCACC |
| Poly(A) tail | AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA…… |
Table 1: Sequences used in the method – the table contains the sequences of the T7 promoter, poly(A) leader, Kozak sequence, poly(A) tail.
| Components | Volume |
| DNase free water: | 38 µL |
| 2X High-fidelity DNA polymerase Master mix: | 50 µL |
| Forward Primer (10 µM): | 4 µl |
| Reverse Primer (10 µM): | 4 µl |
| Luciferase DNA Template (1-10 ng/µL): | 4 µl |
| Total: | 100 µL |
| Source for Fluc DNA template is pGL3-Fluc plasmid | |
| Source for Rluc DNA template is pRL-Rluc plasmid |
Table 2: PCR reaction – the order and the volume of components added in the PCR reaction.
| Step | Temperature | Time | Cycle |
| Initial denaturation | 95 °C | 2 min | (1x Cycle) |
| Denaturation | 95 °C: | 15 s | |
| Annealing | X °C: | 30 s | (25x Cycle) |
| Extension | 72 °C: | T min | |
| Final Extension | 72 °C: | 7 min | (1x Cycle) |
| Hold | 4 °C: | ∞ |
Table 3: PCR Program – the steps for PCR program along with temperature, time and cycle.
| Components | Volume | |
| DNase-RNase free water: | up to 20 µL | |
| NTP Buffer Mix (20 mM of each rNTP): | 2 µL | |
| Cap Analog (40 mM): | 4 µL | |
| Template PCR Product (400 ng)*: | X µL | (PCR Product Concentration dependent) |
| T7-RNA polymerase Mix: | 2 µL | |
| Total: | 20 µL | |
| *T7_12A-Fluc and T7_Kozak-Rluc template used to synthesize 12A-Fluc and Kozak-Rluc mRNA, respectively. |
Table 4: In vitro transcription reaction – the order and the volume of components added for in vitro transcription reaction.
| Steps | Volume/Time |
| Inject Luciferase Assay Substrate (Fluc): | 30 µL |
| Wait / Incubation time: | 2 s |
| Luminescence Measurement (Fluc): | 10 s |
| Stop & Glo Substrate (Rluc): | 30 µL |
| Wait / Incubation time: | 2 s |
| Luminescence Measurement (Rluc): | 10 s |
Table 5: Luciferase Measurement Settings – the steps for luciferase measurement with the recommended volume or time.
| Primers | Sequence |
| T7-12A-Fluc Forward | ATCGACGATAATACGACTCACTATAGGGaaaaaaaaaaaa ATGGAAGACGCCAAAAACATAAAG |
| T7-12A-Fluc Reverse | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTACACGGCGATCTTTCCGC |
| T7-Kozak-Rluc Forward | ATCGACGATAATACGACTCACTATAGGGatcgtagccacc ATGACTTCGAAAGTTTATGATC |
| T7-Kozak-Rluc Reverse | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATTGTTCATTTTT GAGAACTCGCTC |
Table 6: Primers – the primers used in this method with complete sequences.
Supplementary Figure 1: Analysis of raw data – steps for analyzing raw data to get normalized data. Please click here to download this file.
Discussion
All four-core steps are critical to the success of the in vitro transcribed RNA-based luciferase reporter assay. Special attention should be given to primer design, especially for the T7 promoter sequence. T7 RNA polymerase starts transcription from the underlined first G (GGG-5'-UTR-AUG-) in T7 promoter added before the 5'-UTR sequence. Although the transcription start site (TSS) starts from the first G at the 5' end, decreasing the number of G's less than three in T7 promoter region decreased the RNA yield/output from in vitro transcription. During the experiment, we observed that gel purified DNA product was not the best for in vitro transcription as both yield and quality of RNA were lower. We only ran 5-10% of the PCR reaction in 1% agarose gel electrophoresis to determine the size and purified the rest 90-95%, using a PCR purification kit, to be used for in vitro transcription. In the case of non-specific amplification from PCR, cutting the desired sized band from gel and using gel-purified DNA fragment is recommended. As the yield might be low, we suggest increasing the reaction volume for the in vitro transcription reaction. Similar to other transfection-based methods, DNA/RNA may stimulate DNA/RNA sensing pathways that may globally or selectively suppress translation. Therefore, data should be interpreted with cautions, although we did not experience problems potentially caused by this issue in our experiments.
The proposed method is suitable for use in different model systems with some modifications like the method of mRNA delivery, internal control to be used, a suitable time for translation, sample preparation and analysis of data. The main limitation of this method is that it is a reporter assay to quickly test translation regulation by cis elements that do not completely reflect physiological conditions. Therefore, this method should be corroborated by other complementary experiments, if possible.
