Real-time Monitoring of Mitochondrial Respiration in Cytokine-differentiated Human Primary T Cells
Summary
Metabolic adaptation is fundamental for T cells as it dictates differentiation, persistence, and cytotoxicity. Here, an optimized protocol for monitoring mitochondrial respiration in ex vivo cytokine-differentiated human primary T cells is presented.
Abstract
During activation, the metabolism of T cells adapts to changes that impact their fate. An increase in mitochondrial oxidative phosphorylation is indispensable for T cell activation, and the survival of memory T cells is dependent on mitochondrial remodeling. Consequently, this affects the long-term clinical outcome of cancer immunotherapies. Changes in T cell quality are often studied by flow cytometry using well-known surface markers and not directly by their metabolic state. This is an optimized protocol for measuring real-time mitochondrial respiration of primary human T cells using an Extracellular Flux Analyzer and the cytokines IL-2 and IL-15, which differently affect T cell metabolism. It is shown that the metabolic state of T cells can clearly be distinguished by measuring the oxygen consumption when inhibiting key complexes in the metabolic pathway and that the accuracy of these measurements is highly dependent on optimal inhibitor concentration and inhibitor injection strategy. This standardized protocol will help implement mitochondrial respiration as a standard for T cell fitness in monitoring and studying cancer immunotherapies.
Introduction
Correct T cell development and function are essential for the ability of the immune system to recognize and respond to antigens. Mitochondrial oxidative phosphorylation (OxPhos) changes according to the state of the T cell. Naïve T cells predominantly use OxPhos to produce ATP, whereas activated T cells undergo a metabolic transition where glycolysis becomes dominant1. After the effector phase, the small remaining subset of memory T cells reverts to a metabolic state dominated by OxPhos2,3. The changes of OxPhos follow the differentiation of T cells to such a degree that even subsets of T cells can be differentiated by their specific is OxPhos properties1. Conversely, OxPhos is important for T cells' function, and inhibition of OxPhos has been demonstrated to block proliferation and cytokine production of T cells4. Therefore, the ability to quantify the properties of T cell OxPhos in a precise and reproducible manner is a powerful tool for anyone working with T cells.
In this protocol, properties of T cell OxPhos are measured using an extracellular flux analyzer. The core function of this analyzer is to continuously measure the oxygen content of the growth media of the cells to be analyzed. Oxygen removed from the growth media is assumed to be taken up by the cells. By treating the cells with a variety of OxPhos inhibitors or modifiers, a drop in oxygen uptake is associated with the inhibited or modulated function. For example, inhibition of the ATP synthase will lead to a reduced cellular uptake of oxygen that would otherwise be used to produce ATP by oxidative phosphorylation. Other equipment, including the Clark electrode and the Oroboros instrument, offers similar functionality, and each instrument has different advantages and shortcomings. A wide array of cell types can be used for studies in these devices, but one particularly challenging cell type is human primary T lymphocytes5. Due to their small size, poor survival ex vivo, and non-adherent properties, human primary T cells can be challenging to study.
This is a protocol for studying the mitochondrial respiration of human primary T cells by an extracellular analyzer. The protocol is divided into an Optimization run, where optimal concentrations of cell number per well, as well as the optimal concentration of oligomycin and FCCP, are determined. Furthermore, an Assay run, where the optimized conditions are used.
Using blood-derived human PBMCs and ex vivo primary T cell cultures, this protocol demonstrates the importance of optimal inhibitor concentration and the relevance of using separate instead of a sequential injection of mitochondrial inhibitors when working with sensitive cell types. Finally, it is demonstrated that this assay can robustly detect subtle differences in mitochondrial respiration upon polarization with cytokines IL-2 and IL-15.
Protocol
Experiments were carried out under the guidelines from Herlev Hospital and the Capital Region of Denmark.
NOTE: This protocol contains instructions for both an Optimization run and an Assays run. It is clearly written in the text when instructions are for an Optimization run or an Assay run. Run an Optimization run before continuing with the Assay runs
1. Human peripheral blood mononuclear (PBMC) isolation from buffy coats
- PBMC isolation
- Collect buffy coats from the appropriate institution (collected in blood collection bags). Buffy coats originate from healthy donors. Exclude donors that have recently used painkillers.
- Spray the blood collection bag containing the buffy coat with 70% ethanol before transferring to a laminar flow cabinet. Always use sterile techniques and sterile hardware for all steps
NOTE: Ensure that the correct authorizations for the handling of human blood samples are obtained. - Transfer the blood to a sterile 50 mL centrifuge tube.
- Dilute the blood at least 10% with non-supplemented RPMI 1640.
- Pour 20 mL of the preferred density gradient medium in a 50 mL centrifuge tube.
