Rapid Characterization of Genetic Parts with Cell-Free Systems
Summary
Well-characterized genetic parts are necessary for the design of novel genetic circuits. Here we describe a cost-effective, high-throughput method for rapidly characterizing genetic parts. Our method reduces cost and time by combining cell-free lysates, linear DNA to avoid cloning, and acoustic liquid handling to increase throughput and reduce reaction volumes.
Abstract
Characterizing and cataloging genetic parts are critical to the design of useful genetic circuits. Having well-characterized parts allows for the fine-tuning of genetic circuits, such that their function results in predictable outcomes. With the growth of synthetic biology as a field, there has been an explosion of genetic circuits that have been implemented in microbes to execute functions pertaining to sensing, metabolic alteration, and cellular computing. Here, we show a rapid and cost-effective method for characterizing genetic parts. Our method utilizes cell-free lysate, prepared in-house as a medium to evaluate parts via the expression of a reporter protein. Template DNA is prepared by PCR amplification using inexpensive primers to add variant parts to the reporter gene, and the template is added to the reaction as linear DNA without cloning. Parts that can be added in this way include promoters, operators, ribosome binding sites, insulators, and terminators. This approach, combined with the incorporation of an acoustic liquid handler and 384-well plates, allows the user to carry out high-throughput evaluations of genetic parts in a single day. By comparison, cell-based screening approaches require time-consuming cloning and have longer testing times due to overnight culture and culture density normalization steps. Further, working in cell-free lysate allows the user to exact tighter control over the expression conditions through the addition of exogenous components and DNA at precise concentrations. Results obtained from cell-free screening can be used directly in applications of cell-free systems or, in some cases, as a way to predict function in whole cells.
Introduction
A core effort of synthetic biology is to develop genetic tool kits containing well-characterized parts, which can be used to construct genetic circuits1 that carry out useful functions when deployed in microbes or cell-free lysates. Areas in which such genetic circuits have gained traction are sensing2,3,4, human performance5,6, biofuels7,8, materials production9,10, and cellular computing11. Registries of standardized genetic parts have been established12 to catalog new and existing parts into categories such as promoters, operators, coding sequences, and terminators, to name just a few. Efforts such as the iGEM (international Genetically Engineered Machines) competition13 have been instrumental in characterizing and cataloging these genetic parts. Many methods have been developed to facilitate the rapid assembly of these parts into useful genetic circuits14,15. Software has even been developed to automate the composition of well-characterized parts into circuits that achieve a desired function16. However, the assembly of useful genetic circuits with predictable functions rests on the presumption that the genetic tool kits contain well-characterized genetic parts. Due to the necessity of these tool kits toward the advancement of synthetic biology, numerous efforts to better catalog circuits and parts with appropriate characterization data have been described17,18,19,20,21.
One approach to characterizing genetic components makes use of cell-free protein synthesis (CFPS) systems, which reconstitute cellular functions such as transcription and translation ex vivo22. Several studies have demonstrated the potential of CFPS for prototyping genetic components23,24,25,26,27,28,29,30,31,32 whether for direct applications in cell-free systems or to predict the function of genetic constructs in cells, such as the relative activity of parts within a library29, metabolic pathway optimization27, and cellular burden30. Advantages to prototyping in CFPS versus cells highlighted by these studies include avoiding time-consuming cloning, precise control over the concentration of DNA and other reaction components, and the ability to easily mix and match multiple DNA constructs. The advantage of avoiding cloning is especially apparent when using linear DNA templates, which enables new constructs to be assembled by in vitro methods that take hours instead of days33. The ability to manipulate the concentration of DNA constructs and other components simply by pipetting makes the approach even more attractive by enabling high-throughput experimentation powered by liquid handling robots34,35. While successes using CFPS for prototyping have been reported, it is important to note that it remains to be seen under what contexts CFPS results can reliably predict functionality in cells.
Here, we present a method for CFPS prototyping that emphasizes the advantages in speed, throughput, and cost compared to traditional cell-based approaches. The approach is derived from our previous work where we used CFPS to rapidly characterize a library of T7 promoter variants regulated by the transcription factor TetR32, significantly expanding on the small handful of regulated T7 promoter variants that were available in the literature at the time36,37. Others have, since then, further expanded the range of such promoters38. In our method, genetic construct assembly is accelerated by using PCR to amplify template DNA via primers that add variant genetic parts to a reporter gene. Acoustic liquid handling in 384-well plates is used to increase throughput and decrease the volume of materials required. Previous work has demonstrated successful use of acoustic liquid handling at significantly lower volumes39,40 with variability comparable to manual pipetting of larger volumes41. In addition to the method, we provide troubleshooting information and an assessment of potential cost and time savings. Note that while we include a protocol for producing cell-free lysates based on Sun et al.42 here, numerous other commercial kits and protocols43,44 should also work. Similarly, while we demonstrate the method for the characterization of promoter variants32, other parts can be interchanged by PCR amplification, such as riboregulators, Ribosome Binding Sites (RBSs), insulators, protein tags, and terminators. We hope that this methodology can help the synthetic biology community continue to grow the number of characterized parts for the assembly of predictable genetic circuits with useful function.
Protocol
1. Preparation of cell extract
- Preparation of media
- For 2xYT media: Add 16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl to 900 mL of deionized water and adjust the pH to 7.0 with 5 M NaOH. Raise the solution volume to 1 L using deionized water and autoclave or filter sterilize. Alternatively, purchase 2xYT media.
- For S30B buffer: Prepare a solution of 14 mM Mg-glutamate, 60 mM K-glutamate, and 5 mM Tris in 2 L of deionized water. Use 2 M Tris to adjust the pH to 8.2, autoclave, and store at 4 °C. Complete the solution by adding dithiothreitol (DTT) to a final concentration of 1 mM just before use.
- Preparation of cells
- Streak Escherichia coli BL21(DE3) Rosetta2 cells or other cell line of choice (see Cole et al.44 for a recent comprehensive review) onto an LB (Lysogeny Broth) agar plate and incubate at 37 °C for 10-14 h.
