A Semi-Automated and Reproducible Biological-Based Method to Quantify Calcium Deposition In Vitro
Summary
Cardiovascular disease is the leading cause of death worldwide. Vascular calcification contributes substantially to the burden of cardiovascular morbidity and mortality. This protocol describes a simple method to quantify vascular smooth muscle cell-mediated calcium precipitation in vitro by fluorescent imaging.
Abstract
Vascular calcification involves a series of degenerative pathologies, including inflammation, changes to cellular phenotype, cell death, and the absence of calcification inhibitors, that concomitantly lead to a loss of vessel elasticity and function. Vascular calcification is an important contributor to morbidity and mortality in many pathologies, including chronic kidney disease, diabetes mellitus, and atherosclerosis. Current research models to study vascular calcification are limited and are only viable at the late stages of calcification development in vivo. In vitro tools for studying vascular calcification use end-point measurements, increasing the demands on biological material and risking the introduction of variability to research studies. We demonstrate the application of a novel fluorescently labeled probe that binds to in vitro calcification development on human vascular smooth muscle cells and determines the real-time development of in vitro calcification. In this protocol, we describe the application of our newly developed calcification assay, a novel tool in disease modeling that has potential translational applications. We envisage this assay to be relevant in a broader spectrum of mineral deposition research, including applications in bone, cartilage, or dental research.
Introduction
Vascular calcification (VC) is an independent risk factor for cardiovascular morbidity and mortality1,2,3. Long considered a passive chemical process of ectopic mineral deposition, it now appears a modifiable tissue healing response involving the active contribution of various cells including activated vascular smooth muscle cells (hVSMC) as a driver of the disease4,5. In vivo VC can be measured by multislice CT scans as an assessment of atherosclerotic burden6,7,8. Currently, a paradigm shift is underway, wherein VC severity is becoming recognized as a risk factor in cardiovascular disease, type II diabetes, chronic kidney disease, and ageing9,10,11,12,13,14,15.
hVSMCs are the most abundant cell type in the cardiovascular system and a principal actor in the development of VC. In vitro hVSMC-induced calcification is a widely used disease model to study cardiovascular disease16,17. However, most protocols for the detection of in vitro calcification use end-point measurements that can limit data acquisition, require greater use of cellular material, and can slow research. Common methods for the detection of in vitro hVSMC calcification include the o-cresolphthalein assay, which measures solubilized calcium deposition against total protein and requires cell lysis18. Also, Alizarin Red staining is used, which binds directly to calcium deposits on fixed cells or tissue19. To study hVSMC calcification over time with either o-cresolphthalein or Alizarin Red requires batches of replicates per time point, increasing the demand on biological material, and in turn, increasing the chance of variability.
In this paper, we detail the method for the application of a novel assay that utilizes hVSMCs with a fluorescent imaging probe to determine in vitro VC progression as well as function as a singular end-stage calcification assay. We previously demonstrated that this assay is directly comparable to the o-cresolphthalein and Alizarin Red methods and can be used to distinguish between varying culture conditions20. In addition to real-time measurements, this assay may be used to determine the propensity of serum or plasma samples as a surrogate marker for clinical VC development20. This will aid in the application of biological strategies of cardiovascular sciences and disease modeling. A further application of the assay may be as a translational BioHybrid system to assess VC severity or progression from blood constituents such as serum or plasma.
Protocol
1. Cell seeding, maintenance, and calcification induction
- For culturing primary cells, use a laminar airflow cabinet, gloves, and sterile equipment. Disinfect hands and workspace before and after conducting any work. Treat all primary cells and culture media as a potential biohazard, unless proven otherwise. Preferably autoclave surplus cells and media before disposal. Do not chemically inactivate and autoclave since this will liberate toxic fumes.
- Culture hVSMC on uncoated cell culture plates.
- Routinely maintain hVSMC in growth medium consisting of M199 medium supplemented with 10%-20% FBS and 1% Pen/Strep and incubate at 37 °C and 5% CO2.
- Split the cells upon 70%-90% confluency.
