Generation of Multicue Cellular Microenvironments by UV-Photopatterning of Three-Dimensional Cell Culture Substrates
Summary
Traditionally, cell culture is performed on planar substrates that poorly mimic the natural environment of cells in vivo. Here we describe a method to produce cell culture substrates with physiologically relevant curved geometries and micropatterned extracellular proteins, allowing systematic investigations into cellular sensing of these extracellular cues.
Abstract
The extracellular matrix is an important regulator of cell function. Environmental cues existing in the cellular microenvironment, such as ligand distribution and tissue geometry, have been increasingly shown to play critical roles in governing cell phenotype and behavior. However, these environmental cues and their effects on cells are often studied separately using in vitro platforms that isolate individual cues, a strategy that heavily oversimplifies the complex in vivo situation of multiple cues. Engineering approaches can be particularly useful to bridge this gap, by developing experimental setups that capture the complexity of the in vivo microenvironment, yet retain the degree of precision and manipulability of in vitro systems.
This study highlights an approach combining ultraviolet (UV)-based protein patterning and lithography-based substrate microfabrication, which together enable high-throughput investigation into cell behaviors in multicue environments. By means of maskless UV-photopatterning, it is possible to create complex, adhesive protein distributions on three-dimensional (3D) cell culture substrates on chips that contain a variety of well-defined geometrical cues. The proposed technique can be employed for culture substrates made from different polymeric materials and combined with adhesive patterned areas of a broad range of proteins. With this approach, single cells, as well as monolayers, can be subjected to combinations of geometrical cues and contact guidance cues presented by the patterned substrates. Systematic research using combinations of chip materials, protein patterns, and cell types can thus provide fundamental insights into cellular responses to multicue environments.
Introduction
In vivo, cells are subjected to a wide variety of environmental cues that can be of mechanical, physical, and biochemical nature that originate from the extracellular matrix (ECM). Numerous environmental cues have been identified to play vital roles in the regulation of cell behavior, such as proliferation, differentiation, and migration1,2,3,4,5. One of the most widely investigated phenomena is contact guidance, describing adhesion-mediated cell alignment along anisotropic biochemical or topographical patterns present on the extracellular substrate6,7,8,9,10,11. Beyond directing the alignment of cells, contact guidance cues have also been shown to influence other cell properties such as cell migration, organization of intracellular proteins, cell shape, and cell fate12,13,14,15. Additionally, the geometrical architecture of the 3D cellular environment has also been acknowledged for its regulatory influence on cell behavior16,17. In the human body, cells are exposed to a range of curved geometries, ranging from microscale collagen fibers, capillaries, and glomeruli, up to mesoscale alveoli and arteries18,19. Interestingly, recent in vitro studies have shown that cells can sense and respond to such physical cues, from the nano- to mesoscale20,21,22,23.
To date, most studies investigating cell response to environmental cues have been largely performed using experimental setups that isolate single cues. While this approach has allowed tremendous progress in understanding the basic mechanisms behind cellular sensing of environmental cues, it poorly recapitulates the in vivo environment that simultaneously presents multiple cues. To bridge this gap, it is useful to develop culture platforms whereby multiple environmental cues can be independently and simultaneously controlled. This concept has gained increasing traction lately24,25, with studies combining matrix stiffness and ligand density26,27,28,29, substrate stiffness and porosity30, substrate stiffness and 3D microniche volume31, surface topography and contact-guidance cues32,33,34, and nanoscale contact-guidance cues with mesoscale curvature guidance cues23. However, it remains challenging to combine contact-guidance cues with a variety of 3D geometries in a controlled and high-throughput way.
This research protocol addresses this challenge and introduces a method to create cell culture substrates with a controlled combination of patterned adhesive areas of ECM proteins (contact-guidance cues) and substrate curvature (geometrical cues). This approach allows for the dissection of cell response in a biomimetic multicue environment in a systematic and high-throughput manner. The knowledge acquired can aid in further understanding of cell behavior in complex environments and can be used to design instructive materials with properties that steer cell responses into a desired outcome.
3D protein photopatterning
Creation of adhesive areas of ECM proteins (contact-guidance cues) on cell culture materials can be accomplished using a variety of techniques, for example, by deep-ultraviolet (deep-UV) patterning or microcontact printing35,36. Deep-UV patterning makes use of UV-light that is projected through a mask on a polymeric material to degrade passivation polymers at specific locations on the cell culture substrate. The patterned substrate is then incubated with a ligand of interest, resulting in adhesive areas that support cell attachment and culture on predefined locations12,37,38. An alternative way to introduce protein patterns is by means of microcontact printing, where elastomeric stamps containing a desired shape are coated with a protein of choice and are pressed on a cell culture substrate, thereby transferring the protein coating to which cells can adhere35,37,39,40. Unfortunately, since both techniques rely on mask preparation and soft lithography methods, the experiments are time-consuming and labor-intensive, as well as limited in terms of pattern flexibility. In addition, both deep-UV patterning and microcontact printing are most suited for planar materials and are technically difficult, if not impossible, for patterning ligands in a 3D environment.
To improve on these conventional methods, Waterkotte et al. combined maskless lithography, chemical vapor deposition, and thermoforming to generate micropatterned 3D polymeric substrates41. However, this technique relies on the use of thermoformable polymer films and offers low protein-pattern resolution (7.5 µm), while cells have been reported to respond to geometrical protein patterns as small as 0.1 µm2,42. Sevcik et al. described another promising method to nanopattern ECM ligands on substrates containing nano- and micrometer topographies43. Using microcontact printing, ECM proteins were transferred from polydimethylsiloxane (PDMS) stamps to a thermoresponsive poly(N-isopropylacrylamide) (pNIPAM) substrate. Subsequently, the thermoresponsive property of the pNIPAM network allowed them to transfer the two-dimensional (2D) protein pattern to a topographic PDMS substrate (10-100 µm deep grooves), thereby controlling the localization of adhesion sites on topographical features. However, not all possible microtopographies can be patterned since decreased wettability issues make it more difficult to pattern deeper topographic substrates. Trenches with a depth-to-width aspect ratio of 2.4 have been reported to be the ultimate limit to successfully transfer the pattern to the topographic substrate43. Additionally, the flexibility of varying patterns and the resolution of the generated patterns are poor due to the requirement of microcontact printing.
This paper describes a method that overcomes the abovementioned bottlenecks and offers a flexible and high-throughput method to create multicue substrates that can be used for cell culture (see Figure 1). Physiologically relevant geometries (cylinders, domes, ellipses, and saddle surfaces) with curvatures ranging from ĸ = 1/2500 to ĸ = 1/125 µm-1 are predesigned and microfabricated in PDMS chips. Subsequently, contact-guidance cues are created on top of the 3D geometries using a variety of digital pattern designs by employing a photopatterning technique with a resolution as small as 1.5 µm44. To this end, the PDMS chips are initially passivated to prevent cells and proteins from adhering; this passivation layer can then be removed by combination of the photoinitiator 4-benzoylbenzyl-trimethylammonium chloride (PLPP) and UV-light exposure45. A digital mask is designed to specify the locations of UV-exposure, and, thus, the area where the passivation layer is removed. Proteins can subsequently adhere to these areas, enabling cell attachment. Since the patterning is performed using a digital (rather than a physical) mask, a variety of patterns can be created quickly without the hassle and cost associated with designing and fabricating additional photomasks. In addition, a diverse range of ECM proteins (e.g., collagen type I, gelatin, and fibronectin) can be patterned on the substrate. Although this protocol is performed using cell culture chips made of PDMS, the principle can be applied to any other material of interest46.
Protocol
In the studies described in this protocol, primary human keratocytes were used. This research was performed in compliance with the tenets of the Declaration of Helsinki. Primary keratocytes were isolated from leftover human cadaveric corneoscleral tissues from Descemet Membrane Endothelial Keratoplasty surgery, which were obtained from the Cornea Department of the ETB-BISLIFE Multi-Tissue Center (Beverwijk, the Netherlands) after obtaining consent from the next of kin of all deceased donors.
NOTE: See the Table of Materials for details about all materials, reagents, equipment, and software used in this protocol.