- Aspirate 25 mL of the diluted blood into a serological pipette. Set the electric pipette controller to the lowest speed.
- Hold the tube with density gradient medium at a 45° angle and rest the serological pipette tip on the inner side of the 50 mL centrifuge tube. Slowly release 25 mL of the diluted blood from the serological pipette.
- Make sure the diluted blood and the density gradient medium do not mix. Ensure that the diluted blood rests on top of the density gradient medium.
- Repeat this with subsequent density gradient medium tubes until all diluted blood is processed.
- Carefully move the tubes to a centrifuge and centrifuge at 1000 x g for 30 min in a swing-out rotor at room temperature (RT). Ensure that the acceleration and break are at a minimum.
NOTE: After centrifugation, various layers should be visible. The top, clear pink-to-orange layer consists of the blood plasma and platelets. The middle white layer consists of the PBMCs followed by a clear layer that consists of the density gradient medium, and finally a dark red layer at the bottom with red blood cells. - Using a sterile Pasteur pipette, carefully aspirate the PBMC-containing white layer to a 50 mL centrifuge tube.
NOTE: Make sure not to transfer the density gradient medium as this will affect downstream purification. Excess plasma will not affect the results. - Pool all collected PBMCs in the same 50 mL centrifuge tube and top up to 50 mL with RPMI 1640.
- Centrifuge the cells at 500 x g for 5 min at RT (perform the remaining centrifugation steps at this setting unless specified otherwise).
- Aspirate the supernatant and resuspend the cells in 30 mL of RPMI 1640. Count the cells using a hemocytometer or automated cell counter.
- For cryopreservation, resuspend a maximum of 30 million cells per 1 mL of freezing medium.
- Transfer the cells to cryotubes and freeze until -80 °C using a rate-controlled freezing container (-1 °C per min). For long-term storage, move the PBMCs to -140 °C.
NOTE: Limit the time cells are in the freezing medium at RT. At RT, DMSO is highly toxic to cells.
2. Culturing of activated human primary T lymphocytes
- Thawing of cells (Day 1)
- Pre-warm 10 mL of RPMI 1640 per sample to around 37 °C.
- Take the desired number of cell ampules and store them temporarily on dry ice.
- Resuspend the frozen cells in 10 mL of pre-warmed RPMI 1640.
- Centrifuge the cells for 5 min at 500 x g at RT.
- Discard the supernatant and wash the cells again by resuspending in 10 mL of RPMI 1640 and centrifuge as described in 2.1.4.
- Discard the supernatant and resuspend the cells at 2 x 106 cells per mL of X-VIVO 15 medium + 5% human serum (hereafter: T cell medium).
- Plate 2 mL of the cells per well in a 24-well cell culture plate and incubate overnight at 37 °C and 5% CO2.
- Activation of cells (Day 0)
- Wash CD3/CD28 beads by transferring 12.5 µL of beads per 1 million cells to a microcentrifuge tube. Add 12.5 µL of PBS per 12.5 µL of beads.
NOTE: It is important to vortex the vial of beads before use. - Place the microcentrifuge tube on a suitable magnet for 1 min.
- Discard the buffer and resuspend the beads in the original volume of T cell medium (12.5 µL of T cell medium per 12.5 µL of the original volume of beads).
- Add 12.5 µL of beads per million of cells corresponding to a ratio of 1:2 (beads: cells).
- Divide the cells into two conditions with around 5 million cells in each.
- Add the correct volume of cytokines to the conditions as mentioned in Table 1.
- Incubate the cells for 3 days at 37 °C and 5% CO2.
- Wash CD3/CD28 beads by transferring 12.5 µL of beads per 1 million cells to a microcentrifuge tube. Add 12.5 µL of PBS per 12.5 µL of beads.
- Culturing of cells (Day 3 and 5)
- Resuspend the cells and split them by transferring half the volume from each well into a new well. Add the same volume of fresh T cell medium to each well.
- Add new cytokines to each condition as mentioned in Table 1.
3. Extracellular flux assay
- Hydration of the sensor cartridge (Day 0)
- Unpack the sensor cartridge and carefully remove the sensor cartridge from the utility plate.
- Place the sensor cartridge upside-down, taking care not to touch the sensor probes.
- Fill the utility plate with 200 µL of calibrant (see Table of Materials for details) and carefully replace the sensor cartridge back into the utility plate.
- Alternatively, to eliminate any bubble formation, incubate the sensor cartridge in sterile ultrapure water overnight and replace it with a pre-warmed calibrant on the morning of the assay.
- Incubate the sensor cartridge plate at 37 °C in a non-CO2-regulated heating cabinet overnight.