- To split, wash the hVSMCs 2x with PBS. Add trypsin and incubate for 2-5 min at 37 °C. Check for the detachment of cells under the microscope.
- Inhibit the trypsin reaction by adding serum-containing medium. Centrifuge the cells at 350 x g for 4 min and resuspend the pellet in maintenance medium. hVSMCs follow a 1:2 splitting scheme.
- To start the calcification experiment, prepare the cells following instructions for a split (Step 1.4.1). Upon resuspension, count the cells and seed into a 48-well plate at a density 10-15 x 103 cells/cm2. Seed the cells avoiding the use of the outer wells, as recommended in Supplemental Figure 1.
- Let the cells adhere and recover for 24 h (overnight) while incubating at 37 °C and 5% CO2.
- Gently wash the cells 2x with PBS, carefully aspirating all the remaining PBS after the 2nd wash.
- Gently add calcification medium and further incubate at 37 °C and 5% CO2. Calcification medium must contain a calcification stimulus, fluorescently labeled fetuin-A (e.g., with red fluorescent protein [RFP]) (1 µg/mL), and HOECHST 33342 (0.1 µg/mL) nuclear stain or similar live fluorescent nuclear stain. Do not use DAPI, as this is non-permeable.
- Check daily with a light microscope until calcification occurs.
NOTE: For notes on the cell culture and maintenance of hVMSCs, please see Supplemental File 1.
2. Calcification detection via imaging
NOTE: The following protocol provides the general steps to be taken in preparation, imaging, and data analysis. Screenshots supporting the instructions for each step using an automated imaging platform and corresponding image analysis software (see Table of Materials for details) are provided in Supplemental File 2 and Supplemental File 3. Other imaging instruments and image processing tools may be used to apply this protocol. However, repeated imaging at the same location in each well is crucial for meaningful data acquisition. Creating a protocol to image calcification and re-use at every imaging step is necessary for obtaining reproducible results. The first time applying the method, follow the steps below to prepare before the imaging.
- Protocol setup
- Open the software, and then select Protocols and Create New.
- Select Procedure and click Set Temperature. In the pop-up window, set the temperature to 37 °C and the gradient to 1 °C, and click OK. Select the plate type of choice from the dropdown menu. Add the imaging step by clicking Image. Select the inverted imager, and click OK.
- Add three imaging channels and set them to DAPI (377, 447 nm), RFP (531, 593 nm), and brightfield. Tick the Montage box and set the desired number and location of the images. 2 x 2 for a 48-well plate is a common choice. Change the overlap to represent a better coverage of each well. Select which wells are to be imaged. A new window will open where the wells of interest can be selected.
- Click Focus Options to set the focus mode. A new window will open. Set each channel individually. Commonly, wells are autofocused on the "DAPI" channel. Set all other channels to Fixed Focal Height From First Channel and set by clicking OK. Close the window.
- Start the pre-setting data reduction steps by clicking Data Reduction and select Image Preprocessing. Setting up this step reduces the fluorescence background. Accept the default settings by selecting OK. A new set of images will be created with the prefix "TSF". The original images will be preserved.
- Set up a step to count the cells by selecting Cellular Analysis. Select TSF DAPI images from the Channel dropdown menu. Set further details after imaging has been performed. This step can also be considered data reduction; always make sure to keep the original images.
- Prepare the analysis of the RFP (calcification) signal by selecting Statistics. In the pop-up window, label Step and select TSF RFP as the input channel. Tick the Upper Value and Lower Value boxes. Tick the box for Total Area in the lower list. Select None in the Color Effect column. In the pop-up window, click Custom and tick the Background box. This will color code the results from low-high for easy assessment. Press OK.
- For a fair readout, normalize the RFP signal per cell. In doing so, a total area is divided per cell. To set up this step, press Ratio on the left side. In the pop-up window, select for data input 1 RFP Quantification: Total Area from the drop-down menu. Further select Cell Count as data input 2.
- Finally, select a New Data Set Name and press OK and OK again. Select Color Effect as described previously. In the upper-left corner, select File and save the file as protocol (.prt file type).