1. Fabrication of 3D cell culture substrates
- Create a negative glass mold (#1) containing all features of interest, in this case, made from glass using a femtosecond-laser direct-write technique (see Figure 2).
- Carefully place the negative glass mold (#1) on the bottom of a Petri dish.
- Prepare a PDMS prepolymer by placing an empty 50 mL conical tube on a scale, tare the scale, and pour the desired amount of silicone elastomer base into the tube.
- Add curing agent using a Pasteur pipet so that the final ratio of elastomer base and curing agent is 10:1 (w/w).
- Thoroughly mix the components in the conical tube using a spatula.
- Centrifuge at 2,000 × g for 70 s to remove all air bubbles.
- Pour the PDMS prepolymer on top of the negative glass mold (#1) in the Petri dish to cover it completely.
- Place the Petri dish with the negative glass mold (#1) and the PDMS prepolymer in the vacuum desiccator and start the vacuum pump. Once a vacuum is reached, wait for 5 min to remove all bubbles present at the interface between the mold surface and the PDMS prepolymer.
- Remove the vacuum and take the Petri dish out of the desiccator.
- Cure the PDMS prepolymer overnight in the oven at 65 °C.
- Carefully remove the newly cured positive PDMS chip (#2) from the negative glass mold (#1) by lifting the edges of the PDMS using a spatula. If the positive PDMS chip (#2) tends to stick to the negative glass mold (#1), add ethanol or water to the edges of the imprint while lifting.
NOTE: The fluid will run in between the two layers and ease the separation of the negative glass mold (#1) and positive PDMS chip (#2). - Using a blade, cut off the sides of the positive PDMS chip (#2) so that a rectangular chip remains.
- Place the positive PDMS chip (#2) in a desiccator next to a small vial with a droplet of tridecafluoro(1,1,2,2-tetrahydrooctyl)trichlorosilane, a silanization agent, and leave under vacuum overnight.
NOTE: Silanization will make sure the imprint will not bind to other PDMS layers later in the protocol. Other silanization agents and/or methods may also work to ensure that the surfaces of the PDMS chips will not stick to other PDMS layers.
CAUTION: Tridecafluoro(1,1,2,2-tetrahydrooctyl)trichlorosilane is flammable (H226) and causes severe skin burns and eye damage (H314). Wear personal protective equipment, work in a fume hood, and wash hands thoroughly after handling. Keep the silanization agent away from heat, hot surfaces, sparks, open flames and other ignition sources. Store between 15 and 30 °C (P280, P210, P240, P403, P235, P310). - Remove the vacuum and place the positive PDMS chip (#2) on the bottom of a Petri dish. Pour PDMS prepolymer (10:1) on top to produce multiple negative PDMS molds (#3).
- Put the Petri dish under vacuum in the desiccator for 15 min to remove all bubbles.
- Cure the PDMS prepolymer at 65 °C overnight, after which the negative PDMS mold (#3) can be peeled off from of the positive PDMS chip (#2) using a spatula.
- Silanize the final negative PDMS mold (#3) using silanization agent in a vacuum desiccator overnight.
- Produce multiple cell culture chips (#4, see Figure 2) of approximately 5 mm thickness by pouring PDMS prepolymer in the negative PDMS mold (#3), removing bubbles using the desiccator and curing for 3 h at 65 °C. With a razor blade, cut the chip to the final size as visualized in Figure 2. Store the chips at room temperature.
NOTE: Surface roughness may influence the cellular response. If needed, an additional thin layer of PDMS can be used as a coating on the positive PDMS chip (#2) to smoothen the surface. To do so, pour a small droplet of PDMS prepolymer on the chip and spread it over the complete chip using pressurized air. Cure the coated chip at 65 °C for 3 h and continue with step 1.12.
2. Fabrication of flat PDMS samples (control samples)
- Prepare PDMS prepolymer (10:1) according to steps 1.3-1.6.
- Place a glass coverslip on the rounded vacuum post in the center of the spin coater.
- Turn on the vacuum to attach the glass coverslip to the machine and pipet a droplet of PDMS in the middle of the coverslip using a Pasteur pipet.
- Distribute the PDMS prepolymer over the glass substrate using the following protocol to get an approximately 10 µm thick layer.
- Spincoat for 10 s at 0.45 × g, acceleration: 0.2 × g/s.
- Spincoat for 50 s at 44.8 × g, acceleration: 0.54 × g/s.
- Turn off the vacuum, remove the coverslip from the spin coater using tweezers, and place it in a Petri dish. Cure the PDMS overnight in the oven at 65 °C and store at room temperature afterwards.
3. Substrate passivation of 3D cell culture substrates
- Activate the hydroxyl groups on the surface of the PDMS chip (#4) using O2-plasma. Using tweezers, place the chip in the basket of the plasma asher.
- Run an ashing cycle using a power of 20 W for 30 s. Vent the ashing chamber using N2.
- Take the chip out of the basket and place it in a small PDMS container (see Figure 1).
- Using a Pasteur pipet, add 500 µL of Poly-L-lysine (PLL, 0.01%) on the top of the chip so that the complete surface is immersed in the PLL solution. Incubate for 30 min at room temperature.
- Remove 450 µL of the PLL from the cell culture chip with a pipet and rinse the chip surface three times with 500 µL of 0.1 M HEPES buffer (8 < pH < 8.5). Always leave a small volume of liquid on the PDMS chip to avoid drying out of the sample, which will reduce the final pattern quality.
- Make 500 µL of a 50 mg/mL methoxypolyethylene glycol-succinimidyl valerate (mPEG-SVA; MW 5,000 Da) solution in 0.1 M HEPES buffer (8 < pH < 8.5) per cell culture chip and let it incubate on the sample for 60 min. Since the mPEG-SVA has a half-life of 15 min, be sure to prepare the required amount just before use.
NOTE: The mPEG-SVA is dissolved when the solution is completely transparent. - Remove 450 µL of the mPEG-SVA solution using a micropipet, and wash the chip surface five times with phosphate-buffered saline (PBS). Make sure to pipet up and down multiple times per wash to ensure all unbound mPEG-SVA is removed. To prevent the sample from drying out, minimize the time between washing steps and make sure to use an excess (500 µL or more) of PBS for washing.
- Store the samples by immersing them in PBS or continue to the patterning step of the protocol.
NOTE: If desired, passivation can be performed under sterile conditions when working in a culture cabinet and working with sterile solutions and equipment.
4. Storage of patterned cell culture substrates
NOTE: 3D cell culture substrates can be stored during different steps in the process.
- Store cured PDMS cell culture chips under dry conditions at room temperature.
- Store the passivated cell culture chips in one of these two ways:
- Store in PBS at 4 °C for up to 7 days.
- Store in dry conditions for up to several months. To obtain dry samples, remove the PBS and rinse multiple times using double distilled water (ddH2O). Blow-dry using a nitrogen- or air-gun.
5. Design of digital masks used for photopatterning
NOTE: Patterning of 3D substrates can be performed using a single or multiple focal planes (see Figure 3). A single focal plane can be used on features that are not larger than one digital mirror device (DMD, approximately 300 µm x 500 µm) and that are not too tall (50-100 µm). In that case, design a digital pattern using the TIFF-mode. For features that exceed the dimensions of one DMD and are relatively tall, divide the patterning of the substrate into multiple steps. In this case, multiple patterns are designed using the PDF-mode that all individually focus on single focal planes.
- Design a digital mask using design software tools.
- Patterning in TIFF-mode (pixels): Create a completely black artboard of 1,140 x 1,824 pixels (the exact size of 1 DMD, approximately 300 µm x 500 µm) and fill the artboard with the shapes of interest. Export the artboard as an 8-bit TIFF-file.
NOTE: Different grey levels in the pattern design determine the amount of exposure that will be performed on that location. - Patterning in PDF mode (metric units): Create a black artboard of the desired size in mm and fill the artboard with any shape of interest. Divide the pattern into multiple files if the 3D substrate needs to be patterned using multiple focal planes. Save the artboard as a PDF-file.
NOTE: Different grey levels in the pattern design determine the amount of exposure that will be performed on that location.