- Use a single E. coli colony to inoculate 3 mL of 2xYT medium in a 10 mL culture tube. Incubate this tube at 37 °C with shaking at 250 rpm for 8 h.
- Use 50 µL from the 3 mL culture to inoculate 50 mL of 2xYT medium in a 500 mL flask. Incubate this flask at 37 °C with shaking at 250 rpm for 8 h.
- Use 7.5 mL from the 50 mL culture to inoculate each of four 4 L baffled flasks containing 0.75 L of 2xYT medium. Incubate these flasks at 37 °C with shaking at 220 rpm until they have reached an optical density at 600 nm of 2 to 4, after approximately 3-4 h.
- Harvest the cells from each flask by transferring them to 1 L containers and centrifuging at 5,000 x g for 12 min at 4 °C. Discard the supernatant by decanting into a waste container.
- Wash each cell pellet with 150 mL of ice-cold S30B buffer by completely resuspending them using a pipette to disrupt the cell mass, and then collect the cells again by centrifugation at 5,000 x g for 12 min. Discard the supernatant.
- Wash each cell pellet again in 40 mL of ice-cold S30B buffer by completely resuspending them and disrupting the cell mass using a pipette. Transfer the cells to pre-weighed 50 mL conical tubes and collect the cells again by centrifugation at 2,000 x g for 8 min at 4 °C. Discard the supernatant by decanting.
- Weigh the wet cell pellets. Flash-freeze the cell pellets by placing the tubes directly into liquid nitrogen and store at -80 °C.
- Cell lysis
- Thaw the cell pellets on ice.
- Resuspend each cell pellet in 1.4 mL of S30B buffer per 1 g of the cell pellet by vortexing.
- Lyse the cells by French pressure cell at 640 psi at 4 °C. Collect the lysate in microcentrifuge tubes on ice and add 3 µL of 1 M DTT per 1 mL of lysate immediately after lysis.
NOTE: It is best to tap the French press release valve with a small metal rod to maintain even pressure and avoid sudden drops in pressure. - Clear the lysate by centrifugation at 30,000 x g for 30 min at 4 °C and discard the pellet after pipetting the supernatant to a new ice-cold microcentrifuge tube, taking care not to disrupt the pellet.
- Centrifuge the supernatant a second time at 30,000 x g for 30 min at 4 °C. Pipette the resulting supernatant into an ice-cold microcentrifuge tube. Discard the pellet.
- Incubate the supernatant in a 37 °C water bath for 1 h.
- Clear the supernatant by centrifugation at 15,000 x g for 15 min at 4 °C and transfer the resulting supernatant to an ice-cold microcentrifuge tube, taking care not to disrupt the pellet.
- Centrifuge the supernatant a second time at 15,000 x g for 15 min at 4 °C and transfer the resulting supernatant to an ice-cold microcentrifuge tube, taking care not to disrupt any remaining pellet.
- Distribute the supernatant in 100 µL aliquots into 1.5 mL microcentrifuge tubes and flash freeze them by placing directly into liquid nitrogen. Store the supernatant at -80 °C.
2. Linear template preparation
- Primer design
- Choose a core sequence as the PCR template. Include at a minimum a reporter sequence, such as sfGFP (superfolder Green Fluorescent Protein), LacZ, or Spinach aptamer. Include other parts that will be fixed across screened variants, such as terminators, promoters or RBSs, as appropriate for the design.
NOTE: Inclusion of a terminator is not always required for expression from linear DNA in cell-free systems. - For the forward primers, choose a minimum of 20 bp matching the 5ʹ end of the core sequence as the 3ʹ end of the primer. If adding parts to the 5′ end of the construct, design the remainder of the 5ʹ end of the primer to add the genetic parts of interest to the core sequence via PCR amplification (Figure 1A and Figure 2).
NOTE: Since primers above ~60 bp frequently increase dramatically in cost, multiple overlapping primers can be designed to add longer sequences or multiple parts. While multiple primers can be used in a single PCR reaction, performing multiple rounds of PCR is recommended. - For the reverse primers, choose a minimum of 20 bp to match the 3′ end of the core sequence as the 3ʹ end of the primer. If adding parts to the 3′ end of the construct, design the remainder of the 5ʹ end of the primer to add the genetic parts of interest to the core sequence via PCR amplification (Figure 1A and Figure 2). Ensure that the reverse primer's annealing temperature is within 5 °C of the annealing temperature of the entire forward primer.
- Choose a core sequence as the PCR template. Include at a minimum a reporter sequence, such as sfGFP (superfolder Green Fluorescent Protein), LacZ, or Spinach aptamer. Include other parts that will be fixed across screened variants, such as terminators, promoters or RBSs, as appropriate for the design.
- Linear template amplification
- Determine the number of PCR reactions to be performed based on the number of core sequences and calculate the amount of each component required using Table 1.
- Prepare the master mix according to Table 1 and store it on ice. Aliquot 30 or 40 µL (see Table 1) of the master mix into the determined number of PCR tubes and add 10 µL of each variable primer (i.e., primers encoding a part change, see Table 1) at 5 µM to appropriately labeled PCR tubes.
- Place the PCR tubes into the thermocycler and run the following PCR program: 98 °C for 3 min; 30 cycles of 98 °C for 15 s, XX °C for 20 s, 72 °C for YY min; final extension at 72 °C for 10 min. Then, hold the reaction at 4 °C.
Where, XX represents the annealing temperature for the primer with the lower annealing temperature and YY represents the extension time calculated for the length of the amplicon based on the manufacturer's recommendations for the high-fidelity polymerase used. Optimize these conditions as needed for different primers and/or templates. - (Optional) Add 1 µL of DpnI restriction enzyme to digest the original template. Incubate the reaction at 37 °C for 1 h. Perform this step only if the original template is plasmid DNA.
- Analyze 5 µL of each PCR product by gel electrophoresis. Separate the product using a 1% agarose gel at 180 V for 20 min. Check for the correct band size, which will vary with the chosen core sequence and the length of the parts added.