- Daily after the first calcification occurs
- For every time point, repeat these steps. In the startup software, press Read Now and Existing Protocol…. Select the protocol that has been created in the previous step. The program will ask to save the experiment. This can be done at this step but also at any later step manually by clicking the Save button on the upper left.
- A pop-up window will ask to wait for the system to heat. Turn on the CO2 gas-controller and set it to 5%.
- Once the system has reached the set temperature, a prompt will appear to insert a plate and read. Press Cancel. This is because, for each time point, the exposure of each fluorochrome must be adjusted individually.
- Transport the plate from the incubator to the imager in a safe box that resists breaking and spilling and is in line with local biosafety regulations for transporting live cells, in case of an accident. Place the plate into the plate reader.
- After canceling, click on Procedure. In the pop-up window, select Pre-Set Imaging Step. Adjust the focus and exposure on the DAPI channel first. Untick the Auto Exposure box at all channels. Then, click the Microscope icon next to the DAPI channel. A new window will open.
- Select the well to focus on. Use a well with medium to high calcification for this step. If a signal is already visible, first click Autofocus and then Autoexposure. If not, first increase the exposure and repeat Autofocus and Autoexposure. If desired, the exposure can be adjusted manually by selecting the exposure drop-down menu. Check the settings on multiple wells. Once satisfied, save the settings.
- Adjust the exposure in the same manner for all channels.
CAUTION: Do not focus on other channels; only adjust exposure. The focus plain should be the same for all channels. - Close the procedure window. Select the green Read Now button in the top task bar. Save the experiment as indicated by the software.
NOTE: The software will now automatically read all the selected wells in the selected settings.
3. Data analysis
NOTE: For detailed screenshots on how to perform the data analysis using an automated imaging platform and corresponding image analysis software (see Table of Materials for details), please see Supplemental File 4. If using alternative imaging instruments or analysis software, the images should be exported and batch processed ensuring that the exposure, fluorescence threshold, or intensity are adjusted equally for all images in a comparative data set.
- While the system reads the plate, all the images are automatically processed in the background. Adjust the settings for cell count by selecting TSF DAPI images to display. Select the well of choice by double-clicking. A new window will pop up. Select one of the images.
- Select the Analysis tab. In the pop-up window, select Cellular Analysis: Cell Count. Click OK. Deselect the RFP and brightfield channels. Tick the Highlight Objects box.
- Click Options to open the menu. Adjust the size and intensity threshold to include all nuclei but exclude debris. Click OK. Click Apply changes in the lower left to also transfer the settings to all other images.
- In a similar manner as before, select a well and image with a medium to high amount of calcification to best adjust the signal-to-noise ratio; click the Analysis tab in the task menu.
- This time select Image Statistics. Deselect the DAPI and brightfield channel. Click Options to open the menu. Tick the box Threshold Outliers. Adjust the threshold's upper and lower values. Only count the signal if it is above the background and exclude if there are debris/artifacts.
- To set a non-subjective threshold value for the signal above background, select the line tool from the task bar. A window will pop up. Draw a line through a patch of signal, but make sure to also include an area of background.
NOTE: A graph will appear in the pop-up window, showing a peak representing the signal above the background line. The 25% peak height value represents the recommended threshold. It is also recommended to measure multiple signal patches to ensure the selection of the right threshold. While selecting patches of signal, selecting regions with very high signal strength may result in a threshold that neglects medium and or lower signals. It is, therefore, recommended to select patches of medium to low signal above background. Examples of too high, too low, and accurate thresholds are shown in Supplemental File 4. - Once satisfied, click OK and Apply Changes to transfer the settings to all other wells. Check if the threshold is accurately selected in the other wells.
- Once the cell count and RFP thresholds are set, export the data. Select the plate of interest. In the drop-down menu, multiple parameters can be selected for export. Of relevance might be cell count, RFP area, and "area/cell" which is the metric used for comparison between groups. Click the Excel button next to the drop-down menu to export the data directly into a spreadsheet.