- Patterning in TIFF-mode (pixels): Create a completely black artboard of 1,140 x 1,824 pixels (the exact size of 1 DMD, approximately 300 µm x 500 µm) and fill the artboard with the shapes of interest. Export the artboard as an 8-bit TIFF-file.
6. UV-photopatterning of 3D cell culture substrates
- Calibration
NOTE: Calibration of the laser is done to get proper focus on the material of interest. Since the PDMS cell culture substrates are too thick to pattern through, use a glass slide on which the chip is placed upside-down. Since the laser encounters the glass slide first, use glass to calibrate the laser.- Apply fluorescent highlighter on a glass coverslip and place the glass coverslip in the stage of the fluorescent microscope. Ensure that the highlighted surface is facing upwards.
- Switch on the microscope and the PRIMO equipment and open Micro-manager to access Leonardo software under 'plugins'.
- Choose Calibration in the initial menu, followed by selecting the 20x objective both on the microscope and in the software. Click on Next.
- Place the glass slide with fluorescent highlighter in the optical path of the microscope. Switch to fluorescent mode and carefully focus on the PRIMO image that appears, making sure that both the logo and text are in focus. Click on Next to finish the calibration procedure.
- Write down the Z-location of the stage when calibrating and use this location as a reference later in the protocol.
NOTE: The calibration material should match the material that is used for patterning later in the protocol. The above steps describe the calibration needed for the cell culture substrates. For calibration of flat PDMS control samples, apply fluorescent highlighter on top of an additional flat PDMS sample and calibrate using Steps 6.1.2-6.1.5.
- Patterning 3D features using a single focal plane
NOTE: Patterning using a single focal plane is done on 3D features that do not exceed the dimensions of one DMD (approximately 300 µm x 500 µm). A schematic of the focal plane setup is illustrated in Figure 3.- Remove the calibration slide from the stage and place a glass slide containing a droplet (~50 µL) of photoinitiator (PLPP) in the stage.
CAUTION: PLPP is irritating to the eyes, the respiratory system, and the skin (R36-38). Wear personal protective equipment, keep it away from explosive materials, never add water to this product, take precautionary measures against static discharges, and avoid shock and friction (S26-36). - Place the PDMS cell culture substrate upside-down in the droplet of photoinitiator. Ensure that the 3D features on the surface of the cell culture substrate face the glass slide and are fully submerged in PLPP to ensure proper patterning.
- Select Pattern in the software.
- Switch to brightfield mode on the microscope and move the stage to the feature of interest.
- Focus on the top or bottom of convex and concave structures respectively (Figure 3).
- Select PRIMO to insert a pattern of choice. Observe the pattern preview in orange on top of the live brightfield image.
- Adjust the patterning settings according to the feature (location, angle, repetitions) and select a dose of 1,000 mJ/mm2.
- Click on Lock and switch the microscope to fluorescent mode.
- Click on the Play button on the right bottom of the screen to start the patterning. Once finished, observe the pattern displayed in green.
- Once finished with all features on a cell culture chip, remove the chip from the patterning slide and store in PBS at 4 °C.
- Remove the calibration slide from the stage and place a glass slide containing a droplet (~50 µL) of photoinitiator (PLPP) in the stage.
- Patterning 3D features using multiple focal planes
NOTE: Patterning using multiple focal planes is done on 3D features that are larger than one DMD (approximately 300 µm x 500 µm) or are relatively tall. In this case, the 3D features need to be patterned in multiple steps, that is, the pattern design should be adapted according to the example in Figure 3.- Remove the calibration slide from the stage and place a glass slide containing a droplet (~50 µL) of photoinitiator (PLPP) in the stage.
- Place the PDMS cell culture chip upside-down in the droplet of photoinitiator using tweezers.
- Select Pattern in the software. Switch to brightfield mode on the microscope and move the stage to the feature of interest.
- Focus on the area of the chip on the right location. Since patterning is performed in a single focal plane and the features are larger than a single DMD, use multiple focal planes and thus multiple rounds of patterning per 3D substrate to ensure sufficient pattern resolution along the complete height and width of the feature (see Figure 3). For example, for a feature with a height of 150 µm, pattern the feature in three rounds (± 1 focal plane per 50 µm of Z-travel), focusing around 25 µm, 75 µm, and 125 µm from the bottom of the feature. Make sure that the bottom of the feature is around the value written down in step 6.1.5.
- Select PRIMO to insert the pattern of choice. Observe the pattern preview shown in orange on top of the live brightfield image.
- Adjust the patterning settings according to the feature (location, angle) and select a dose of 1,000 mJ/mm2.
- Click on Lock and switch the microscope to fluorescent mode.
- Click on the Play button on the right bottom of the screen to start the patterning. Once finished, observe the pattern displayed in green.
- Repeat steps 6.3.4-6.3.8 on the feature of interest when multiple focal planes are used.
- Once finished with all features on a cell culture chip, remove the chip from the patterning slide and store it in PBS at 4 °C.
NOTE: If desired, apply the UV-photopatterning under sterile conditions, making use of glass-bottom Petri dishes and prepare all substrates in sterile culture cabinets. Store the patterned samples in PBS for up to a couple of weeks. When washed with ddH2O and dried using pressurized air, samples can be stored up to a few months at 4 °C.
7. Protein incubation
NOTE: It is advised to use freshly protein-incubated substrates for cell culture. Only proceed to this part of the protocol if the cell seeding (step 8) is done directly afterwards.
- Transfer the patterned cell culture chip to sterile PDMS containers in the culture cabinet.
- Wash the patterned cell culture chips 3x with an excess of sterile PBS if the patterning was not performed under sterile conditions.
- Prepare a fresh protein solution in PBS.
- Fibronectin: add 2 mL of PBS using a micropipet to a vial of 20 µg of rhodamine-labeled fibronectin to obtain a concentration of 10 µg/mL. Pipet gently to avoid the formation of protein clumps and protect from light.
- Gelatin: thaw a 200 µL aliquot of dissolved gelatin-fluorescein. Protect from light.
- Add 200-500 µL of the protein solution to the cell culture chip using a micropipet. Adjust the incubation time and temperature depending on the protein of choice: Fibronectin: 5 min at room temperature, Gelatin: 15 min at 37 °C. Make sure to cover the sample (e.g., with aluminum foil).
- Remove the protein solution and wash 5x with 500 µL of sterile PBS. Make sure to pipet the PBS up and down multiple times above all relevant features of the cell culture chip to remove any unbound protein.
NOTE: During the washing steps, it is crucial that the sample never dries out. When removing the protein solution or PBS during washing, add new PBS immediately to prevent protein clumps from forming (see Figure 4). Convex features are especially sensitive to drying out as they are elevated above the substrate surface. - Optional: review the protein patterns under a fluorescence microscope. Keep the samples sterile and immersed in PBS.
8. Cell seeding
NOTE: This protocol uses human primary keratocytes and human dermal fibroblasts. The keratocytes were harvested from human corneal tissue from patients, in line with Dutch guidelines for secondary use of materials, and previously characterized as keratocytes47. These cells are cultured in DMEM supplemented with 5% Fetal Bovine Serum (FBS), 1% penicillin/streptomycin (P/S), and 1 mM L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (vitamin C) at 37 °C for a maximum of four passages. Human dermal fibroblasts were purchased and cultured in DMEM supplemented with 10% FBS and 1% P/S at 37 °C for a maximum of 15 passages. For seeding of both keratocytes and dermal fibroblasts on the photopatterned cell culture chip, 20,000 cells per chip were used.
- Detach the cells of interest (e.g., using trypsin) and prepare 1 mL of a cell suspension of ± 10,000-50,000 cells/mL in culture medium per chip. Adjust the exact number of cells added per substrate depending on the cell size and the desired readout.
- Remove the PBS from the cell culture chip and add 1 mL of the cell suspension.
- Gently transport the cell culture chip with the cells to the incubator and incubate for 60 min at 37 °C.
- Check the adhesion of the cells on the patterned cell culture chip under a brightfield microscope. Look for elongated cell morphologies in the case of line patterns (see Figure 5). If cells have also started adhering outside the patterned area, remove these by pipetting medium up and down directly above the cells on the substrate.