- Purify the linear template using a commercial PCR purification kit or by the preferred PCR cleanup method. If multiple bands were present by gel electrophoresis analysis, either optimize the PCR conditions or purify the correct molecular weight bands using a commercial gel extraction kit as per the manufacturer's recommendation.
- Quantify each DNA template using a spectrophotometer. Assess the DNA template quality by checking that the 260 nm/280 nm ratio is approximately 1.8.
- (Optional) Again separate a portion of the DNA template using a 1% agarose gel at 180 V for 20 min and ensure that any unwanted bands were removed during template purification.
- Use purified DNA templates immediately or store at -20 °C.
3. Purified protein preparation
- Protein expression
- For each protein to be expressed, assemble an appropriate expression construct. Codon-optimize the gene for expression in E. coli. Insert the gene into a pET-22b expression vector or other appropriate expression vector via the preferred plasmid assembly method. Transform the expression plasmid into BL21(DE3) Rosetta2 expression cells or other appropriate cell line.
- For each protein, use a single colony to inoculate 3 mL of LB medium in a 10 mL culture tube. Incubate these tubes at 37 °C with shaking at 250 rpm overnight.
- Inoculate a 2 L flask containing 750 mL of LB medium with 1 mL of the overnight culture. Incubate these flasks at 37 °C with shaking at 250 rpm until they reach an OD600 of 0.6-1.0.
- Induce protein expression by adding 0.75 mL of 1 M isopropyl β-D-1-thiogalactopyranoside (IPTG) in water to each flask and continue to incubate these flasks at 37 °C with shaking at 250 rpm for 4 h.
- Harvest the cells from each flask, using a 1 L centrifuge bottle, by centrifugation at 5,000 x g for 12 min. Discard the supernatant.
- Transfer the pellets to a 50 mL conical tube and weigh each pellet. Flash-freeze the cells in liquid nitrogen and store them at -80 °C or proceed to step 3.2.
NOTE: 2-5 g of cells per 0.75 mL are expected to result from this step.
- Protein purification by nickel affinity column chromatography
- Prepare the lysis buffer by combining 50 mM Tris-Cl, 500 mM NaCl, and 5 mM imidazole. Adjust to pH 8.0.
- Prepare the wash buffer by combining 50 mM Tris-Cl, 500 mM NaCl, and 25 mM imidazole. Adjust to pH 8.0.
- Prepare the elution buffer by combining 50 mM Tris-Cl, 500 mM NaCl, and 250 mM imidazole. Adjust to pH 8.0.
- Prepare the dialysis buffer by combining 50 mM NaHPO4, 100 mM NaCl, and 2% DMSO. Adjust to pH 7.5.
- Thaw the cell pellets by placing the tubes in room temperature water. Add 5 mL of the lysis buffer per 1 g of the cell pellet and resuspend by vortexing.
- Lyse the cells by sonication. Separate the cell homogenate so that there is no more than 30 mL per 50 mL conical tube and keep each tube on ice. Lyse the cells using a sonicator with a 0.16 cm diameter probe in 15 s rounds with 30 s breaks, 10 times.
NOTE: Avoid foaming, as this denatures the protein. The formation of foam can be avoided by keeping the sonicator tip at least 2/3 submerged in the lysate while it is operational. Other methods of cell lysis besides sonication are also possible44. - Clear the lysate by centrifugation at 15,000 x g for 30 min at 4 °C and place the supernatant into a new 50 mL conical tube.
- Add 1 mL of nickel-nitrilotriacetic acid (Ni-NTA) resin for each 5 mL of supernatant. Divide the cell lysate/Ni-NTA slurry so that there is no more than 36 mL per 50 mL conical tube. Incubate at 4 °C on a tube rotator at 10 rpm for 1 h.
- Load the resin by decanting the cell lysate/Ni-NTA slurry into a 2 mL bed volume chromatography column and collect the eluant if needed for further analysis, otherwise, discard. Wash the resin with 10 resin bed volumes of wash buffer.
- Collect the protein by adding three resin bed volumes of elution buffer to the column and concentrate the volume to 1.5 mL using a centrifugal concentrator with the appropriate molecular weight cut-off membrane for each protein.
- Dialyze the protein against 2 L of dialysis buffer at 4 °C for 1 h. Dialyze the protein again against 2 L of dialysis buffer overnight at 4 °C.
- Quantify the protein using its molar extinction coefficient and absorbance at 280 nm. Analyze the protein for purity by separating it using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Store the protein at -80 °C.
4. Cell-free protein synthesis
- Preparation of CFPS reaction mixture
- Prepare the Supplement Mix by following the Amino Acid Solution Preparation, Energy Solution Preparation, and Buffer Preparation steps in Sun et al.42. Store separately or combined at -80 °C in aliquots. Ensure that final concentrations match those described in Sun et al.42 in the Experimental Execution of a TX-TL Reaction section.
- Prepare an additive to protect linear DNA from degradation. If using GamS33,45, prepare via steps in section 3 above, or obtain from a commercial vendor; for other approaches, check the corresponding literature46,47,48. Alternatively, use a CFPS system that does not require additives49.
- Prepare T7 polymerase, repressor proteins, and other additives using the steps in section 3 above or obtain from a commercial vendor.
- Determine the number of CFPS reactions to be performed and calculate the amount of each component required using Table 2. Modify the concentrations of the components, including adding or removing components as needed, and adjust the amount of water such that the final volume of each reaction mixture is always 10 µL. Similarly, modify the master mix to facilitate dispensing of other components by acoustic liquid handling as desired (see the Discussion section).
- Thaw all the components on ice and prepare a master mix by mixing each component as calculated above. Mix all the components thoroughly by pipette. Pay careful attention to avoid precipitation, especially for the amino acid mixture. Keep the master mix on ice.
- Chill a 384-well plate on ice and distribute the master mix in 9 µL aliquots into each well.
NOTE: It is possible to distribute these components by acoustic liquid handling, though care should be taken to ensure proper dispensing (see the Discussion section for troubleshooting).
- Distribution of additional components by acoustic liquid handling
- Calculate the amount of repressor protein (and other optional components) required for all the CFPS reactions.