NOTE: It is very important to use a non-CO2 regulated cabinet since excess CO2 will influence the sensor cartridge. Make sure that the Flux analyzer (see Table of Materials for details) is turned on at least a day before use to allow it to warm up to 37 °C.
- Cell coating and preparation of mitochondrial inhibitors (Day 1)
- Prepare a coating solution (see Table of Materials) containing NaHCO3 (pH 8.3, 0.1 M, 1128 µL), Cell-Tak (1 mg/mL, 48 µL) and NaOH (1.0 M, 24 µL).
NOTE: Coating solution should always be made and used fresh. - Open a fresh XF cell culture plate and add 12 µL of the freshly prepared coating solution to each well. Ensure even distribution of coating solution in the bottom of all the wells.
- Incubate the plate at RT with the lid on for 30 min and discard the remaining liquid solution from all the wells.
- Wash the plate with 200 µL of sterile water and discard the liquid.
- Wash the plate with 200 µL of cell culture grade sterile PBS and discard the liquid.
- Leave the plate at RT and let it dry for at least 30 min.
- Prepare a coating solution (see Table of Materials) containing NaHCO3 (pH 8.3, 0.1 M, 1128 µL), Cell-Tak (1 mg/mL, 48 µL) and NaOH (1.0 M, 24 µL).
- Plate T cells in the XF cell culture plate (Day 1)
- Prepare 50 mL of assay media by mixing suitable XF RPMI media with glucose, pyruvate, and glutamine according to experimental setup (Recommended levels: 4 mM glucose, 1 mM pyruvate and 3 mM glutamine).
- Heat to 37 °C in a non-CO2 regulated incubator and set pH at 7.4. Ensure that there is enough media for plating cells and preparing oligomycin and FCCP solutions (section 3.4).
- Design a plate layout with an increasing number of cells per well for Optimization run or Assay run. Use four 4 wells, filled with media and injected with media, for background measurements.
- Count T cells prepared in section 2.3 (by the preferred method) and pipette the correct number of cells to each well of the XF cell culture plate coated with the coating solution, according to the plate layout.
NOTE: The final volume of each well may vary but must be enough to cover the bottom of the well. - Centrifuge the XF cell culture plate at 1000 x g at RT for 10 min to adhere the T cell to the coated surface.
- Wash the cells with 200 µL of assay media, discard the media and add 180 µL of assay media.
NOTE: Visually inspect the wells using an inverted light microscope to ensure that the cells are attached and evenly distributed across the well surface. - Incubate the XF cell culture plate in the non-CO2 regulated heating cabinet for 30 min to ensure that the plate's temperature is 37 °C.
- Loading sensor cartridge with oligomycin and FCCP for Optimization run (Day 1)
NOTE: If oligomycin and FCCP concentrations were already optimized, continue at section 3.5.- Prepare working solutions of oligomycin and FCCP in assay media (prepared in step 3.3.1) as described in steps 3.4.2 and 3.4.3.
- Oligomycin working solutions: prepare 5 µM solution (2 mL of assay media + 10 µL of 1 mM oligomycin stock) and 3 µM solution (8 mL of assay media + 24 µL of 1 mM oligomycin stock).
- FCCP working solution: Prepare 2 µM solution (2 mL of assay media + 13.2 µL of 300 µM FCCP stock) and a 1.3 µM solution (8 ml of assay media + 34.6 µL of 300 µM FCCP stock)
- Load the working solutions of either oligomycin or FCCP into the injection ports of the sensor cartridge (Table 2).
NOTE. It is important that no injection ports contain air only. If, for any reason, not all injection ports are used, empty ports need to be filled with assay media. - Gently knock the edges of the plate on the table to remove potential bubbles in the injection ports.
- Loading sensor cartridge with oligomycin, FCCP, and antimycin A for Assay run (Day 1)
- Prepare solutions of oligomycin, FCCP in assay media (prepared in step 3.3.1) according to the optimal concentrations identified in a previous Optimization run. Also, prepare a 20 µM antimycin A solution.
- Load 20 µL of either oligomycin or FCCP into injection port A of the sensor cartridge according to plate layout. Add 22 µL of 20 µM antimycin A to injection port B of all wells. The resulting concentration of antimycin A once injected into the well will be 2 µM.
- Setting up experimental protocol in Flux analyzer software.
- Assign the groups and plate map for each condition per plate layout.
- Design the protocol according to the injection strategy (Table 3 and Figure 1).
NOTE: When running Assay runs, injection C and D can be omitted. - Save the assay set up, fill out the information required to be included for the assay, and press Start.
- The flux analyzer will ask for the sensor cartridge prepared in section 3.5. Remove the lid and insert the sensor cartridge as directed by the analyzer.