NOTE: Cells attached to the patterned area will remain attached, while cells outside the patterned areas will detach. - Remove the cell culture chip from the PDMS container and use sterile tweezers to put it in a 6-well plate filled with ~5 mL of culture medium.
- Culture the cells for the desired length of time. Replace the cell culture medium every 2-3 days. To ensure the visualization of the protein pattern after cell culture, reduce the amount of exposure of the sample to light during culture.
9. Staining, image acquisition, and analysis
- Fixation and staining
- After the desired length of culture, remove almost all the medium and wash three times with excess PBS. Next, incubate with 3.7% formalin for 15 min at room temperature followed by three washing steps with PBS for 5 min per wash at room temperature. Never let the sample dry out.
CAUTION: Formalin is harmful if swallowed or if inhaled (H302, H332), may cause an allergic skin reaction (H317), is suspected of causing genetic defects (H341), and may cause cancer (H350). Wear personal protective equipment and work in a fume hood. - Stain the cell culture substrate with the desired staining agents or antibodies. To reduce the volumes of staining agents, place the cell culture chips upside-down in a droplet of staining solution pipeted on a glass slide.
- Store the samples in PBS (short-term) or attached to a coverslip using mounting medium (long-term) at 4 °C.
- After the desired length of culture, remove almost all the medium and wash three times with excess PBS. Next, incubate with 3.7% formalin for 15 min at room temperature followed by three washing steps with PBS for 5 min per wash at room temperature. Never let the sample dry out.
- Image acquisition
- Place the stained sample upside down in a droplet of PBS on a glass slide. Put the sample in the stage of a confocal microscope.
- Depending on the required level of detail, make Z-stacks with a suitable objective (10x, 20x, or 40x) and Z-spacing to ensure proper image acquisition.
- Image analysis and visualization
- Open the raw image files in image analysis software and check that the image properties (e.g., dimensions, resolution) are correct.
- Adjust the brightness and contrast per channel if needed.
- Crop the region of interest containing the pattern and cells.
- Optional: perform a deconvolution step if needed.
- Create a 3D render of the Z-stack using 3D rendering software.
- Optimize the contrast and gain settings for each individual channel.
- To make an image of the 3D render, create a snapshot and export as .TIFF file.
- To create a movie of the 3D render, set the start and final frame as well as the timeframe before registering the movie. Export as .avi.
Representative Results
By means of the described protocol, 3D PDMS cell culture substrates can be UV-photopatterned to create precise and high-throughput adhesive areas suitable for cell attachment. In this way, cells are subjected to both relevant substrate geometries and adhesive ligand patterns simultaneously. Cell properties such as orientation, cell area, and number of focal adhesions can easily be monitored and used to better understand cell behavior in complex, in vivo-like environments.
To verify the patterning events on the 3D PDMS substrates, atomic surface compositions of the material in different stages of the protocol have been measured using atomic X-ray photoelectron spectroscopy (XPS)48. In summary, the XPS measurements showed the presence of PEG chains with an increased carbon signal on passivated samples, which was reduced after photopatterning. Incubation with fibronectin resulted in an increase of carbon signal, again indicating successful protein adhesion on the surface of the cell culture chip. Next, pattern resolution and alignment on 3D features were characterized on a variety of circle-patterned, concave pits (ĸ = 1/250 µm-1, ĸ = 1/1,000 µm-1, and ĸ = 1/3,750 µm-1, see Figure 6). From the maximum-intensity projections, it can be concluded that the protein pattern was successfully patterned on all three 3D features. The intensity profile in Figure 6A shows high pattern resolution with sharp transitions between patterned and non-patterned areas. Additionally, consistent protein intensity across the complete pattern in the pit was obtained.
The concave pit with ĸ = 1/250 µm-1 was patterned using the single focal plane method (one pattern), whereas the pits with ĸ = 1/1,000 µm-1 and ĸ = 1/3,750 µm-1 were patterned using two and three focal planes (patterns), respectively. As can be seen in the maximum-intensity projections in Figure 6, both methods result in perfect alignment of the patterns on top of the features. No misaligned transitions between the two different focal planes and patterns can be observed.
Using the described protocol, a wide range of protein pattern designs can be applied to a variety of geometries (see Figure 7 and Video 1). To illustrate the versatility of this method, semicylinders (convex and concave), a saddle surface, and pit were patterned using lines and circles of various widths. The photopatterned materials can subsequently be used for cell culture (see Figure 7, Figure 8, Video 2, Video 3, and Video 4). An example of dermal fibroblasts cultured on a patterned (fibronectin lines, red, 5 µm wide, and 5 µm gaps) concave semicylinder is shown in Figure 8, Figure 9, and Video 4. During the experiment, cells sense and adhere to the multicue cell culture substrate and remain viable over time. As can be seen from the immunofluorescent staining in Figure 8, cells form focal adhesions (vinculin clusters) mainly on the fibronectin lines.
Another example study making use of these cell culture materials was recently published by our group48. In this study, human myofibroblasts and endothelial cells were subjected to the combination of contact guidance cues and geometrical topographies. In vivo, both types of cells experience curvature- and contact-guidance cues in native tissues such as in the human vasculature. By subjecting the cells in vitro to an environment that combines both environmental cues, the in vivo situation can be recapitulated, providing a deeper understanding of the role of the microenvironment on cell behavior. Human myofibroblasts were shown to align with contact guidance cues (parallel fibronectin lines) on concave cylindrical substrates48. However, on convex structures with increasing curvatures, the geometrical cues overruled the biochemical cues, suggesting that myofibroblasts can sense both the degree and sign of curvature. Interestingly, endothelial cells could only adhere to the concave multicue substrates and not to the convex PDMS substrates. On concave, protein-patterned substrates, the endothelial cells are oriented in the direction of the contact guidance cue. This fundamental in vitro knowledge has physiological relevance in the field of vascular tissue engineering and can eventually aid in the design of smart tissue engineering constructs.
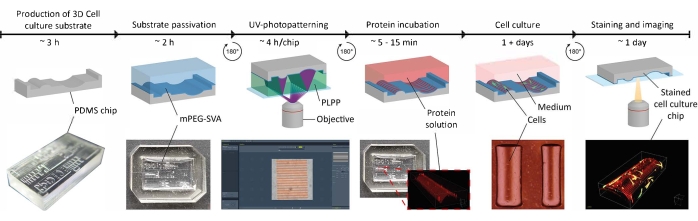
Figure 1: The experimental timeline of applying contact guidance cues on 3D cell culture substrates. First, positive cell culture chips are produced from a negative PDMS mold containing a range of geometries. Uncured PDMS is poured into the mold and cured for 3 h at 65 °C. Subsequently, the PDMS is treated with O2-plasma and incubated with PLL and mPEG-SVA (blue, labeled) to passivate the surface of the cell culture substrate. After washing, the substrate is flipped upside-down in a droplet of photoinitiator (PLPP, green, labeled) and UV-photopatterned using the LIMAP approach. Here, a digital mask with a user-defined pattern is used to cleave the passivation layer at defined locations. Next, a protein solution (red, labeled) can be incubated and will only adhere to the locations where the passivation layer is removed. Cells seeded on the substrate are subjected to both geometry and protein patterns, which enables research into cell behavior in complex, in vivo-mimicking environments. Abbreviations: PDMS = polydimethylsiloxane; mPEG-SVA = methoxypolyethylene glycol-succinimidyl valerate; PLPP = 4-benzoylbenzyl-trimethylammonium chloride; LIMAP = Light-Induced Molecular Adsorption of Proteins. Please click here to view a larger version of this figure.