- Thaw the repressor protein on ice and distribute it into an acoustic liquid handling source plate or other appropriate plate. Ensure that the appropriate amount of dead volume required for the type of source plate used is included.
- Distribute the repressor protein in 1 µL volumes into the appropriate wells via the liquid handler. For more information on distribution troubleshooting, see the Discussion section.
- Standard curves
- Include a serial dilution of the purified reporter (see section 3 for protein purification)41 or appropriate chemical standard50 on the plate to enable comparison of results with other studies and other labs. Choose a range of concentrations appropriate for the reporter used and expected expression range of the experiments.
- Running CFPS reactions
- Pre-warm the plate reader to 30 °C. Set the plate reader to read at settings appropriate for the reporter used in the core sequence without shaking steps.
NOTE: While 30 °C is used here, 29 °C and 37 °C are also commonly used and work well with this protocol. Other temperatures may be preferred for alternative cell-free reaction preparations. For read intervals, 10 min is sufficient to achieve a good resolution for the representative data presented here; however, other resolutions may be better depending on the reporter protein and the particular CFPS recipe. - (Optional) Run a test reaction first to set the appropriate gain or sensitivity setting to capture the change in fluorescence without signal overflow.
- Seal the 384-well plate with an impermeable plastic sealable lid to prevent evaporation. If possible, on the instrument, set a 1 °C vertical temperature gradient to limit condensation on the seal. Place the 384-well plate on the plate holder and begin reading.
- Pre-warm the plate reader to 30 °C. Set the plate reader to read at settings appropriate for the reporter used in the core sequence without shaking steps.
Representative Results
To demonstrate the utility of our methods, we present results that describe the effects of proximity of the tetO sequence to the T7 promoter on the regulation of T7 RNAP-driven expression. The full results and their implications can be found in the work of McManus et al.32. The workflow is described in Figure 1. Fifteen linear templates, varying only in the distance of the T7 promoter relative to the tetO sequence, were prepared by PCR-amplifying the sfGFP reporter with primers designed to add each promoter variant (Figure 2) as described in section 2 of the protocol. CFPS reaction components and reactions were prepared following the protocol. The expression of sfGFP was measured from each template with a titration of 12 different concentrations of the TetR protein, in triplicate, using an acoustic liquid handler. At 36 CFPS reactions per template and 15 templates, a total of 540 reactions for the entire set of T7-tetO combinations were performed. The entire evaluation was carried out on two plates in two plate readers. Analysis of this data showed that the T7 RNAP downregulates T7-driven expression equally up through 13 bp downstream from the start of the T7 transcript (Figure 3). This result has implications for the future design of regulatable T7-driven gene circuits by describing a putative window for an effective repression of T7 by other repressors. Comparison of results from the protocol described here with DNA prepared by traditional cloning revealed a small but statistically significant difference in the degree of TetR repression between formats. We hypothesized that non-specific binding of TetR to the vector DNA could explain the observed difference. Experimental results showed that addition of linear vector DNA to reactions with linear template DNA reduced the difference to non-statistical significance, though it did not rule out contributions from other factors, such as differences in periodicity of the DNA helix for linear vs. circular formats, which, in turn, could affect TetR binding. Depending upon the application, the use of linear template may require additional validation.
We further include representative data on potential issues with accurate dispensing using acoustic liquid handling (Figure 4). A solution of 1x phosphate buffered saline (PBS), pH 7.4 containing 0.25 mM tartrazine dye was used to evaluate two methods of programming an acoustic liquid handler to dispense volumes >1 µL. Following liquid dispensing, the destination plate was sealed and centrifuged at 1,500 x g for 1 min, and the absorbance at 425 nm measured with a plate reader. Representative results of nine experiments are shown and demonstrate more consistent dispensing across the series of eight destination wells when the 5 µL transfer is divided into separate 1 µL dispenses. Based on these observations, it is recommended that transfers >1 µL be broken down into multiple transfers of ≤1 µL. See the Discussion section for more details on troubleshooting this important aspect of the protocol.
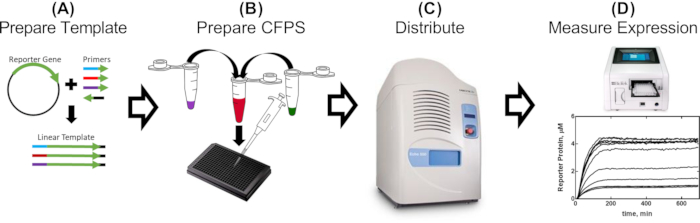
Figure 1: Single-day workflow for the evaluation of promoter parts in cell-free extract. (A) A reporter is PCR-amplified using primers containing genetic parts to be evaluated (2-5 h). (B) The cell-free reaction mix is prepared as detailed in the protocol and distributed into a 384-well plate with the PCR-amplified templates (30 min). (C) Acoustic liquid handling is used to distribute additional components, which can include repressor proteins, effector molecules, and any other conditional effectors (10 min). (D) Reporter protein expression from each reaction is measured in a plate reader (2-16 h, depending on the CFPS recipe and construct). Please click here to view a larger version of this figure.
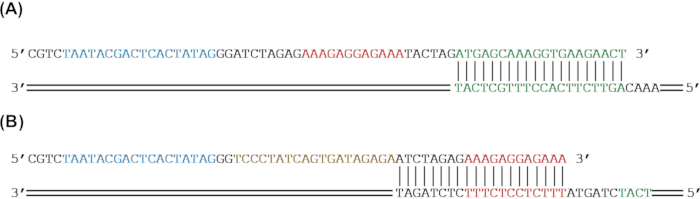
Figure 2: Primer design for adding genetic parts to a reporter gene by PCR amplification. (A) The sfGFP reporter gene (green) will be amplified to add an RBS (red) and a T7 promoter (blue) by PCR. (B) The sfGFP (green) and an RBS (red) will be amplified to add a tetO sequence (gold) and a T7 promoter (blue) by PCR. Please click here to view a larger version of this figure.