NOTE: The assay will calibrate and check sensors indicated by checkmarks. After successful calibration, the analyzer will request for the XF cell culture plate prepared in section 3.3. The utility plate is ejected from the analyzer and replaced with the XF cell culture plate to start the assay.
Representative Results
A correct determination of OxPhos properties is an indispensable tool when studying T cells. However, if the assay conditions have not been optimized, there is a substantial risk of misleading or erroneous results. In this protocol, there is a strong focus on the optimization of cell number per well and concentrations of oligomycin and FCCP to be used. In the described setup, oligomycin and FCCP are added incrementally to the same well, increasing the concentration of the mitochondrial modulators. The optimal concentration of oligomycin and FCCP can be determined from the resulting OCR curves of the wells as the concentration where a plateau is reached.
In the representative run, oligomycin is added in an increasing concentration and inhibiting ATP synthase (Complex V of the electron transport chain), resulting in decreased mitochondrial respiration. A plateau in OCR is reached after the accumulative concentration of the wells reached 1 µM. From this concentration and increasing concentrations OCR was not reduced further (Figure 1A). For wells treated with an incremental concentration of the uncoupler FCCP, OCR levels increased as expected until reaching a plateau after 0.2 µM of FCCP was added, indicating that at this concentration full uncoupling was obtained (Figure 1B). An optimization of cells plated per well is important for a correct and reproducible assay. If the used cell number is too low, then the level of oxygen removed from the assay media by the cells is too low to be correctly measured by the analyzer. On the other hand, if the number of cells per well is too high, the oxygen consumption of the cells can become so high that the system cannot replenish the oxygen levels of the assay media after each measurement, leading to an increasingly hypoxic environment and erroneous OxPhos characterization.
In the representative run, cells were seeded at a density of 200,000 and 400,000 cells per well (Figure 2A–C). For a run with 200,000 cells, the initial OCR is approximately half of a run with 400,000 cells per well. For FCCP treatment, maximal OCR is 61.6 pmol/min (200,000 cells) versus 190,4 pmol/min (400,000 cells). Following oligomycin treatment, the OCR in the run with 200,000 cells collapses into single-digit OCR (6.4 pmol/min). This is lower than the OCR of the run, with 400,000 cells per well treated with oligomycin (25.8 pmol/min, respectively).
Therefore, from the optimization run, it is clear that a cell number of 400,000 cells per well was required for future assays, using 1 µM oligomycin and 0.2 µM FCCP. In the classical setup recommended by the manufacturer, oligomycin and FCCP are added sequentially with the final addition of antimycin A. For T cells, this is not the optimal approach as the oligomycin treatment can be seen to limit the uncoupling after FCCP treatment (Figure 3A,B). In this presented method, it is recommended to run each condition in duplicate wells and treat one well with oligomycin and the other with FCCP, with a final addition of antimycin A for both wells. By using this approach, the oligomycin treatment does not affect OCR after FCCP treatment. This approach allows for the determination of the same mitochondrial properties as the classical setup, where drugs are added in sequence (Figure 3C)
Finally, it was investigated whether the effects of cytokines IL-2 and IL-15 could be differentiated on the metabolism of ex vivo cultured human primary T cells. Indeed, IL-15 cultured cells possessed higher maximal respiration and spare respiratory capacity, as has been shown before1 (Figure 4A–E). Basal respiration and ATP production were not affected. Taken together, this data shows that the mitochondrial respiration of ex vivo cultured human primary T cells can successfully be analyzed using the extracellular flux analyzer.
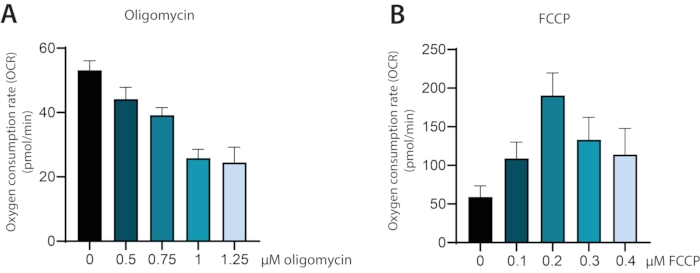
Figure 1: Oxygen consumption rate (OCR) measured during titration of inhibitors oligomycin and FCCP in ex vivo cultured human primary T cells. (A) OCR during stepwise titration of oligomycin from 0-1.25 µM final concentration. (B) OCR during stepwise titration of FCCP from 0-0.5 µM final concentration. Please click here to view a larger version of this figure.