Compared to DNA modification, the roles of RNA modifications are less well understood. However, with the discovery of enzymes that write, read and erase RNA modifications30,31,32,33,34,35, it is now possible to study the influence of RNA modification in gene expression30,31,32,33,34,35. The in vitro transcribed RNA-based luciferase reporter assay may be modified to incorporate different RNA modifications and used to test their effects on RNA translation. For example, this method can incorporate different cap analogs that have various modifications30,31. Additionally, supplementing an internal RNA modifying enzyme during or after in vitro transcription can possibly incorporate internal RNA modification. Addition of a modification to cap 0, cap 1, and an internal RNA modification will provide a tool to study the roles of these RNA modifications in translation.
The in vitro transcribed RNA-based luciferase reporter assay has great potential and broad application in understanding basic biology about RNA translation. Different mechanisms for the initiation of translation, including cap-dependent initiation, cap-independent initiation, re-initiation and internal initiation such as IRES can be studied using this method. On top of these advantages, this assay can be employed to test translation regulation by cis-elements at 5'-UTR and 3'-UTR in an mRNA. The described protocol uses the PCR product, which provides the advantage to avoid lengthy cloning and quickly examine the effects of RNA elements on translation. To minimize potential errors during PCR, high fidelity polymerase and low PCR cycle number should be used. Alternatively, if a template is used frequently, the desired 5-'UTR and luciferase ORF can be cloned into a plasmid as the template of in vitro transcription. Together, the protocol consolidates transcription and mRNA capping in a single reaction and utilizes conventional transfection and analysis that make in vitro transcribed RNA-based luciferase reporter assay a user-friendly, quick, and straightforward method to study mechanisms of mRNA translation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The project was funded by the National Institutes of Health (AI128406 www.nih.gov) to ZY and in part by Johnson Cancer Research Center (http://cancer.k-state.edu) in the form of Graduate Student Summer Stipend to PD.
Materials
| 2X-Q5 Master mix | New England Biolabs | M0492 | High-Fidelity DNA Polymerase used in PCR |
| 3´-O-Me-m7G(5')ppp(5')G RNA Cap Structure Analog | New England Biolabs | S1411L | Anti reverse Cap analog or ARCA |
| Corning 96 Well Half-Area white flat bottom polystyrene microplate | Corning | 3693 | Opaque walled 96 well white plate with solid bottom |
| Dual-Luciferase Reporter Assay System | Promega | E1960 | Dual-Luciferase Assay Kit (DLAK) |
| E.Z.N.A. Cycle Pure Kit | OMEGA BIO-TEK | D6492 | PCR purification kit |
| GloMax Navigator Microplate Luminometer | Promega | GM2010 | Referred as multimode plate reader luminometer |
| HiScribe T7 Quick High Yield RNA synthesis Kit | New England Biolabs | E2050S | In-Vitro transcription kit |
| Lipofectamine 2000 | Thermo Fisher Scientific | 11668019 | Cationic lipid transfection reagent |
| NanoDrop2000 | Thermo Fisher Scientific | ND-2000 | Used to measure DNA and RNA concentration |
| Opti-MEM | Thermo Fisher Scientific | 31985070 | Reduced serum media |
| Purelink RNA Mini Kit | Thermo Fisher Scientific | 12183018A | RNA purification kit |
| Vaccinia Capping System | New England Biolabs | M2080 | Capping system |
References
- Crick, F. H. On protein synthesis. Symposia of the Society for Experimental Biology. 12, 138-163 (1958).
- Crick, F. Central Dogma of Molecular Biology. Nature. 227, 561-563 (1970).
- Sonenberg, N., Hinnebusch, A. G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 136, 731-745 (2009).
- Spriggs, K. A., Bushell, M., Willis, A. E. Translational Regulation of Gene Expression during Conditions of Cell Stress. Molecular Cell. 40, 228-237 (2010).
- Shchelkunov, S. N., Marennikova, S. S., Moyer, R. W. . Orthopoxviruses Pathogenic for Humans. , (2005).
- Gale, M., Tan, S. L., Katze, M. G. Translational Control of Viral Gene Expression in Eukaryotes. Microbiology and Molecular Biology Reviews. 64, 239-280 (2000).
- Walsh, D., Mathews, M. B., Mohr, I. Tinkering with Translation: Protein Synthesis in Virus-Infected Cells. Cold Spring Harbor Perspectives in Biology. 5, a012351 (2013).