NOTE: In the dropdown menu, the calculated values for the cell count, the total area of RFP signal (reflecting the area of calcification), as well as the normalized ratio of total area of RFP signal per cell can now be selected individually and exported to a spreadsheet. Display the results as the ratio of total area of RFP signal per cell and visualize (e.g., bar graphs). Apply an unpaired Student's t-test to compare two groups or an unpaired ANOVA to compare different groups at the same time point. Comparing the same group at different time points may require paired statistical analysis.
Representative Results
The outcome includes original images of HOECHST-stained nuclei, RFP-labeled calcification, and brightfield images. Different stages of calcification ranging from low (Figure 2) to high (Figure 3) may be detected and analyzed. Calcification can usually be spotted as black speckles using light microscopy (Figure 2D and Figure 3B, arrows indicate calcification), which are useful for primary assessment and to determine when to start imaging. For improved signal-to-noise ratio, the processed RFP images should be analyzed to quantify calcification (Figure 2F and Figure 3D, arrows indicate calcification). Finally, data may be presented as a bar graph comparing two or more conditions at one time point, accompanied by representative images (Figure 4A,B,C). Data should be displayed normalized to cell count (e.g., as calcification area per cell). Data may also be displayed as time-series data showing the same condition at various time points (Figure 4D).
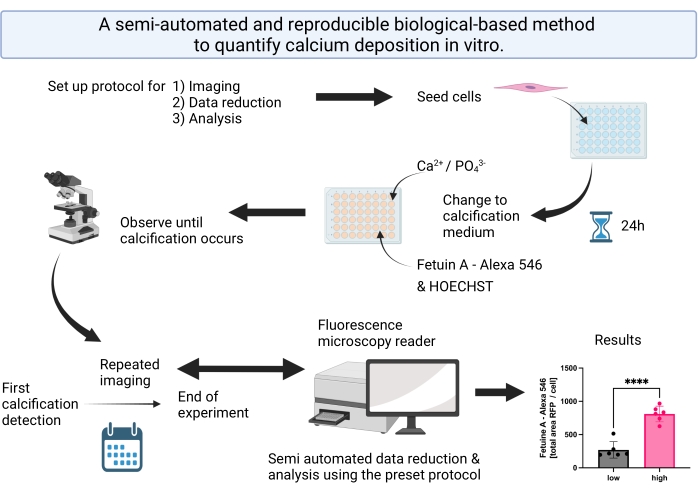
Figure 1: Visual abstract summarizing the steps for the semi-automated calcification detection and analysis. Please click here to view a larger version of this figure.
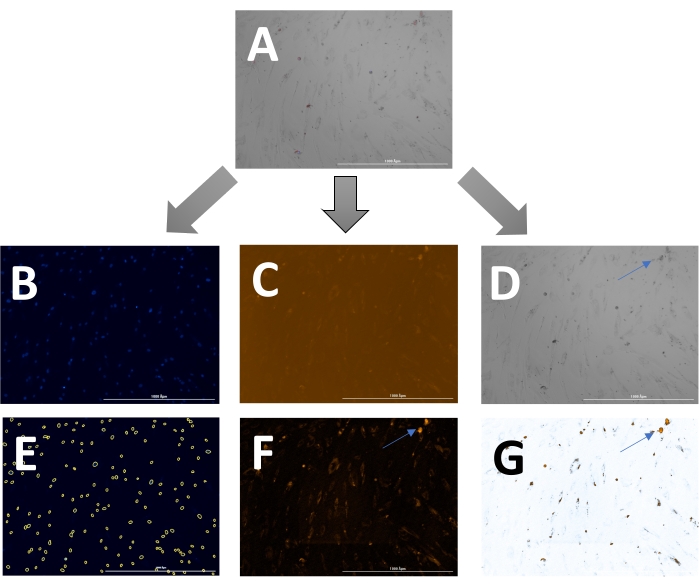
Figure 2: Example of early-stage calcification. (A)The overlay image can be displayed and analyzed as (B) separate DAPI (nuclei), (C) RFP (calcification), and (D) brightfield images. (E) Nuclei are identified by the software and can be highlighted as yellow circles to adjust the settings. (F) For analysis of the RFP signal, images are pre-processed to reduce background signal and, (G) subsequently, a threshold can be set to measure the signal. Arrows indicate calcification in the (D) brightfield and (F & G) transformed RFP images. Please click here to view a larger version of this figure.