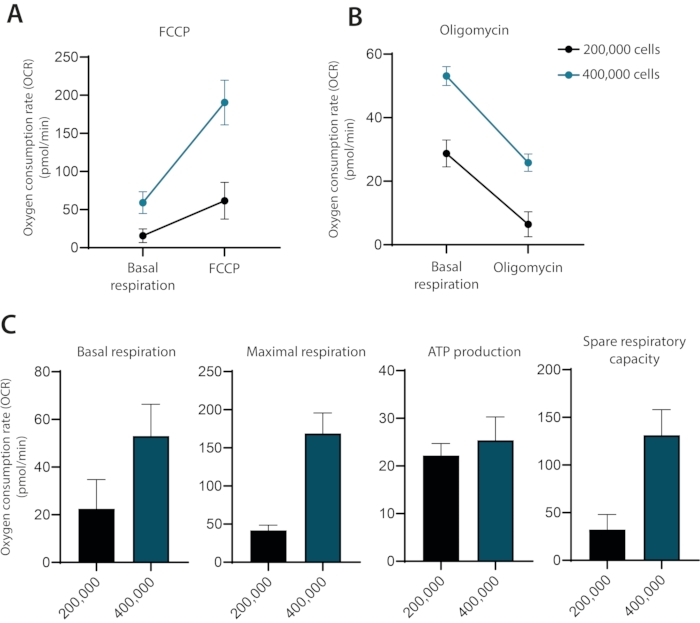
Figure 2: The influence of cell concentration on OCR measurements in ex vivo cultured human primary T cells. OCR measurements of ex vivo cultured human primary T cells with 200,000 or 400,000 cells per well after injection of either (A) FCCP or (B) Oligomycin. (C) Basal respiration, maximal respiration, ATP production, and spare respiratory capacity of human primary T cells using 200,000 or 400,000 cells per well. Representative of three independent experiments. Please click here to view a larger version of this figure.
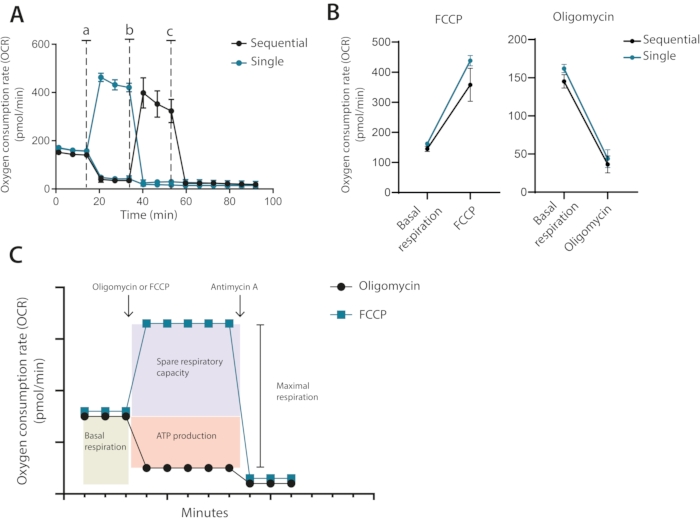
Figure 3: Single or sequential injection of mitochondrial modulators. (A) Representative OCR measurements during baseline and after injection of oligomycin and FCCP (a,b), or antimycin A (c) as single individual injections or as sequential injections. (B) OCR values before injection (basal respiration) or after injection of oligomycin or FCCP as single injections or sequential injections. (C) Schematic representation of injection and measurement strategy. Representative of one independent experiment Please click here to view a larger version of this figure.
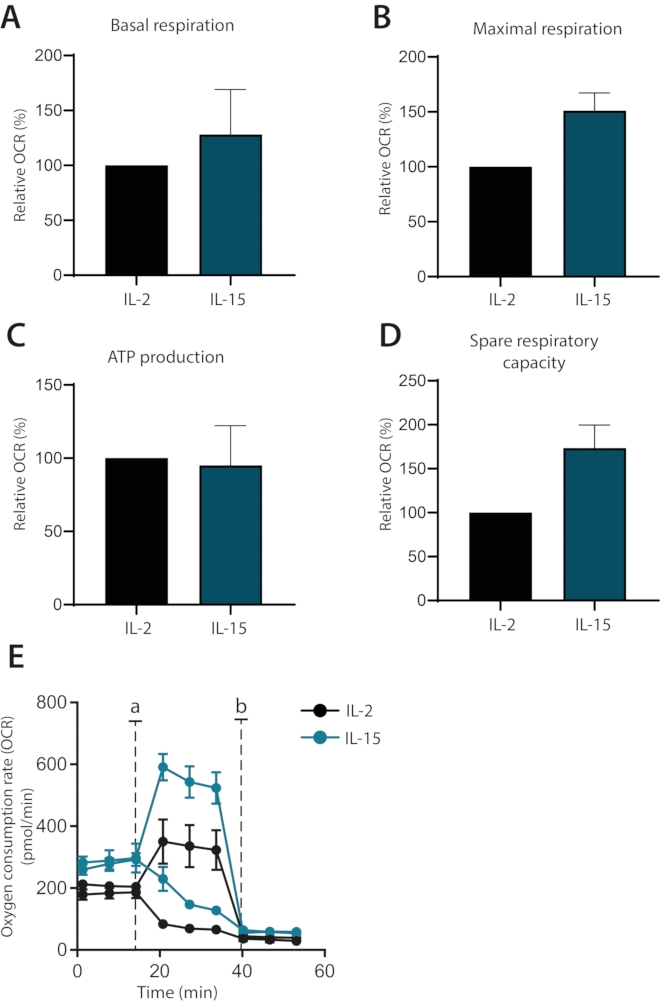
Figure 4: Differences in mitochondrial respiration in cytokine-differentiated human primary T cells. (A–D) Basal respiration, maximal respiration, ATP production, and spare respiratory capacity of human primary T cells cultured with IL-2 or IL-15 for seven days (n = 3). (E) Representative plots of (A–D), with injections of Oligomycin or FCCP (a) or antimycin A (b). Please click here to view a larger version of this figure.