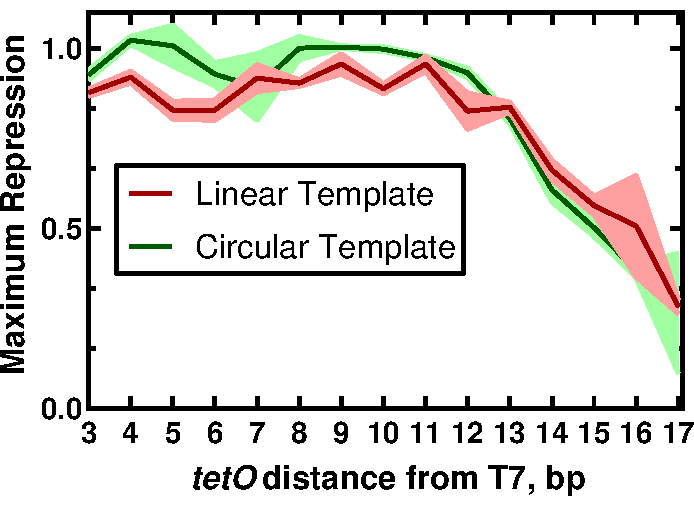
Figure 3: The effect of tetO position on the regulation of a T7-driven expression. Normalized maximum repression values for linear and circular template as a function of tetO position. Traces represent the mean and standard deviations for three replicates. This figure has been modified from McManus et al.32 under a Creative Commons CC-BY license. Please click here to view a larger version of this figure.
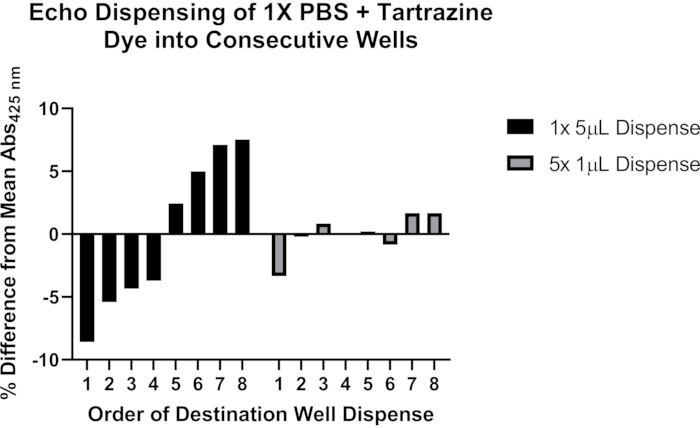
Figure 4: Using tartrazine dye to validate liquid dispensing with an acoustic liquid handler. Black bars indicate dispensing 5 µL of tartrazine solution from a single source well into each of the eight consecutive destination wells of a 384-well plate using a single programming command. Gray bars indicate dispensing 1 µL from a single source well into each of eight consecutive destination wells using a single programming command, and then repeating this step four times for a total of 5 µL dispensed in each destination well. Please click here to view a larger version of this figure.
| Component Name | Volume for 1 reaction (µL) | Volume for 110% of X number of reactions (µL) |
| Q5 PCR Premix | 25 | |
| Water | 4 | |
| Template (1–3 ng/µL) | 1 | |
| (if fixed1) Forward Primer (5 µM) | 0 or 10 | |
| (if fixed1) Reverse Primer (5 µM) | 0 or 10 | |
| Master Mix Total: | 30 or 40 | |
| (if variable1) Forward Primer (5 µM) | 0 or 10 | |
| (if variable1) Reverse Primer (5 µM) | 0 or 10 |
Table 1: Worksheet for the preparation of reagents for PCR reactions. Values in the rightmost column can be filled in by users depending on the intended number of reactions. 1Variable primers contain a specific part to be added in the PCR reaction and can be the forward primer, reverse primer, or both. Fixed primers do not add a part and can be the forward primer or reverse primer but not both.
| Component Name | Volume for 1 reaction (µL) | Volume for 110% of X number of reactions (µL) |
| Cell Extract | 4.2 | |
| Supplement Mix | 3.3 | |
| GamS Protein (207 µM) | 0.15 | |
| Template DNA (20 nM) | 1 | |
| T7 Polymerase (13 mg/mL) | 0.12 | |
| Water | 0.73 (this number may vary) | |
| Master Mix Total: | 9 | |
| Repressor Protein: | 1 |
Table 2: Worksheet for the preparation of reagents for CFPS reactions. Values in the rightmost column can be filled in by users depending on the intended number of reactions.
Supplementary Table. Please click here to download this Table.
Discussion
The protocols described here provide a cost-effective and rapid means to screen genetic parts via the expression of a reporter protein by CFPS. Well-characterized genetic parts are crucial to the design of predictable genetic circuits with useful function. This methodology increases throughput and decreases the time needed to screen new genetic parts by removing the requirement to work in living cells, while retaining functionality that mirrors the cellular environment by retaining the metabolic process of protein expression in the cell lysate. Our protocol can be performed in 1 day after receipt of primers (~2.5-6 h for reaction preparation, 2-16 h for CFPS reaction; Figure 1), compared to at least 3 days for traditional cloning (1 day each for construct assembly and transformation, sequence verification of clones, and culturing of cells for assessment). We further estimate that the cost per construct using linear DNA is roughly one-third of the traditional cloning ($78 vs. $237; Supplementary Table 1) methods. Commercial synthesis services currently quote a minimum of 4 business days depending on size, though they would have similar costs to our method if linear fragments are screened directly in CFPS ($78 vs. $91); we have not verified this approach. The cost to evaluate a part with CFPS is small compared to the generation of the template DNA ($0.05/reaction22 vs. $78 per template), though it should be noted that the startup costs for bulk reagents and lysis equipment is at least several thousand dollars. The use of an acoustic liquid handler only marginally improves costs by enabling smaller volumes down to 0.5 µL40; the more significant advantage is the reduction of time to prepare reactions (~10 min vs. up to 1 h, depending on the number of reactions), especially when preparing a large number of reactions raises concerns of prepared reaction sitting for extended times before incubation.