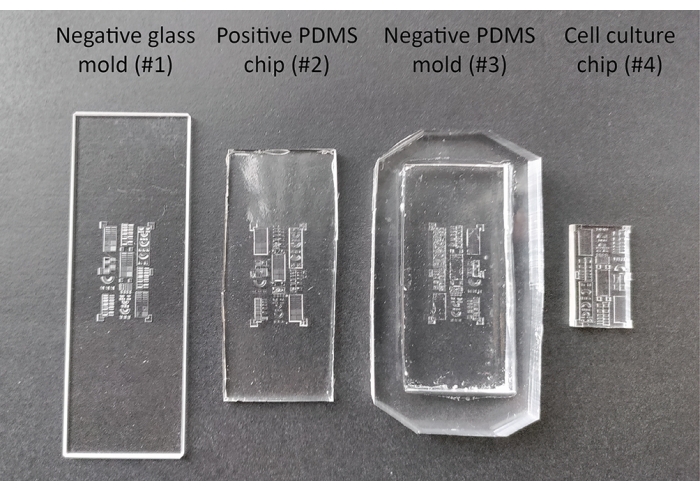
Figure 2: Different stages during the production and passivation of the 3D cell culture substrate. The negative glass mold (#1) is designed with computer assisted design software and produced using a femtosecond-laser direct-write technique. This mold is utilized to produce the intermediate positive PDMS chip (#2) and negative PDMS mold (#3), which are subsequently used to produce the final cell culture chip (#4). Abbreviation: PDMS = polydimethylsiloxane. Please click here to view a larger version of this figure.
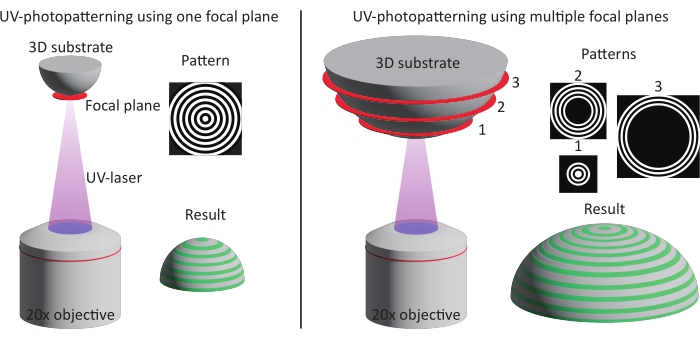
Figure 3: Schematic illustration of the two patterning methods. Left: UV-photopatterning is performed on smaller features (approximately one DMD) using a single focal plane and pattern. As a result, the complete feature is patterned in one go. Right: When larger features are used (larger than one DMD), the patterning is divided over multiple focal planes and patterns. Abbreviation: DMD = digital mirror device. Please click here to view a larger version of this figure.
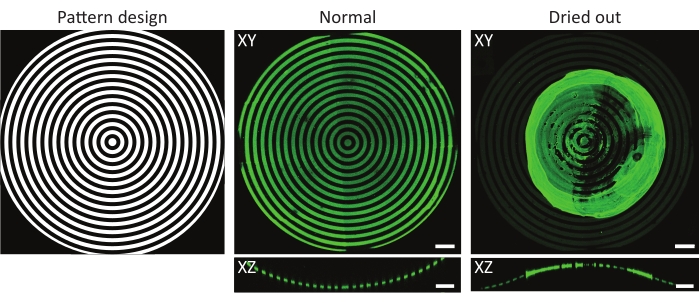
Figure 4: A typical example of a normal and dried-out protein-incubated substrate. Maximum-intensity projections (XY) and orthogonal views (XZ) of normal and dried-out protein-incubated substrates. When washing a patterned cell culture substrate after incubation with a protein solution, it is crucial to always keep the sample wet. Although the pattern is identical in all images on the features (ĸ = 1/1,000 µm-1), the gelatin-fluorescein (green) aggregated forming a major clump when the sample was left to dry for a few seconds. If the sample always remains wet, correct protein patterns can be observed. Scale bars = 100 µm. Please click here to view a larger version of this figure.
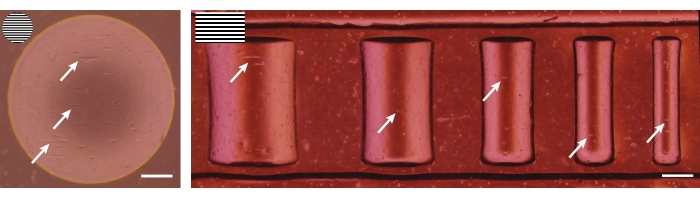
Figure 5: Brightfield images after seeding. Primary keratocytes (left) and dermal fibroblasts (right) 4 h after seeding on 3D geometrical features (concave pit of ĸ = 1/1,000 µm-1 and semicylinders of ĸ = 1/500, 1/375, 1/250, 1/175, and 1/125 µm-1). The top-left inserts represent the line pattern used for the patterning of the geometries. White arrows indicate spreading cells that already show alignment. Scale bars = 250 µm. Please click here to view a larger version of this figure.
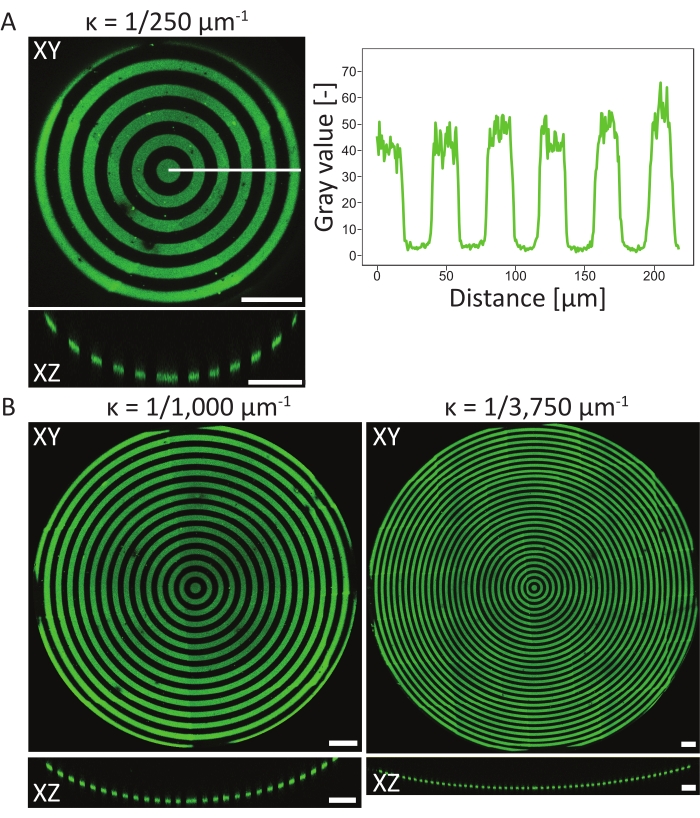
Figure 6: Characterization of circular patterns on concave pits. (A) The maximum-intensity projection (XY) and orthogonal view (XZ) of aconcave pit (ĸ = 1/250 µm-1) patterned using LIMAP (linewidth: 20 µm, gap width: 20 µm) and incubated with gelatin-fluorescein (green). The intensity profile along the white line is plotted against the distance, showing a consistent pattern quality and resolution. (B) Additional patterning performed on concave pits with ĸ = 1/1,000 µm-1 and ĸ = 1/3750 µm-1, showing flexibility in terms of geometrical features that can be used for patterning. Again, both the maximum intensity projections (XY) and orthogonal views (XZ) are visualized. Scale bars = 100 µm. Please click here to view a larger version of this figure.
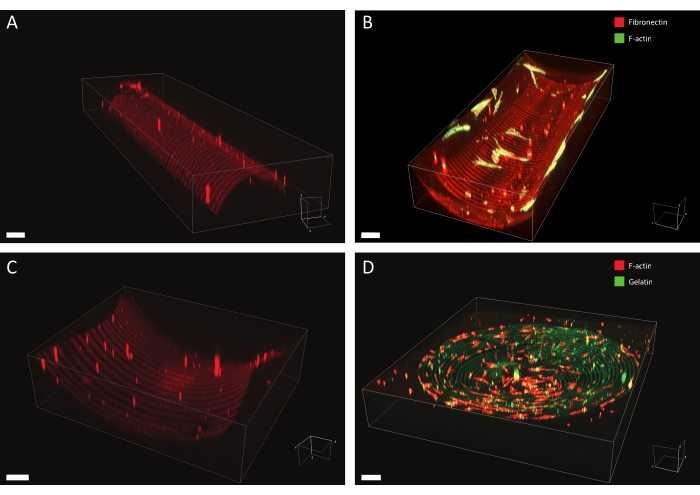
Figure 7: 3D microscopy data of patterned structures. Typical examples of 3D patterned cell culture materials after photopatterning and cell culture, visualized using 3D rendering software. (A) Convex semicylinder patterned with 10 µm wide lines (rhodamine-fibronectin, red) and 10 µm wide gaps. Scale bar = 5 µm. (B) Dermal fibroblasts stained for F-actin (green) cultured on concave semicylinder patterned with 20 µm wide lines (rhodamine-fibronectin, red) and 20 µm wide gaps. Scale bar = 5 µm. (C) Saddle surface patterned with 20 µm wide lines (rhodamine-fibronectin, red) and 20 µm wide gaps. Scale bar = 5 µm. (D) Concave pit patterned with concentric circles of 20 µm wide lines (gelatin-fluorescein, green) and 20 µm wide gaps. The F-actin cytoskeleton of the human keratocytes is stained using phalloidin and visualized in red. Scale bar = 200 µm. Please click here to view a larger version of this figure.