| Cytokine | [Stock] | Dilution factor | [Final] | |
| Condition 1 | IL-2 | 3 x 106 U/mL | 30,000 | 100 U/mL |
| Condition 2 | IL-15 | 2 x 105 U/mL | 2,000 | 100 U/mL |
Table 1: Preparation of cytokine cultures used to guide metabolic changes in T cells.
| Oligomycin | FCCP | |||||
| Working sol. | Final conc. | Vol. | Working sol. | Final conc. | Vol. | |
| Port A | 5.0 μM | 0.50 μM | 20 μL | 2.0 μM | 0.2 μM | 20 μL |
| Port B | 3.0 μM | 0.75 μM | 22 μL | 1.30 µM | 0.3 μM | 22 μL |
| Port C | 3.0 μM | 1.0 μM | 24 μL | 1.30 µM | 0.4 μM | 24 μL |
| Port D | 3.0 μM | 1.25 μM | 27 μL | 1.30 µM | 0.5 μM | 27 μL |
Table 2: Strategy for preparation of mitochondrial inhibitors and modulators. Preparation concentrations, working concentrations, and injection strategies for an Optimization run.
| Action | Details | Measurement details |
| Baseline | Baseline measurements | 3 measurements |
| Injection | Port A injection | 3 measurements |
| Injection | Port B injection | 3 measurements |
| Injection | Port C injection | 3 measurements |
| Injection | Port D injection | 3 measurements |
| Measure | Additional measurements | Optional |
Table 3: Protocol design of an Optimization run with titration of mitochondrial inhibitors and modulators using 4 injections with 3 measurements in each.
| Element of mitochondrial oxidative phosphorylation | Explanation | ||
| Basal respiration | Basal respiration is a baseline measure of the rate of oxygen consumed by the stimulated T cells before addition of mitochondrial inhibitors. It is a measure of the oxygen consumption used to meet cellular ATP demand resulting from mitochondrial proton leak. As such, it provides an overview of the proton current generated to supply ATP synthesis and proton leak. However, it is also a measure that can be altered depending on the substrates present in the growth media, stimulation of cells prior to assay and other extrinsic factors. The basal respiration is therefore a measure used to compare two or more different cell types and/or different treatments believed to affect the cellular metabolic state of the cells. Basal respiration is calculated as the difference in OCR before adding any mitochondrial modulators (oligomycin or FCCP) and after adding antimycin A |
||
| Maximal respiration | This measure is the maximal rate of oxygen that can be consumed for oxidative phosphorylation. The rate of oxygen consumed by oxidative phosphorylation is determined both by the ability of the electron transport chain to pump protons across the inner mitochondrial membrane, and the ability of the ATP synthase to use the proton gradient to phosphorylate ATP from ADP. The speed of the ATP synthase is limited by free ADP substrate and thereby by the general energetic state of the cell. When treating the cells with the mitochondrial uncoupler FCCP, protons can freely traverse back across the inner mitochondrial membrane. This mimics a situation where the cells experience an unsaturable energy demand, and the maximal respiration is therefore a measure of the maximal rate of oxygen that can be consumed by the electron transport chain. Maximal respiration is calculated as the difference in OCR of cells treated with FCCP and cells treated with antimycin A |
||
| ATP turnover | ATP-linked respiration is measured as the difference in OCR after inhibition of the ATP synthase using oligomycin. The oxygen that would otherwise be consumed for phosphorylation of ADP by oxidative phosphorylation will no longer be used as this process is arrested. ATP-linked respiration is therefore relative to ATP produced by oxidative phosphorylation. Changes in AT-linked respiration is a response of the mitochondria to an altered ATP demand of the cell ATP-linked respiration is calculated as the difference in OCR before adding any mitochondrial modulators (oligomycin or FCCP) and after adding oligomycin |
||
| Spare respiratory capacity | The spare respiratory capacity is a measure of a theoretical extra capacity to produce ATP as a response to an increased energetic demand. It is defined as the difference between basal respiration and maximal respiration. Changes in spare respiratory capacity can be an indicator of mitochondrial and cell fitness and flexibility. | ||
Table 4: Explanation of the various components of mitochondrial respiration that are studied using the flux analyzer
Discussion
Detailed and correct quantification of oxidative phosphorylation is an indispensable tool when describing the energy states of T cells. The state of mitochondrial fitness can be directly related to T cell activation potential, survival, and differentiation1,5. With this protocol, it is possible to determine the various properties of oxidative phosphorylation (see Table 4 for a detailed explanation). Precise quantification of these properties of oxidative phosphorylation offers a detailed insight into the energy states of T cells. However, to obtain reliable results, great care must be taken when setting up the experiment.