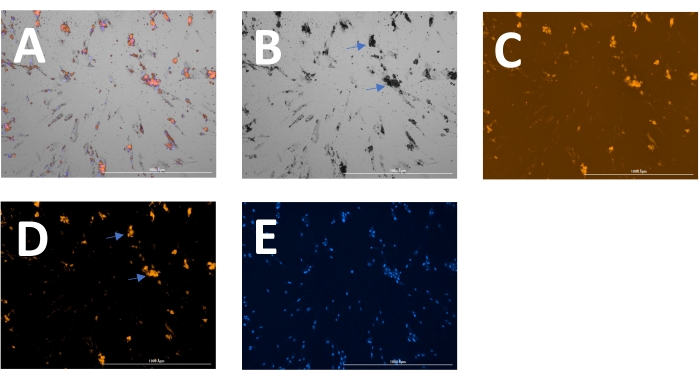
Figure 3: Example of later-stage calcification. (A) The overlay image can be displayed and analyzed as separate (B) brightfield, (C) RFP (calcification), and (E) DAPI (nuclei) images. (D) For analysis of the RFP signal, images are pre-processed to reduce background signal. Arrows indicate calcification in the (B) brightfield and (D) transformed RFP images. Please click here to view a larger version of this figure.
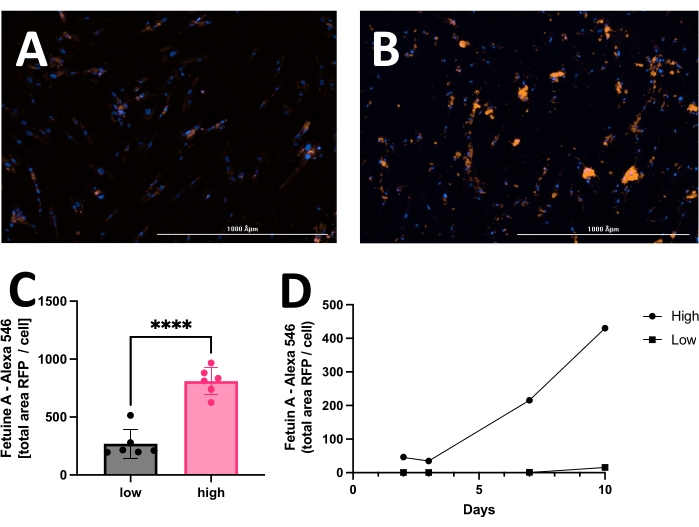
Figure 4: Representative comparison between low and high calcification of hVSMC after 14 days in culture with calcification medium. Commonly, data may be displayed as a bar graph and analyzed employing the unpaired Student's t-test. Representative images of (A) low calcification and (B) high calcification. The red signal (RFP) reflects calcification, and the blue signal (HOECHST) displays nuclei. (C) Calcification presented as fetuin A-RFP positive signal (total area) per cell. (D) Example of a calcification assay measured over time. Please click here to view a larger version of this figure.
Supplemental Figure 1: Example of a range of wells used for a calcification experiment with hVSMCs. The outer ring of the wells is not used for the calcification experiment but filled with liquid. Please click here to download this File.
Supplemental File 1: Notes on the cell culture and maintenance of hVMSC. Please click here to download this File.
Supplemental File 2: Automated imaging protocol. Please click here to download this File.
Supplemental File 3: Image analysis protocol. Please click here to download this File.
Supplemental File 4: Data analysis protocol. Please click here to download this File.
Discussion
In this manuscript, we describe a semi-automated method for in vitro calcification determination. For this method, three critical steps of hVSMC calcification should be optimized. First, cellular density is critical for hVSMC calcification development. Low densities of hVSMCs will result in slow or no calcification and cell death due to the lack of cell-to-cell contact and the stress that is induced under calcifying conditions21. High cellular densities result in over-confluency, after which cells become senescent22 and calcification development stops. It is critical to seed roughly 70% confluency in the well plate that will be used for subsequent calcification development, ensuring proliferative capacity and cellular connections of hVSMCs.