- Cao, S., Dhungel, P., Yang, Z. Going against the Tide: Selective Cellular Protein Synthesis during Virally Induced Host Shutoff. Journal of Virology. 91, e00071-e00017 (2017).
- Pelletier, J., Kaplan, G., Racaniello, V. R., Sonenberg, N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5’ noncoding region. Molecular and Cellular Biology. 8, 1103-1112 (1988).
- Pelletier, J., Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 334, 320-325 (1988).
- Guo, L., Allen, E., Miller, W. A. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 6, 1808-1820 (2000).
- Simon, A. E., Miller, W. A. 3′ Cap-Independent Translation Enhancers of Plant Viruses. Annual Review of Microbiology. 67, 21-42 (2013).
- Marom, L., et al. Diverse poly(A) binding proteins mediate internal translational initiation by a plant viral IRES. RNA Biology. 6, 446-454 (2009).
- Liu, B., Qian, S. B. Translational reprogramming in cellular stress response. Wiley Interdisciplinary Review. RNA. 5, 301-305 (2014).
- Ahn, B. Y., Moss, B. Capped poly(A) leaders of variable lengths at the 5’ ends of vaccinia virus late mRNAs. Journal of Virology. 63, 226-232 (1989).
- Ahn, B. Y., Jones, E. V., Moss, B. Identification of the vaccinia virus gene encoding an 18-kilodalton subunit of RNA polymerase and demonstration of a 5’ poly(A) leader on its early transcript. Journal of Virology. 64, 3019-3024 (1990).
- Schwer, B., Visca, P., Vos, J. C., Stunnenberg, H. G. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell. 50, 163-169 (1987).
- Yang, Z., Martens, C. A., Bruno, D. P., Porcella, S. F., Moss, B. Pervasive initiation and 3′ end formation of poxvirus post-replicative RNAs. Journal of Biological Chemistry. 287, 31050-31060 (2012).
- Dhungel, P., Cao, S., Yang, Z. The 5’-poly(A) leader of poxvirus mRNA confers a translational advantage that can be achieved in cells with impaired cap-dependent translation. PLOS Pathogens. 13, e1006602 (2017).
- Jha, S., et al. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature. 546, 651-655 (2017).
- Dai, A., et al. Ribosome Profiling Reveals Translational Upregulation of Cellular Oxidative Phosphorylation mRNAs during Vaccinia Virus-Induced Host Shutoff. Journal of Virology. 91, e01858-e01816 (2017).
- De Silva, F. S., Moss, B. Origin-independent plasmid replication occurs in vaccinia virus cytoplasmic factories and requires all five known poxvirus replication factors. Journal of Virology. 2, 23 (2005).
- Yang, Z., Bruno, D. P., Martens, C. A., Porcella, S. F., Moss, B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proceedings of the National Academy of Sciences. 107, 11513-11518 (2010).
- Yang, Z., Bruno, D. P., Martens, C. A., Porcella, S. F., Moss, B. Genome-Wide Analysis of the 5′ and 3′ Ends of Vaccinia Virus Early mRNAs Delineates Regulatory Sequences of Annotated and Anomalous Transcripts. Journal of Virology. 85, 5897-5909 (2011).
- Lorenz, T. C. Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies. Journal of Visualized Experiments. , (2012).
- Dieffenbach, C. W., Lowe, T. M., Dveksler, G. S. General concepts for PCR primer design. PCR Methods and Applications. 3, S30-S37 (1993).
- Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. . PCR Protocols: A Guide to Methods and Applications. , (2012).
- Baklanov, M. M., Golikova, L. N., Malygin, E. G. Effect on DNA Transcription of Nucleotide Sequences Upstream to T7 Promoter. Nucleic Acids Research. 24, 3659-3660 (1996).
- Thornton, J. A. Splicing by Overlap Extension PCR to Obtain Hybrid DNA Products. The Genetic Manipulation of Staphylococci: Methods and Protocols. , 43-49 (2017).
- Akichika, S., et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase. Science. , (2018).
- Sun, H., Zhang, M., Li, K., Bai, D., Yi, C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Research. 1, (2018).
- Boulias, K., et al. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. bioRxiv. , 485862 (2018).
- Dominissini, D., Rechavi, G. N4-acetylation of Cytidine in mRNA by NAT10 Regulates Stability and Translation. Cell. 175, 1725-1727 (2018).
- Wei, J., et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Molecular Cell. 71, 973-985 (2018).
- Fu, Y., Dominissini, D., Rechavi, G., He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nature Reviews Genetics. 15, 293-306 (2014).