In this protocol, the three following optimization steps are advised-first, optimization of cell numbers. The ability of a flux analyzer to correctly measure oxygen concentrations follows a sigmoid curve; changes in oxygen that are too small or too large will fall outside the operating interval of the machine and will therefore not be measured correctly. This necessitates an optimization of cell numbers to be used throughout the entire experiment. If too few cells are assayed, the changes in oxygen consumption are too low to be correctly measured. If too many cells, there is a risk of oxygen depletion in the assay media. An initial run is therefore recommended, with cell numbers ranging from 100,000-400,000 cells per well. When plotting cell number versus basal respiration, the optimal cell count will be in the linear range of the curve. When optimizing the setup, please be aware that there can be an exponential difference in mitochondrial activity between resting and activated cells, and therefore needs to be optimized accordingly.
Second, titration of inhibitor concentrations. When treating the cells with oligomycin and FCCP, it is important to identify the optimal concentrations of the inhibitors to be used. A too low concentration will result in a suboptimal inhibition and an incorrect measurement of mitochondrial respiration. It is common that people use the highest recommended concentrations of the inhibitors to ensure that a full inhibition is obtained. This is also problematic as too high concentrations of the inhibitors can have pleiotropic effects. Uncouplers like FCCP also exert their effects on membranes other than the mitochondrial, resulting in a range of undesired effects, including plasma membrane depolarization, mitochondrial inhibition, and cytotoxicity. In this protocol, titration of oligomycin and FCCP is done simultaneously with cell number optimization. During an optimization run, increasing concentrations of oligomycin or FCCP are added using the four available substrate ports. In the resulting OCR diagram, the optimal concentration can be visually determined as the concentration at which the OCR reaches a steady plateau. Once the concentration of oligomycin and FCCP have been titrated, these concentrations are to be used throughout the experiment.
Third, sequential versus single individual addition of inhibitors. Classical Seahorse assays are typically conducted with the sequential addition of the first oligomycin followed by the addition of FCCP. In T cells and other sensitive cells, such a sequential addition can result in an erroneous quantification of maximal respiration. In turn, the measured levels of spare respiratory capacity will report lower than they are. Grave examples of this include values of spare respiratory capacity that are negative. This is, of course, not biologically possible and is caused by a pre-sensitization of the mitochondria by oligomycin treatment. In this protocol, it is instead recommended that cells are only treated with either oligomycin or FCCP (see Figure 3C for illustrative comparison).
Finally, this optimized protocol is used to show how IL-15-supplemented human primary T cell cultures can be clearly distinguished from IL-2-supplemented cells based on their mitochondrial respiration. IL-15-cultured cells possess higher maximal respiration and spare respiratory capacity, a metabolic state linked to memory T cells1,6. These observations are in line with previous studies which link IL-15 to memory T cell subsets8. In addition, a difference in basal respiration but not in ATP production when compared to IL-2-cultured cells was observed. This indicates that these cells use their glycolytic capacity to comply with basal metabolic demands, a pathway associated with more differentiated cells. Taken together, it is shown that a human memory T cell model can be established in vitro by using IL-15 supplementation. Using an IL-15-rich environment to promote the development of memory cells has previously been demonstrated and further supports the findings8.