While rapid and cost-effective, the limitations on when CFPS prototyping adequately predicts in vivo function remain to be seen. For example, any cross-reactivity with genomic DNA will not be detected due to removal of the host genome during the production of the CFPS system. Also, component concentrations can be 1-2 orders of magnitude lower in CFPS than in cells51, which is likely to affect the behavior of some parts as a result of different macromolecular crowding conditions. Further, the ability of linear DNA to predict in vivo function may be limited, for example, when DNA secondary structure plays an important role. A final limitation is that constructs are not sequence-verified before testing for functions. There may be cases where the part characterized is not actually aligned with the intended theoretical sequence. All of these limitations can be mitigated by validating a subset of the parts screened by this method in the intended in vivo application.
We originally developed this methodology to investigate the effects of changing the operator position on hybrid T7-tetO promoters32. We have presented the protocols here in a more generic format, such that they can be applied to promoters, operators, ribosome binding sequences, insulators, and terminators. These genetic parts can be added to the 5ʹ or 3ʹ end of the reporter gene by PCR using primers for each design, obviating the need for synthesis or cloning of each variant to test. The resulting PCR products serve as template DNA for evaluation via the expression of a reporter protein. In our work, the affinity purification protocol provided here was used for TetR and GamS. The same procedure can be used for the expression and purification of other repressors, activators, polymerases, sigma factors, and other proteins cognate to a genetic part of interest, although modifications may be needed for the desired protein being expressed. Purification and titration of these proteins into CFPS reactions enables a more detailed characterization of a particular genetic part. Finally, numerous alternative CFPS protocols exist and each should be amenable to the parts screening portion of methodology. As an example, we do not include a dialysis step in this protocol, which others have found to be important for expression from native bacterial promoters22. Varying the concentrations of underlying constituent components of the CFPS is also possible. The use of liquid handling enhances the ability to test the myriad conditions by increasing throughput and decreasing the materials required34,35.
One area that can require significant troubleshooting is optimization of the acoustic liquid handler. Acoustic liquid handler dispensing should be optimized for each component being transferred and it is strongly recommended to run controls to verify proper distribution and reproducibility before collecting data. The ideal source plate type and liquid class setting will depend on the specific liquid to be dispensed and its components. It is not recommended to use amine-coated plates to dispense DNA, as the amine coating may interact with the DNA. It should also be noted that the ability to dispense higher concentrations of certain components may depend on the acoustic liquid handler model. A test liquid transfer may be conducted by dispensing onto a foil plate seal to visualize successful droplet formation; however, this test provides limited information and droplets from different settings may appear identical. The use of a water-soluble dye, such as tartrazine, may be used to more accurately verify the correct volume is dispensed with a given setting or workflow (see Representative Results). Optimal programming of liquid transfers can also influence the accuracy and consistency of data generated; for transfers >1 µL from one source well to one destination well, we have found that sequential transfers of ≤1 µL should be programmed to reduce systematic well-to-well variability (Figure 4). Lastly, theoretical and actual source well dead volumes can vary dramatically depending on the source plate type, liquid class setting, and components of the specific liquid; using the acoustic liquid handler survey function to assess the well volumes prior to running a program may help gauge how accurately the instrument is able to measure a particular liquid.
CFPS reaction performance can vary when comparing results between different users, batches of materials, plate readers, and laboratories41. For instances where such comparisons are required while prototyping genetic circuits, we recommend including internal control reactions with standard constitutive promoters in each reaction plate to help normalize results across experimental setups. The method of DNA preparation can also contribute majorly to CFPS activity; the inclusion of an ethanol precipitation step is recommended. In addition, the optimal reaction composition can vary by the batch of extract34. Optimal magnesium glutamate and potassium glutamate concentrations, in particular, have been shown to vary by batch42 or with the promoter or reporter protein used24. Concentrations of these components should be optimized by screening across several concentrations of each component per genetic construct and per cell extract preparation to determine the optimal conditions for protein expression. Finally, best practices for consistent CFPS reaction performance include thorough mixing, careful pipetting, and consistency in the preparation of each reagent component.
Beyond characterization of individual parts, the same method can be used to screen combinations of parts that form complex circuits, such as logic circuits16 or oscillators52,53. This method can also be applied to screening and optimizing biosensors for applications in epidemiological diagnostics54,55,56,57 or hazard detection and quantification3,58,59. The application of AI-driven techniques such as active learning34 can also be paired with the high-throughput nature of this method to drive rapid exploration of complex biological design spaces. Ultimately, we envision this approach supporting accelerated development times for new genetic designs in synthetic biology.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was made possible by the Office of the Secretary of Defense's Applied Research for the Advancement of Science and Technology Priorities program. We thank Scott Walper (Naval Research Laboratory) for providing the stock of sfGFP used, and Zachary Sun and Abel Chiao (Tierra Biosciences) for fruitful discussions related to prototyping with cell-free systems and related troubleshooting of acoustic liquid handling.