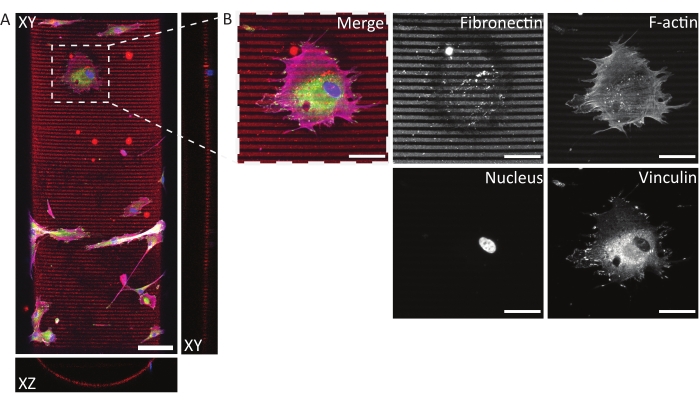
Figure 8: Immunofluorescent staining of human dermal fibroblasts on a photopatterned, concave semicylinder. (A) Maximum-intensity projection (XY) and orthogonal sections (XZ and YZ) of human dermal fibroblasts cultured for 24 h on a patterned (fibronectin lines, red, 5 µm wide, and 5 µm gaps) concave semicylinder. Cells are stained for F-actin (magenta), vinculin (green), and nuclei (blue). Scale bar = 100 µm. (B) Zoom-in of a cell adhering to the multicue environment. Scale bars = 50 µm. Please click here to view a larger version of this figure.
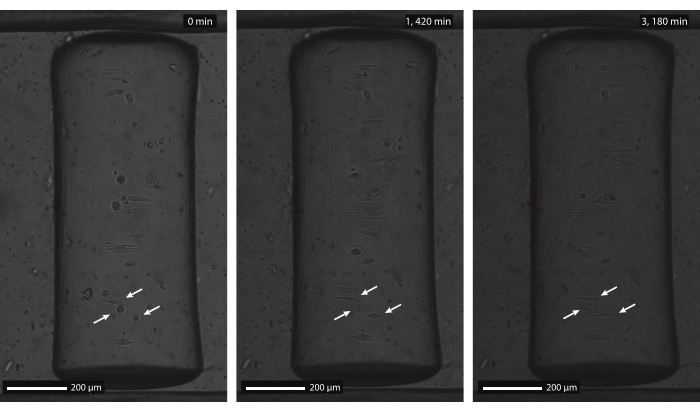
Figure 9: Brightfield timelapse images of human dermal fibroblasts on a patterned, concave cylinder. The concave semicylinder (ĸ = 1/250 µm-1) was patterned with parallel lines (5 µm wide and 5 µm gaps) and incubated with rhodamine-fibronectin before cell seeding. The timelapse imaging is started 1 h after initial cell seeding (left, 0 min), when cells are still rounded and non-adherent (arrows). After approximately 24 h (middle, 1,420 min), cells adhered to the multicue substrate and show an alignment response according to the contact guidance pattern. Both the alignment response and cell viability are maintained throughout the entire culture duration (right, 3,180 min). Scale bars = 200 µm. Please click here to view a larger version of this figure.
Video 1: Pattern example on a 3D cylindrical substrate. 3D representation of a convex cylinder patterned with rhodamine-fibronectin (red). Please click here to download this Video.
Video 2: 3D representation of dermal fibroblasts cultured on a patterned, 3D cylindrical substrate (ĸ = 1/500 µm-1). Dermal fibroblasts cultured for 24 h on a patterned (fibronectin lines, red, 10 µm wide, and 10 µm gaps) convex semicylinder. Cells are stained for F-actin (magenta), vinculin (green), and nuclei (blue). Please click here to download this Video.
Video 3: 3D representation of human keratocytes cultured on a patterned, 3D pit (ĸ = 1/3,750 µm-1). 3D representation of human keratocytes cultured for 24 h in a concave, patterned pit (gelatin circles, green, 20 µm wide, and 20 µm gaps). Cells are stained for F-actin (red). Please click here to download this Video.
Video 4: Brightfield timelapse imaging of human dermal fibroblasts on a patterned, concave cylinder. The concave semicylinder (ĸ = 1/250 µm-1) was patterned with parallel lines (5 µm wide and 5 µm gaps) and incubated with rhodamine-fibronectin before cell seeding. The timelapse imaging is started 1 h after initial cell seeding, when cells show initial adherence to the multicue environment. During the complete timelapse, cells predominantly orient along the contact guidance cues, while cell viability is maintained. Please click here to download this Video.
Discussion
Nowadays, cell behavior is often studied on flat culture substrates that lack the complexity of the native cell microenvironment. 3D environments such as scaffolds and hydrogels are used as an alternative. Although these cell culture environments improve the in vivo relevance, both systematic studies of cell behavior and the feasibility of readout methods remain challenging. To systematically investigate cell behavior on representative culture substrates, consistent multicue substrates that allow for microscopic readout are needed. Therefore, in this protocol, we describe a method to create multicue cell culture substrates with physiologically relevant geometries and patterned ECM proteins. The main challenge in combining environmental cues such as tissue geometry and contact-guidance cues on in vitro platforms is largely of a technological nature. Conventional methods to apply contact-guidance cues (e.g., soft lithography, deep-UV patterning, and microcontact printing35,36) to cell culture materials have been optimized for planar substrates. The need for 3D cell culture materials combined with contact guidance cues highlighted several technological challenges, such as poor pattern alignment, resolution, and flexibility. To overcome these challenges, a high-throughput, maskless, light-based patterning method can be used45,49. Here, an optical microscope enables precise pattern alignment and a resolution in the order of micrometers (see Figure 6). Further, the use of a digital mask enables researchers to study cell behavior on a wide range of patterns without the need to fabricate labor-intensive physical masks.
The UV-photopatterning approach can be used in combination with a variety of 3D geometries (e.g., cylinders, saddles, domes, pits) produced from a range of materials48. The 3D cell culture substrates used in this study are made from PDMS; however, other materials can be used as well. This might require different steps to produce the final cell culture substrate containing the features of interest. Since cells have been shown to be sensitive to surface roughness of cell culture materials, it is important to create the cell culture chips with a smooth surface so that the observed cells’ response can be fully attributable to the 3D geometry and contact-guidance cues50,51. Measurement methods such as optical profilometry, scanning electron microscopy, or atomic force microscopy can be used to measure surface roughness. After fabrication of the cell culture material, one can select a patterning method based on one or multiple focal planes dependent on the specific dimensions of the feature of interest (see Figure 3). Typically, a single focal plane is used to pattern an area within a Z-range of approximately 50 µm. The pattern resolution was shown to be consistent using this rule of thumb (see Figure 6). However, a downside of this method is the increased patterning time with the introduction of multiple focal planes and patterns. In our hand, using multiple focal planes, 3D geometrical features up to 16 mm x 16 mm x 0.17 mm (X x Y x Z) have been successfully patterned with high pattern quality.
Additionally, it is important to mention that the height (Z-axis) of geometrical features that can be used in combination with this protocol is limited. Since both the UV-photopatterning and many cellular readouts rely on a microscopy setup, the working distance of the objectives determines the maximum height of a feature. In our hand, geometries exceeding a height of 300 µm can still be UV-photopatterned, and readouts have been performed using a confocal microscope with 40x objectives. Thus, mechanobiological research ranging from intracellular to cellular and tissue scales is possible using the described protocol.