In this method, oligomycin, FCCP, and antimycin A have been used to quantify the properties of OxPhos. Other compounds exist with similar effects, which potentially would be better suited for T cells. An example would be to use the uncoupler BAM15 instead of FCCP to decrease depolarization of the mitochondrial membrane and to avoid cytotoxicity9. In this method, these compounds have not been considered, as oligomycin, FCCP, and antimycin A have been the recommended mitochondrial modulators for Seahorse experiments for the last decade. The use of these compounds is therefore recognized by reviewers and other researchers working with OxPhos. More experienced users of the Seahorse flux analyzer are encouraged to use these alternative compounds, but the use of these is outside the scope of this paper.
Monitoring mitochondrial OxPhos is an essential tool for understanding T cell function and improving cancer immunotherapies. As previously mentioned, IL-15 expanded cells – with a less differentiated memory phenotype – were shown to improve responses to CAR T cell therapies, as they were less exhausted and had an increased antitumor activity10. This optimized protocol could be an effective tool to study the quality of T cells in both preclinical and clinical settings. In conclusion, this protocol implements steps for optimizing cell numbers and inhibitor concentrations for the use of ex vivo cultured human primary T cells in metabolic assays.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Kasper Mølgaard and Anne Rahbech received grants from Tømmermester Jørgen Holm og Hustru Elisa f. Hansens Mindelegat. Kasper Mølgaardalso received a grant from Børnecancerfonden.
Materials
| 24-well tissue culture plate | Nunc | 142485 | |
| Anti-CD3xCD28 beads | Gibco | 11161D | |
| Antimycin A | Merck | A8674 | |
| Carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone (FCCP) | Sigma-Aldrich | C2920 | |
| Cell-Tak | Corning | 354240 | For coating |
| Dimethyl sulfoxide (DMSO) | Sigma Aldrich | D9170 | |
| Human Serum | Sigma Aldrich | H4522 | Heat inactivated at 56 °C for 30 min |
| IL-15 | Peprotech | 200-02 | |
| IL-2 | Peprotech | 200-15 | |
| Lymphoprep | Stemcell Technologies | 07801 | |
| Oligomycin | Merck | O4876 | |
| PBS | Thermo Fisher | 10010023 | |
| RPMI 1640 | Gibco-Thermo Fisher | 61870036 | |
| Seahorse Calibrant | Agilent Technologies | 102416-100 | |
| Seahorse XF 1.0 M glucose solution | Agilent Technologies | 103577-100 | |
| Seahorse XF 100 mM pytuvate solution | Agilent Technologies | 103578-100 | |
| Seahorse XF 200 mM glutamine solution | Agilent Technologies | 103579-100 | |
| Seahorse XF RPMI medium, pH7.4 | Agilent Technologies | 103576-100 | XF RPMI media |
| Seahorse XFe96 Analyser | Agilent Technologies | Flux analyzer | |
| Seahorse XFe96 cell culture microplates | Agilent Technologies | 102416-100 | XF cell culture plate |
| Seahorse XFe96 sensor cartridge | Agilent Technologies | 102416-100 | |
| Sodium Bicarbonate concentrate 0.1 M (NaHCO3) | Sigma Aldrich | 36486 | |
| Sodium Hydroxide solution 1 N (NaOH) | Sigma Aldrich | S2770-100ML | |
| X-VIVO 15 | Lonza | BE02-060F | |
| T cell beads magnet DynaMag-2 Magnet | Thermo Fisher | 12321D | |
| Seahorse wave | Flux analyzer software |
References
- vander Windt, G. J. W., et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 36 (1), 68-78 (2012).
- Krauss, S., Brand, M. D., Buttgereit, F. Signaling takes a breath–new quantitative perspectives on bioenergetics and signal transduction. Immunity. 15 (4), 497-502 (2001).
- vander Windt, G. J. W., et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences of the United States of America. 110 (35), 14336-14341 (2013).
- Chang, C. -. H., et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 153 (6), 1239-1251 (2013).
- vander Windt, G. J. W., Chang, C. -. H., Pearce, E. L. Measuring bioenergetics in T cells using a Seahorse extracellular flux analyzer. Current Protocols in Immunology. 113, 1-14 (2016).
- Buck, M. D., O’Sullivan, D., Pearce, E. L. T cell metabolism drives immunity. Journal of Experimental Medicine. 212 (9), 1345-1360 (2015).
- Rivadeneira, D. B., Delgoffe, G. M. Antitumor T-cell reconditioning: Improving metabolic fitness for optimal cancer immunotherapy. Clinical Cancer Research. 24 (11), 2473-2481 (2018).
- Cieri, N., et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood. 121 (4), 573-584 (2013).
- Kenwood, B. M., et al. Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Molecular Metabolism. 3 (2), 114-123 (2013).
- Alizadeh, D., et al. IL15 enhances CAR-T cell antitumor activity by reducing mTORC1 activity and preserving their stem cell memory phenotype. Cancer Immunology Research. 7 (5), 759-772 (2019).