Materials
| 2x YT medium | Sigma-Aldrich | Y2377-250G | Alternative to making 2xYT media |
| Acetic Acid | J.T. Baker | 9508-01 | S30 Buffer B |
| Agar | Bacto | 214010 | For plating cells |
| Chromatography column (5 cm diameter) | BIO-RAD | 731-1550 | Used for protein purification. |
| Destination plate | Thermo Scientific Nunc plate | 142761 | For CFPS reactions |
| DMSO | Sigma-Aldrich | D2650 | For dialysis buffer |
| DpnI | NEB | R0176L | For digestion of plasmid templates |
| DTT | Roche | 20871723 | S30 Buffer B |
| E. coli BL21(DE3) Rosetta2 | Novagen | 70954 | Cell line used for production of lysate and purified proteins |
| Echo acoustic liquid handler | Labcyte | 525 | Acoustic liquid handler |
| French pressure cell | Thermo Spectronic | FA-078 | For lysing cells for CFPS |
| Imidazole | Sigma-Aldrich | 56750 | For buffers |
| Impermeable plastic sealable lid | Thermo | 232702 | Plate seal |
| IPTG | RPI | I56000-25.0 | Used for protein induction. |
| K-Glu | Sigma-Aldrich | g1501-500G | S30 Buffer B |
| Labcyte Echo source plate | Labcyte | PL-05525 | For use with Echo acoustic liquid handler |
| Mg-Glu | Sigma-Aldrich | 49605-250G | S30 Buffer B |
| NaCl | Sigma-Aldrich | S7653-250G | For buffers |
| NaHPO4 | Sigma-Aldrich | 71505 | For dialysis buffer |
| NaOH | Mallinckrodt Chemicals | 7708-10 | For making 2xYT media. Currently not produced by Mallinckrodt. Alternate: Sigma-Aldrich S0899 |
| Ni-NTA resin | Invitrogen | R901-15 | For production of purified proteins |
| PCR H2O | Ambion | AM9937 | PCR of linear templates |
| Plate Reader | BioTek | H10 | Plate reader used |
| Q5 PCR Master Mix | NEB | M0494S | PCR of linear templates |
| QIAquick Gel Extraction Kit | Qiagen | 28606 | PCR of linear templates |
| QIAquick PCR Purification Kit | Qiagen | 28004 | PCR of linear templates |
| QSonica Ultrasonic Processor | Qsonica | Q700 | Cell disruption during protein purification |
| RTS Amino Acid Sampler | biotechrabbit | BR1401801 | Updated supplier from Sun et al. |
| Tris | MP | 819623 | S30 Buffer B |
| Tris-Cl | Sigma-Aldrich | T5941 | For buffers |
| Tryptone | Fluka | T7293 | For making 2xYT media |
| Yeast Extract | Bacto | 212750 | For making 2xYT media |
References
- Purnick, P. E. M., Weiss, R. The second wave of synthetic biology: from modules to systems. Nature Reviews. Molecular Cell Biology. 10 (6), 410-422 (2009).
- Saltepe, B., Kehribar, E. &. #. 3. 5. 0. ;., Su Yirmibeşoǧlu, S. S., Şafak Şeker, U. &. #. 2. 1. 4. ;. Cellular biosensors with engineered genetic circuits. ACS Sensors. 3 (1), 13-26 (2018).
- Bereza-Malcolm, L. T., Mann, G., Franks, A. E. Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach. ACS Synthetic Biology. 4 (5), 535-546 (2015).
- Mimee, M., et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science. 360 (6391), 915-918 (2018).
- Isabella, V. M., et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nature Biotechnology. 36 (9), 857-867 (2018).
- Healy, C. P., Deans, T. L. Genetic circuits to engineer tissues with alternative functions. Journal of Biological Engineering. 13, 39 (2019).
- Kaushik, A., Tiwari, S., Dev Jayant, R., Marty, A., Nair, M. Towards detection and diagnosis of Ebola virus disease at point-of-care. Biosensors and Bioelectronics. 75, 254-272 (2016).
- Jagadevan, S., et al. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnology for Biofuels. 11, 185 (2018).
- Keating, K. W., Young, E. M. Synthetic biology for bio-derived structural materials. Current Opinion in Chemical Engineering. 24, 107-114 (2019).
- Smanski, M. J., et al. Synthetic biology to access and expand nature’s chemical diversity. Nature Reviews Microbiology. 14 (3), 135-149 (2016).
- Xiang, Y., Dalchau, N., Wang, B. Scaling up genetic circuit design for cellular computing: advances and prospects. Natural Computing. 17 (4), 833-853 (2018).
- Galdzicki, M., Rodriguez, C., Chandran, D., Sauro, H. M., Gennari, J. H. Standard biological parts knowledgebase. PLoS One. 6 (2), 17005 (2011).
- Kelwick, R., Bowater, L., Yeoman, K. H., Bowater, R. P. Promoting microbiology education through the iGEM synthetic biology competition. FEMS Microbiology Letters. 362 (16), (2015).
- Torella, J. P., et al. Unique nucleotide sequence-guided assembly of repetitive DNA parts for synthetic biology applications. Nature Protocols. 9 (9), 2075-2089 (2014).
- Halleran, A. D., Swaminathan, A., Murray, R. M. Single day construction of multigene circuits with 3G assembly. ACS Synthetic Biology. 7 (5), 1477-1480 (2018).
- Nielsen, A. K., et al. Genetic circuit design automation. Science. 352 (6281), (2016).
- Canton, B., Labno, A., Endy, D. Refinement and standardization of synthetic biological parts and devices. Nature Biotechnology. 26 (7), 787-793 (2008).
- Boehm, C. R., Bock, R. Recent advances and current challenges in synthetic biology of the plastid genetic system and metabolism. Plant Physiology. 179 (3), 794-802 (2019).
- Takahashi, M. K., et al. Characterizing and prototyping genetic networks with cell-free transcription-translation reactions. Methods. 86, 60-72 (2015).
- Chappell, J., Watters, K. E., Takahashi, M. K., Lucks, J. B. A renaissance in RNA synthetic biology: New mechanisms, applications and tools for the future. Current Opinion in Chemical Biology. 28, 47-56 (2015).
- Endy, D. Foundations for engineering biology. Nature. 438 (7067), 449-453 (2005).
- Silverman, A. D., Karim, A. S., Jewett, M. C. Cell-free gene expression: an expanded repertoire of applications. Nat. Reviews. Genetics. 21 (3), 151-170 (2020).
- Dopp, J. L., Rothstein, S. M., Mansell, T. J., Reuel, N. F. Rapid prototyping of proteins: Mail order gene fragments to assayable proteins within 24 hours. Biotechnology and Bioengineering. 116 (3), 667-676 (2019).
- Garamella, J., Marshall, R., Rustad, M., Noireaux, V. The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synthetic Biology. 5 (4), 344-355 (2016).
- Kopniczky, M. B., et al. Cell-free protein synthesis as a prototyping platform for mammalian synthetic biology. ACS Synthetic Biology. 9 (1), 144-156 (2020).
- Kelwick, R., et al. Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synthetic Biology. 3 (1), (2018).
- Karim, A. S., et al. In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nature Chemical Biology. 16 (8), 912-919 (2020).