Another factor that needs to be taken into account is the risk of samples drying out during or after the UV-photopatterning49. This is especially relevant when using 3D geometries, as convexities are often exposed outside cell culture materials. As shown in Figure 4, this might result in uneven patterns with protein aggregates forming on top of the feature of interest. The washing of the cell culture chips following protein incubation and during cell culture is critical for the proper coating of 3D geometries. Therefore, it is advised to always leave small volumes of a working solution (PBS, PLPP, protein solution, cell culture medium) on top of the cell culture chip.
So far, several protein coatings (fibronectin, collagen type I and IV, gelatin, FNC) and cell types (human bone marrow stromal cells, human myofibroblasts, human endothelial cells, human keratocytes, and dermal fibroblasts) have been used in combination with the described photopatterning approach on structured cell culture materials. As shown in a previous study48, the optimization of protein incubation parameters is key for the systematic investigation in new cell types. Therefore, before performing a new experiment with new proteins or cells, it is advised to test a range of protein concentrations, incubation temperatures, and incubation times. By comparing cell morphology after initial adhesion on homogeneous, patterned flat areas with cell morphology under ‘normal’ cell culture conditions, an optimized set of experimental parameters can be obtained. Additionally, each cell type might require a different amount of time after seeding to show a recognizable adhesion morphology on a specific multicue environment (see Figure 5). To this end, it is crucial to optimize the time required per cell type to present contact events on the patterned area during the washing in step 8.4. For example, we observed that on line patterns, human keratocytes show elongated morphologies within the first 30 min after seeding, while endothelial cells and dermal fibroblasts require multiple hours before showing a change in adhesion morphology. The experimental parameters required for protein incubation (step 7) and cell seeding (step 8) might thus be dependent on the protein and cell type of choice.
The presented approach to apply contact guidance cues on 3D geometries can aid in creating a deeper understanding of cell behavior in complex multicue environments. This can include investigations into intracellular components, such as focal adhesions and nuclei, and can also involve experiments performed on a larger, cell, or tissue scale using the proposed method. Eventually, it is anticipated that the knowledge gained can be used in the design of tissue engineering applications, where complex cellular environments are designed to steer cell behavior towards a desired outcome.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Nello Formisano (MERLN Institute for Technology-Inspired Regenerative Medicine) for providing human primary keratocytes. This work has been supported by the Chemelot InSciTe (project BM3.02); the European Research Council (grant 851960); and the Ministry of Education, Culture and Science for the Gravitation Program 024.003.013 "Materials Driven Regeneration". The authors would like to thank Alvéole for their correspondence, help, and troubleshooting.
Materials
| Anti-vinculin antibody, mouse monoclonal IgG1 | Sigma | V9131 | Dilution: 1/600 |
| Bovine Serum albumin, Fraction V | Roche | 10735086001 | |
| DMEM, high glucose, pyruvate | Gibco | 41966029 | |
| DMEM/F-12 + GlutaMAX (1x) | Gibco | 10565018 | |
| DMi8 epifluorescent microscope | Leica Microsystems | ||
| Ethanol | Biosolve | 0005250210BS | |
| Fetal Bovine Serum | Serana | 758093 | |
| Fiji/ImageJ, version v1.53k | www.imageJ.nih.gov | ||
| Fluorescent highlighter | Stabilo | 4006381333627 | |
| Fluorescin-labeled gelatin | Invitrogen | G13187 | Concentration: 0.01% |
| Formaldehyde solution | Merck | F8775 | |
| Glass coverslips 24 x 60 mm, #1 | VWR | 631-1575 | |
| Glass coverslips, ø = 32 mm, #1 | Menzel-Gläser | ||
| HCX PL fluotar L 20X/0.40na microscope objective | Leica | 11506242 | |
| HEPES | Gibco | 15630080 | |
| Human dermal fibroblasts | Lonza | CC-2511 | |
| Human primary keratocytes | MERLN Institute for Technology-Inspired Regenerative Medicine | ||
| Illustrator, Version 26.0.1 | Adobe | ||
| Laboratory oven | Carbolite | ||
| L-Ascorbic acid 2-phosphate sesquimagnesium salt hydrate | Sigma-Aldrich | A8960 | |
| Leica Application Suite X software, version 3.5.7.23225 | Leica Microsystems | ||
| Leonardo software, version 4.16 | Alvéole | ||
| Micro-manager, version 1.4.23 | Open imaging | ||
| Mowiol 4-88 | Sigma-Aldrich | 81381 | mounting medium |
| mPEG-succinimidyl valerate MW 5,000 Da | Laysan Bio | MPEG-SVA-5000 | Concentration: 50 mg/mL |
| Negative glass mold | FEMTOprint | ||
| NucBlue Live Readyprobes Reagent (Hoechst 33342) | Invitrogen | R37605 | 2 drops/mL |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140163 | |
| Petri dish (ø=100 mm) | Greiner Bio-one | 664160 | |
| Phalloidin Atto 647N | Sigma | 65906 | Dilution: 1/250 |
| Phosphate Buffered Saline | Sigma | P4417 | |
| Plasma asher | Emitech | K1050X | |
| PLPP (photoinitiator) | Alvéole | ||
| Poly-L-lysine, sterile-filtered | Sigma-Aldrich | P4707 | Concentration: 0.01% |
| PRIMO | Alvéole | ||
| Rhodamine-labeled fibronectin | Cytoskeletn, Inc. | FNR01 | Concentration: 10 µg/mL |
| Secondary antibody with Alexa 488, Goat anti-mouse IgG1 (H) | Molecular Probes | A21121 | Dilution: 1/300 |
| Secondary antibody with Alexa 555, Goat anti-mouse IgG1 (H) | Molecular Probes | A21127 | Dilution: 1/300 |
| Spin coater | Leurell Technologies Corporation | model WS-650MZ-23NPPB | |
| SYLGARD 184 Silicone Elastomer Kit | DOW | 1673921 | |
| TCS SP8X confocal microscope | Leica Microsystems | ||
| tridecafluoro(1,1,2,2-tetrahydrooctyl)trichlorosilane | ABCR | AB111444 | |
| TrypLE Express Enzyme (1x), no phenol red | Gibco | 12604013 | |
| Trypsin-EDTA (0.05%), phenol red | Gibco | 25300054 |
References
- Wang, Y., Wang, G., Luo, X., Qiu, J., Tang, C. Substrate stiffness regulates the proliferation, migration, and differentiation of epidermal cells. Burns. 38, 414-420 (2012).
- Viswanathan, P., et al. 3D surface topology guides stem cell adhesion and differentiation. Biomaterials. 52, 140-147 (2015).
- Peyton, S. R., et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnology and Bioengineering. 108 (5), 1181-1193 (2011).
- Vining, K. H., Mooney, D. J. Mechanical forces direct stem cell behaviour in development and regeneration. Nature Reviews Molecular Cell Biology. 18 (12), 728-742 (2017).
- Chaudhuri, O., Cooper-White, J., Janmey, P. A., Mooney, D. J., Shenoy, V. B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature. 584, 535-546 (2020).
- Guido, S., Tranquillo, R. T. A methodology for the systematic and quantitative study of cell contact guidance in oriented collagen gels. Correlation of fibroblast orientation and gel birefringence. Journal of Cell Science. 105 (2), 317-331 (1993).
- Teixeira, A. I., Abrams, G. A., Bertics, P. J., Murphy, C. J., Nealey, P. F. Epithelial contact guidance on well-defined micro- and nanostructured substrates. Journal of Cell Science. 116, 1881-1892 (2003).
- Driscoll, M. K., Sun, X., Guven, C., Fourkas, J. T., Losert, W. Cellular contact guidance through dynamic sensing of nanotopography. ACS Nano. 8 (4), 3546-3555 (2014).
- Buskermolen, A. B. C., et al. Cellular contact guidance emerges from gap avoidance. Cell Reports Physical Science. 1 (5), 100055 (2020).