Secondly, the cell culture media that will be used for calcification induction requires optimization. Within vascular calcification research, a variety of conditions and media compositions have been reported to calcify hVSMCs23,24. We believe the method is suitable to detect all kinds of in vitro-mediated calcification, and it has been used to detect calcium, phosphate, or calcium-phosphate stimulated deposits. Regardless of the mode for calcification induction, optimization of the calcifying media is pivotal. In the presented protocol, we optimized calcification induction by using M199 with a total calcium concentration of 4.5 mM Ca2+ with 2.5% FBS.
Lastly, local differences in plates during calcification have been observed. It is critical to apply random loading of technical replicates to prevent sample bias. Additionally, loading of the outer well lanes should be avoided as these wells always calcify more rapidly in the 48-well setting. This is potentially caused by the dysregulation of intraplate humidity, wherein not using the outermost wells and loading these wells with large volumes of liquid helps control for this.
While the calcification assay itself can require multiple optimization steps to ensure reproducibility with a particular setup, once up and running this becomes straightforward. Calcification assays can be run simultaneously and repeatedly under established conditions without the need for further optimization. hVSMC-mediated calcification can be challenging and require experimentation before the robustness of assays has been achieved. A researcher should be able to determine the optimal timing before starting regular imaging, which can take up to 1 week. A fixed imaging schedule can be established from the start of the experiment, although this may produce many images without differential read-outs, use a relatively large amount of data storage for images, and be time-consuming in analysis.
The procedure described in this protocol is one way to perform the analysis of the calcification assay. For other purposes, the procedure should be adjusted accordingly. Our analysis using the referenced automated imaging platform ensures reproducibility, although analysis can be performed using any live cell and temperature- and CO2-controlled imaging device. Additionally, the corresponding commercial software packages are optimized for high content cellular screening and analysis, ideally suited to measuring calcification propensity over time. Other software solutions, such as the freeware ImageJ, can provide image analysis and quantify calcification development as well.
The imaging of late-stage calcification plates can be difficult due to issues with autofocusing should the culture encounter floating debris, resulting in a reduced number of sharp images and replicates. Image analysis should be adjusted accordingly, and some solutions have been developed in the software employed in this protocol to improve and simplify analysis.
Cellular heterogeneity plays a crucial role in the quantification of calcification in vitro. In this platform, we use hVSMCs as biosensors for the development of calcification. Primary hVSMCs are derived from various donors with different underlying vascular pathologies; therefore, this assay is still subject to high variability due to the heterogeneity of VSMCs batches. A possible solution is the use of immortalized cell lines or the use of hVSMCs derived from pluripotent stem cells.
Another limitation is that the quantification method is still sensitive to subjectivity. Subjectivity in calcification assays arises because of end-point assays, which were only available until recently. Researchers must decide when to stop the experiment and measure calcification, increasing the subjectivity of the assay. We believe the method introduced in this manuscript is superior as we measure over time and can compare calcification development in a certain period. Linked to this, the illumination setting needs to be adjusted for every time point separately due to the decrease in signal over time. Due to this, the amount of signal represented in a picture is still swayed by an individual's opinion. It is crucial that illumination is performed with the highest signal-to-noise ratios so that post-image analysis can be performed as objectively as possible.
Although we see the subjectivity of this assay as a limitation, we believe that it is superior to other in vitro calcification methods. Unlike the existing methods, our semi-automated calcification assay has the advantage that images can be analyzed anonymously, thereby providing an independent blinded opinion. Additionally, the images can be analyzed at a later stage with the same defined settings across a data set, thereby reducing subjectivity.