- Takahashi, M. K., et al. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free Transcription-Translation (TX-TL) systems. ACS Synthetic Biology. 4 (5), 503-515 (2015).
- Chappell, J., Jensen, K., Freemont, P. S. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Research. 41 (5), 3471-3481 (2013).
- Borkowski, O., et al. Cell-free prediction of protein expression costs for growing cells. Nature Communications. 9 (1), 1457 (2018).
- Dudley, Q. M., Karim, A. S., Nash, C. J., Jewett, M. C. In vitro prototyping of limonene biosynthesis using cell-free protein synthesis. Metabolic Engineering. 61, 251-260 (2020).
- McManus, J. B., Emanuel, P. A., Murray, R. M., Lux, M. W. A method for cost-effective and rapid characterization of engineered T7-based transcription factors by cell-free protein synthesis reveals insights into the regulation of T7 RNA polymerase-driven expression. Archives of Biochemistry and Biophysics. 674, 108045 (2019).
- Sun, Z. Z., Yeung, E., Hayes, C. A., Noireaux, V., Murray, R. M. Linear DNA for rapid prototyping of synthetic biological circuits in an escherichia coli based TX-TL cell-free system. ACS Synthetic Biology. 3 (6), 387-397 (2014).
- Borkowski, O., et al. Large scale active-learning-guided exploration for in vitro protein production optimization. Nature Communications. 11 (1), 1872 (2020).
- Caschera, F., et al. High-throughput optimization cycle of a cell-free ribosome assembly and protein synthesis system. ACS Synthetic Biology. 7 (12), 2841-2853 (2018).
- Iyer, S., Karig, D. K., Norred, S. E., Simpson, M. L., Doktycz, M. J. Multi-input regulation and logic with T7 promoters in cells and cell-free systems. PLoS One. 8 (10), 78442 (2013).
- Karig, D. K., Iyer, S., Simpson, M. L., Doktycz, M. J. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Research. 40 (8), 3763-3774 (2012).
- Jung, J. K., et al. Cell-free biosensors for rapid detection of water contaminants. Nature Biotechnology. 38 (12), 1451-1459 (2020).
- Bailey, J., Eggenstein, E., Lesnick, J. Miniaturization and rapid processing of TXTL reactions using acoustic liquid handling. Labcyte Technical Note. , 1-12 (2018).
- Marshall, R., Garamella, J., Noireaux, V., Pierson, A. High-throughput microliter-sized cell-free transcription-translation reactions for synthetic biology applications using the echo 550 liquid handler. Labcyte Application Note. , 1-6 (2018).
- Cole, S. D., et al. Quantification of interlaboratory cell-free protein synthesis variability. ACS Synthetic Biology. 8 (9), 2080-2091 (2019).
- Sun, Z. Z., et al. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. Journal of Visualized Experiments: JoVE. (79), e50762 (2013).
- Dopp, J. L., Tamiev, D. D., Reuel, N. F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnology Advances. 37 (1), 246-258 (2019).
- Cole, S. D., Miklos, A. E., Chiao, A. C., Sun, Z. Z., Lux, M. W. Methodologies for preparation of prokaryotic extracts for cell-free expression systems. Synthetic and Systems Biotechnology. 5 (4), 252-267 (2020).
- Sitaraman, K., et al. A novel cell-free protein synthesis system. Journal of Biotechnology. 110 (3), 257-263 (2004).
- Marshall, R., Maxwell, C. S., Collins, S. P., Beisel, C. L., Noireaux, V. Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription-translation systems. Biotechnology and Bioengineering. 114 (9), 2137-2141 (2017).
- Yim, S. S., Johns, N. I., Noireaux, V., Wang, H. H. Protecting linear DNA templates in cell-free expression systems from diverse bacteria. ACS Synthetic Biology. 9 (10), 2851-2855 (2020).
- Zhu, B., et al. Increasing cell-free gene expression yields from linear templates in Escherichia coli and Vibrio natriegens extracts by using DNA-binding proteins. Biotechnology and Bioengineering. 117 (12), 3849-3857 (2020).
- Tuckey, C., Asahara, H., Zhou, Y., Chong, S. Protein synthesis using a reconstituted cell-free system. Current Protocols in Molecular Biology. 108, 1-22 (2014).
- Hoffman, R. A., Wang, L., Bigos, M., Nolan, J. P. NIST/ISAC standardization study: Variability in assignment of intensity values to fluorescence standard beads and in cross calibration of standard beads to hard dyed beads. Cytometry. Part A. 81 (9), 785-796 (2012).
- Karzbrun, E., Shin, J., Bar-Ziv, R. H., Noireaux, V. Coarse-grained dynamics of protein synthesis in a cell-free system. Physical Review Letters. 106 (4), 048104 (2011).
- Rossi, N. A., Dunlop, M. J. Making waves with synthetic oscillators. Cell Systems. 6 (4), 406-407 (2018).
- Niederholtmeyer, H., et al. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife. 4, 09771 (2015).
- Chang, H., Voyvodic, P. L., Structurale, C. D. B. Microbially derived biosensors for diagnosis, monitoring and epidemiology. Microbial Biotechnology. 10 (5), 1031-1035 (2017).
- Pardee, K., et al. low-cost detection of Zika virus using programmable biomolecular components. Cell. 165 (5), 1255-1266 (2016).
- Pardee, K., et al. Portable, on-demand biomolecular manufacturing. Cell. 167 (1), 248-259 (2016).
- Pardee, K., et al. Paper-based synthetic gene networks. Cell. 159 (4), 940-954 (2014).
- Liu, X., Germaine, K. J., Ryan, D., Dowling, D. N. Whole-cell fluorescent biosensors for bioavailability and biodegradation of polychlorinated biphenyls. Sensors. 10 (2), 1377-1398 (2010).
- Gautam, P., Suniti, S., Amrita, K., Madathil, D., Nair, B. A Review on Recent Advances in Biosensors for Detection of Water Contamination. International Journal of Environmental Sciences. 2 (3), 1565-1574 (2012).