- Thrivikraman, G., et al. Cell contact guidance via sensing anisotropy of network mechanical resistance. Proceedings of the National Academy of Sciences of the United States of America. 118 (29), 1-11 (2021).
- Buskermolen, A. B. C., et al. Entropic forces drive cellular contact guidance. Biophysical Journal. 116 (10), 1994-2008 (2019).
- Vignaud, T., et al. Reprogramming cell shape with laser nano-patterning. Journal of Cell Science. 125 (9), 2134-2140 (2012).
- Pouthas, F., et al. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. Journal of Cell Science. 121 (14), 2406-2414 (2008).
- Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M., Ingber, D. E. Geometric control of cell life and death. Science. 276 (5317), 1425-1428 (1997).
- Théry, M., et al. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proceedings of the National Academy of Sciences of the United States of America. 103 (52), 19771-19776 (2006).
- Huang, G., et al. Functional and biomimetic materials for engineering of the three-dimensional cell microenvironment. Chemical Reviews. 117 (20), 12764-12850 (2017).
- Callens, S. J. P., Uyttendaele, R. J. C., Fratila-Apachitei, L. E., Zadpoor, A. A. Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials. 232, 119739 (2020).
- Yilmaz, C. O., Xu, Z. S., Gracias, D. H. Curved and Folded Micropatterns in 3D Cell Culture and Tissue Engineering. Methods in Cell Biology. 121, (2014).
- Silver, F. H., Freeman, J. W., Seehra, G. P. Collagen self-assembly and the development of tendon mechanical properties. Journal of Biomechanics. 36 (10), 1529-1553 (2003).
- Werner, M., Kurniawan, N. A., Bouten, C. V. C. Cellular geometry sensing at different length scales and its implications for scaffold design. Materials. 13 (4), 963 (2020).
- Di Cio, S., Bøggild, T. M. L., Connelly, J., Sutherland, D. S., Gautrot, J. E. Differential integrin expression regulates cell sensing of the matrix nanoscale geometry. Acta Biomaterialia. 50, 280-292 (2017).
- Fioretta, E. S., Simonet, M., Smits, A. I. P. M., Baaijens, F. P. T., Bouten, C. V. C. Differential response of endothelial and endothelial colony forming cells on electrospun scaffolds with distinct microfiber diameters. Biomacromolecules. 15 (3), 821-829 (2014).
- Werner, M., Kurniawan, N. A., Korus, G., Bouten, C. V. C., Petersen, A. Mesoscale substrate curvature overrules nanoscale contact guidance to direct bone marrow stromal cell migration. Journal of The Royal Society Interface. 15 (145), 20180162 (2018).
- Ruprecht, V., et al. How cells respond to environmental cues – insights from bio-functionalized substrates. Journal of Cell Science. 130 (1), 51-61 (2017).
- Bao, M., Xie, J., Huck, W. T. S. Recent advances in engineering the stem cell microniche in 3D. Advanced Science. 5 (1800448), 1-16 (2018).
- Zhan, X. Effect of matrix stiffness and adhesion ligand density on chondrogenic differentiation of mesenchymal stem cells. Journal of Biomedical Materials Research – Part A. 108 (3), 675-683 (2020).
- Jiang, T., et al. Untangling the response of bone tumor cells and bone forming cells to matrix stiffness and adhesion ligand density by means of hydrogels. Biomaterials. 188, 130-143 (2019).
- Choi, J. S., Harley, B. A. C. The combined influence of substrate elasticity and ligand density on the viability and biophysical properties of hematopoietic stem and progenitor cells. Biomaterials. 33, 4460-4468 (2012).
- Rape, A. D., Zibinsky, M., Murthy, N., Kumar, S. A synthetic hydrogel for the high-throughput study of cell-ECM interactions. Nature Communications. 6 (8129), 1-9 (2015).
- Camarero-Espinosa, S., et al. 3D printed dual-porosity scaffolds: the combined effect of stiffness and porosity in the modulation of macrophage polarization. Advanced Healthcare Materials. 11 (2101415), 1-16 (2022).
- Bao, M., et al. Cellular volume and matrix stiffness direct stem cell behavior in a 3D microniche. ACS Applied Materials and Interfaces. 11 (2), 1754-1759 (2019).
- Charest, J. L., Eliason, M. T., García, A. J., King, W. P. Combined microscale mechanical topography and chemical patterns on polymer cell culture substrates. Biomaterials. 27, 2487-2494 (2006).
- Bilem, I., et al. Interplay of Geometric Cues and RGD/BMP-2 crosstalk in directing stem cell fate. ACS Biomaterials Science and Engineering. 3 (10), 2514-2523 (2017).
- Nam, K. -. H., et al. Multiscale cues drive collective cell migration. Scientific Reports. 6, 29749 (2016).
- Alom Ruiz, S., Chen, C. S. Microcontact printing: A tool to pattern. Soft Matter. 3 (2), 168-177 (2007).
- Azioune, A., Carpi, N., Tseng, Q., Théry, M., Piel, M. Protein Micropatterns. A Direct Printing Protocol Using Deep UVs. Methods in Cell Biology. 97, (2010).
- Kane, R. S., Takayama, S., Ostuni, E., Ingber, D. E., Whitesides, G. M. Patterning proteins and cells using soft lithography. Biomaterials. 20 (23-24), 2363-2376 (1999).
- Azioune, A., Storch, M., Bornens, M., Théry, M., Piel, M. Simple and rapid process for single cell micro-patterning. Lab on a Chip. 9 (11), 1640 (2009).
- Offenhäusser, A., et al. Microcontact printing of proteins for neuronal cell guidance. Soft Matter. 3 (3), 290-298 (2007).
- Ricoult, S. G., Sanati Nezhad, A., Knapp-Mohammady, M., Kennedy, T. E., Juncker, D. Humidified microcontact printing of proteins: universal patterning of proteins on both low and high energy surfaces. Langmuir. 30 (40), 12002-12010 (2014).
- Waterkotte, B., et al. Biofunctional Micropatterning of Thermoformed 3D Substrates. Advanced Functional Materials. 24 (4), 442-450 (2014).
- Lehnert, D., et al. Cell behaviour on micropatterned substrata: Limits of extracellular matrix geometry for spreading and adhesion. Journal of Cell Science. 117 (1), 41-52 (2004).
- Sevcik, E. N., Szymanski, J. M., Jallerat, Q., Feinberg, A. W. Patterning on topography for generation of cell culture substrates with independent nanoscale control of chemical and topographical extracellular matrix cues. Current Protocols in Cell Biology. 75, 1-25 (2017).
- Micropatterning: Surface functionalization. Alvéole Available from: https://www.alveolelab.com/technology/micropatterning-surface-functionalization/ (2022)
- Strale, P. O., et al. Multiprotein printing by light-induced molecular adsorption. Advanced Materials. 28 (10), 2024-2029 (2016).
- van Gaal, R. C., Miltenburg, R. P. R. S., Kurniawan, N. A., Bouten, C. V. C., Dankers, P. Y. W. Renal epithelial cell responses to supramolecular thermoplastic elastomeric concave and convex structures. Advanced Materials Interfaces. 8 (1), 2001490 (2021).
- Foster, J. W., Gouveia, R. M., Connon, C. J. Low-glucose enhances keratocyte-characteristic phenotype from corneal stromal cells in serum-free conditions. Scientific Reports. 5, 1-16 (2015).
- Van Der Putten, C., et al. Protein micropatterning in 2.5D: an approach to investigate cellular responses in multi-cue environments. ACS Applied Materials and Interfaces. 13 (22), 25589-25598 (2021).
- Melero, C., et al. Light-induced molecular adsorption of proteins using the primo system for micro-patterning to study cell responses to extracellular matrix proteins. Journal of Visualized Experiments. (152), e60092 (2019).
- Hou, Y., et al. Surface roughness gradients reveal topography-specific mechanosensitive responses in human mesenchymal stem cells. Small. 16 (10), 1905422 (2020).
- Bourkoula, A., et al. Roughness threshold for cell attachment and proliferation on plasma micro-nanotextured polymeric surfaces: The case of primary human skin fibroblasts and mouse immortalized 3T3 fibroblasts. Journal of Physics D: Applied Physics. 49 (30), 304002 (2016).