The current methods of calcification determination rely on end-point measurements or lack the vascular component25,26,27. Within the clinic, tools such as computed tomography, intravascular ultrasound, and magnetic resonance imaging are expensive and a burden for patients. Biomarker research has proven its use but does not reflect the calcification burden of patients. We believe that this semi-automated assay uses not only one single biomarker but a collection of circulating components, reflecting a patient's cardiovascular status. This can be used to measure a calcification response as a biosensor. Potential further applications of the described method include patients' serum screening for in vitro calcification development as a surrogate marker for personalized development of vascular calcification. The platform has demonstrated its sensitivity toward dialysis and vitamin K treatment, in addition to both metabolic and non-metabolic diseases that are linked to patients with poor cardiovascular status and prognosis20. Since the assay's principle is based on the detection of calcium crystals, we hypothesize that it might also be relevant in other research areas where mineralization may be of relevance, such as osteoarthritis, osteoporosis, bone regenerative medicine, or dental research.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was funded by the European Union's Horizon 2020 research and innovation programs under the Marie Sklodowska-Curie grant agreement No 722609 and 764474, NWO ZonMw (MKMD 40-42600-98-13007). This research was supported by BioSPX. WJ-D received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) TRR219-project ID 322900939 and project ID 403041552
Materials
| Calcium chloride, 93%, anhydrous | Thermo Fisher Scientific | 349615000 | |
| Costar 6-well Clear TC-treated well plates | Corning | 3516 | |
| Cytation 3 System | BioTek, Abcoude, The Netherlands | ||
| Fetal Bovine Serum | Merck | F7524-100ML | |
| Fetuin-A-Alexa Fluor-546 | Prepared in-house | ||
| Gen5 Software v3.10 | BioTek | ||
| Gibco Medium 199 | Thermo Fisher Scientific | 11150059 | |
| Hoechst 33342, Trihydrochloride | Thermo Fisher Scientific | H3570 | |
| PBS (10X), pH 7.4 | Thermo Fisher Scientific | 70011044 | |
| Penicillin-Streptomycin | Thermo Fisher Scientific | 15140122 | |
| Trypsin-EDTA (0.05%), phenol red | Thermo Fisher Scientific | 25300062 |
References
- Taylor, A. J., Bindeman, J., Feuerstein, I., Cao, F., Brazaitis, M., O’Malley, P. G. Coronary calcium independently predicts incident premature coronary heart disease over measured cardiovascular risk factors: mean three-year outcomes in the Prospective Army Coronary Calcium (PACC) project. Journal of the American College of Cardiology. 46 (5), 807-814 (2005).
- Arad, Y., Goodman, K. J., Roth, M., Newstein, D., Guerci, A. D. Coronary calcification, coronary disease risk factors, C-reactive protein, and atherosclerotic cardiovascular disease events the St. Francis Heart Study. Journal of the American College of Cardiology. 46 (1), 158-165 (2005).
- Detrano, R., et al. Coronary calcium as a predictor of coronary events in four racial or ethnic groups. New England Journal of Medicine. 358 (13), 1336-1345 (2008).
- Schurgers, L. J., Akbulut, A. C., Kaczor, D. M., Halder, M., Koenen, R. R., Kramann, R. Initiation and propagation of vascular calcification is regulated by a concert of platelet- and smooth muscle cell-derived extracellular vesicles. Frontiers in Cardiovascular Medicine. 5, 36 (2018).
- Jaminon, A., Reesink, K., Kroon, A., Schurgers, L. The role of vascular smooth muscle cells in arterial remodeling: focus on calcification-related processes. International Journal of Molecular Sciences. 20 (22), 5694 (2019).
- Mollet, N., et al. Coronary plaque burden in patients with stable and unstable coronary artery disease using multislice CT coronary angiography. La Radiologia Medica. 116 (8), 1174-1187 (2011).
- Galal, H., Rashid, T., Alghonaimy, W., Kamal, D. Detection of positively remodeled coronary artery lesions by multislice CT and its impact on cardiovascular future events. The Egyptian Heart Journal. 71 (1), 26 (2019).
- Benedek, T., Gyöngyösi, M., Benedek, I. Multislice computed tomographic coronary angiography for quantitative assessment of culprit lesions in acute coronary syndromes. The Canadian Journal of Cardiology. 29 (3), 364-371 (2013).
- Raggi, P. Cardiovascular calcification in end stage renal disease. Cardiovascular Disorders in Hemodialysis. 149, 272-278 (2005).
- Raggi, P. Coronary artery calcification predicts risk of CVD in patients with CKD. Nature Reviews Nephrology. 13 (6), 324-326 (2017).
- Durham, A. L., Speer, M. Y., Scatena, M., Giachelli, C. M., Shanahan, C. M. Role of smooth muscle cells in vascular calcification: implications in atherosclerosis and arterial stiffness. Cardiovascular Research. 114 (4), 590-600 (2018).
- Yahagi, K., et al. Pathology of human coronary and carotid artery atherosclerosis and vascular calcification in diabetes mellitus. Arteriosclerosis, Thrombosis, and Vascular Biology. 37 (2), 191-204 (2017).
- Harper, E., Forde, H., Davenport, C., Rochfort, K. D., Smith, D., Cummins, P. M. Vascular calcification in type-2 diabetes and cardiovascular disease: Integrative roles for OPG, RANKL and TRAIL. Vascular Pharmacology. 82, 30-40 (2016).
- Lacolley, P., Regnault, V., Segers, P., Laurent, S. Vascular smooth muscle cells and arterial stiffening: relevance in development, aging, and disease. Physiological Reviews. 97 (4), 1555-1617 (2017).
- Pescatore, L. A., Gamarra, L. F., Liberman, M. Multifaceted mechanisms of vascular calcification in aging. Arteriosclerosis, Thrombosis, and Vascular Biology. 39 (7), 1307-1316 (2019).
- Herrmann, J., Babic, M., Tölle, M., vander Giet, M., Schuchardt, M. Research models for studying vascular calcification. International Journal of Molecular Sciences. 21 (6), 2204 (2020).
- Bowler, M. A., Merryman, W. D. In vitro models of aortic valve calcification: solidifying a system. Cardiovascular Pathology: The Official Journal of the Society for Cardiovascular Pathology. 24 (1), 1-10 (2015).
- Gitelman, H. J. An improved automated procedure for the determination of calcium in biological specimens. Analytical Biochemistry. 18 (3), 521-531 (1967).
- Furmanik, M., et al. Endoplasmic reticulum stress mediates vascular smooth muscle cell calcification via increased release of Grp78 (glucose-regulated protein, 78 kDa)-loaded extracellular vesicles. Arteriosclerosis, Thrombosis, and Vascular Biology. 41 (2), 898-914 (2021).
- Jaminon, A. M. G., et al. Development of the BioHybrid assay: combining primary human vascular smooth muscle cells and blood to measure vascular calcification propensity. Cells. 10 (8), 2097 (2021).
- Reynolds, J. L., et al. Human vascular smooth muscle cells undergo vesicle-mediated calcification in response to changes in extracellular calcium and phosphate concentrations: a potential mechanism for accelerated vascular calcification in ESRD. Journal of the American Society of Nephrology: JASN. 15 (11), 2857-2867 (2004).
- Wang, X. -. R., Zhang, J. -. J., Xu, X. -. X., Wu, Y. -. G. Prevalence of coronary artery calcification and its association with mortality, cardiovascular events in patients with chronic kidney disease: a systematic review and meta-analysis. Renal Failure. 41 (1), 244-256 (2019).
- Willems, B. A., et al. Ucma/GRP inhibits phosphate-induced vascular smooth muscle cell calcification via SMAD-dependent BMP signalling. Scientific Reports. 8 (1), 4961 (2018).
- Furmanik, M., et al. Reactive oxygen-forming Nox5 links vascular smooth muscle cell phenotypic switching and extracellular vesicle-mediated vascular calcification. Circulation Research. 127 (7), 911-927 (2020).
- Virtanen, P., Isotupa, K. Staining properties of alizarin red S for growing bone in vitro. Acta Anatomica. 108 (2), 202-207 (1980).
- Yang, H., Curinga, G., Giachelli, C. M. Elevated extracellular calcium levels induce smooth muscle cell matrix mineralization in vitro. Kidney International. 66 (6), 2293-2299 (2004).
- Pasch, A., et al. Nanoparticle-based test measures overall propensity for calcification in serum. Journal of the American Society of Nephrology: JASN. 23 (10), 1744-1752 (2012).

